Open Access Article
Open Access ArticleMonooxygenase-catalyzed regioselective hydroxylation for the synthesis of hydroxyequols†
Takafumi Hashimotoa,
Daiki Nozawaa,
Katsuyuki Mukaib,
Akinobu Matsuyamab,
Kouji Kuramochia and
Toshiki Furuya *a
*a
aDepartment of Applied Biological Science, Faculty of Science and Technology, Tokyo University of Science, 2641 Yamazaki, Noda, Chiba 278-8510, Japan. E-mail: tfuruya@rs.tus.ac.jp
bDaicel Corporation, 2-18-1 Konan, Minato-ku, Tokyo 108-8230, Japan
First published on 15th July 2019
Abstract
Monooxygenases exhibiting high activity and differing regioselectivity for the dietary isoflavone metabolite equol were discovered among enzymes in the HpaBC family by a genome mining approach. These enzymes enabled the one-step product-selective synthesis of 3′- and 6-hydroxyequols from equol and molecular oxygen.
(S)-Equol is a biologically active molecule synthesized from the dietary soy isoflavone daidzein by the gut microflora. This molecule has recently attracted much attention due to reports that it has various health-benefiting properties. For example, (S)-equol is a powerful antioxidant, exhibiting much greater antioxidant activity than the biosynthetic precursor daidzein.1 (S)-Equol also exhibits estrogenic activity.2 Interestingly, the binding affinity of (S)-equol for estrogen receptor β (ERβ) is higher than that for ERα. In addition, (S)-equol specifically binds 5α-dihydrotestosterone and sequesters it from the androgen receptor.2 Recent reports indicated that these properties of (S)-equol ameliorate menopause symptoms and metabolic syndrome, enhance bone health, and prevent skin aging.3 Although (S)-equol has various health-benefiting properties, only 30–50% of individuals in most populations metabolize daidzein to (S)-equol in the gut.4 Several microorganisms that convert daidzein to (S)-equol have been isolated, resulting in the marketing of (S)-equol produced via microbial fermentation as a nutritional supplement.4
In addition to (S)-equol, various analogues have exhibited high potential for use as nutritional supplements or pharmaceuticals. For example, dehydroequol (or phenoxodiol) is currently undergoing phase III clinical trials for the treatment of ovarian cancer.5 Hydroxyequols are also promising candidate biologically active molecules. The biological activities of polyphenols differ based on the number and position of hydroxy groups in the structure, and hydroxylated analogues often exhibit enhanced biological activity.6 For example, compared with resveratrol, piceatannol (a hydroxylated analogue of resveratrol) exhibits more-pronounced effects on melanogenesis inhibition, collagen synthesis, and vasorelaxation.7 Among hydroxyequols, 5-hydroxyequol reportedly exhibits greater antioxidant activity than (S)-equol.8 5-Hydroxyequol also preferentially binds ERα over ERβ, similar to the natural estrogen 17β-estradiol in terms of selectivity.9 In addition, as hydroxylated analogues are often generated via oxidative metabolism of exogenous compounds in mammals, analysis of the biological activity (including toxicity) of synthetic analogues is important.10 Indeed, (S)-equol is metabolized to 3′-hydroxyequol and 6-hydroxyequol as the primary products in rat and human liver microsomes.11 However, the biological activities of 3′-hydroxyequol and 6-hydroxyequol have yet to be elucidated, in part due to the difficulty of preparing these commercially unavailable compounds. It was reported that 3′-hydroxyequol was synthesized from 2-hydroxybenzyl acetate and aryl-substituted enol ether through Diels–Alder reaction followed by reductive cleavage of the methoxy group.12 Oxidants and/or oxidation catalysts that hydroxylate (S)-equol might provide a relatively easy and cost-effective approach for the synthesis of hydroxyequols. Although reactions using such compounds are difficult to carry out using chemical methods, biocatalytic methods could enable regioselective hydroxylation in just one step. To date, however, there have been no reports describing enzymes that exhibit such activity for (S)-equol.
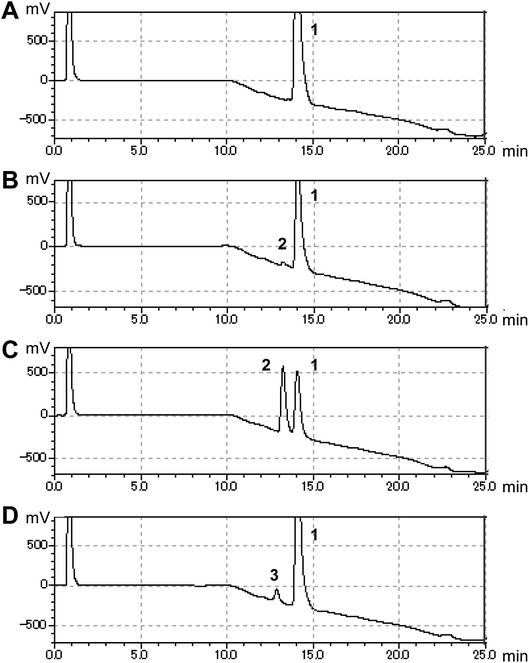
In this study, we searched for enzymes that can convert (S)-equol to hydroxyequols. The two-component flavin-dependent monooxygenase HpaBC is a candidate for such activity. Enzymes in the HpaBC family consist of a flavin-dependent monooxygenase (HpaB) and an NAD(P)H:flavin oxidoreductase (HpaC) and catalyze hydroxylation reactions using molecular oxygen as an innocuous oxidant under ambient conditions.13 HpaBC monooxygenases play a physiological role in oxidative metabolism of 4-hydroxyphenylacetate. The HpaBC monooxygenase from Pseudomonas aeruginosa PAO1 (HpaBCpa) reportedly catalyzes the hydroxylation of a variety of aromatic compounds, including p-coumaric acid and resveratrol to the corresponding o-diphenol compounds.7,13 We first explored the catalytic potential of HpaBCpa for equol hydroxylation using whole-cell assays with HpaBCpa-expressing E. coli. Reaction conditions previously optimized for resveratrol were employed, with some modifications.7 When (S)-equol (10 mM) was incubated with the transformed E. coli cells in a microtube containing 250 μL of reaction mixture, high-performance liquid chromatography (HPLC) analysis exhibited a very small product peak (retention time, 13.2 min) in addition to the substrate peak (14.0 min) (Fig. 1). The product peak was not detected in the reaction with E. coli cells carrying the empty vector without the hpaBCpa gene (Fig. 1). However, due to low yield, we could not identify the product.
We next searched for (S)-equol hydroxylases using a genome mining approach. A BLAST search of microbial genome sequences performed using the amino acid sequence of P. aeruginosa HpaBpa identified numerous HpaB homologues. Interestingly, the genome sequences of Gram-negative Photorhabdus luminescens subsp. laumondii TTO1 and Gram-positive Rhodococcus opacus B-4 were found to contain three hpaB homologues encoding gene products exhibiting moderate amino acid identity (51–72%) with P. aeruginosa HpaBpa (Fig. S1†). It was reported that three amino acid residues, Arg100, Tyr104, and His142, were involved in binding the hydroxy group of the substrate 4-hydroxyphenylacetate in the crystal structure of HpaB from Thermus thermophilus (HpaBtt).14 These amino acids are conserved in the HpaB homologues (Fig. S2†). In contrast, although Ser197 and Thr198 of HpaBtt reportedly interacted with the carboxy group of 4-hydroxyphenylacetate, these amino acids are not conserved (Fig. S2†). The divergent amino acid sequences of these putative gene products indicated that they were promising candidates for (S)-equol hydroxylation. We therefore focused on the HpaB homologues of P. luminescens (HpaBpl-1, HpaBpl-2, and HpaBpl-3) and R. opacus (HpaBro-1, HpaBro-2, and HpaBro-3) (Table S1†). As a control, we examined HpaB from E. coli BL21(DE3) (HpaBec), as this enzyme has been extensively studied (Table S1†).15
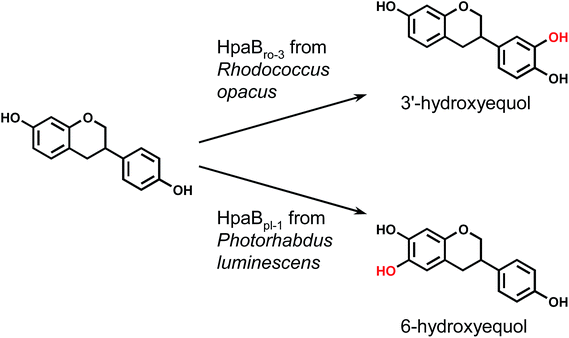
Each hpaB homologue was cloned and co-expressed with hpaCpa in E. coli (Table S1†). SDS-PAGE analysis revealed major bands corresponding to HpaBpa, HpaBec, HpaBpl-2, HpaBro-1, HpaBro-2, and HpaBro-3 (ca. 50–60 kDa) in the soluble fractions of the transformed E. coli cells (Fig. S3†), whereas HpaBpl-1 and HpaBpl-3 were expressed primarily in the insoluble fractions (Fig. S3†). Whole-cell assays using HpaBro-3-expressing E. coli and (S)-equol as the substrate revealed a large peak (retention time, 13.2 min) upon HPLC analysis (Fig. 1C and S4†). The product corresponding to this peak was confirmed to be formed from (S)-equol via one-oxygen addition based on determination of its mass. Analyses using 1H nuclear magnetic resonance (NMR), 13C NMR, and two-dimensional NMR spectroscopic data (Fig. S5–S8†) identified the product as (S)-3′-hydroxyequol (Scheme 1). The C-3′ position of (S)-equol was regioselectively hydroxylated without any detectable by-products by E. coli cells expressing HpaBro-3.
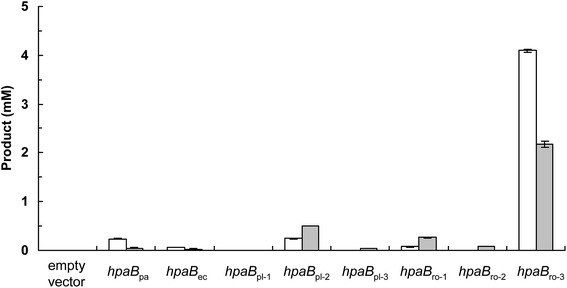
The production of (S)-3′-hydroxyequol was compared among the HpaB homologues (Fig. 2). The highest catalytic activity was exhibited by E. coli cells expressing HpaBro-3, with 4.1 mM of (S)-3′-hydroxyequol produced from 10 mM of (S)-equol in 24 h, representing 17 times more product than was produced by E. coli cells expressing HpaBpa. (S)-Equol was also converted to (S)-3′-hydroxyequol by E. coli cells expressing HpaBec, HpaBpl-2, and HpaBro-1, although the production was low. As equol has a chiral center, both (R)-equol and (S)-equol are present. Both enantiomers are currently being developed as nutraceutical and pharmacological agents.16 We also examined catalytic activities of the HpaB homologues with respect to (R)-equol (Fig. 2). Efficient catalysis of the hydroxylation of (R)-equol in addition to (S)-equol was observed with E. coli cells expressing HpaBro-3, with 2.2 mM of (R)-3′-hydroxyequol produced from 10 mM of (R)-equol in 24 h. Interestingly, HpaBro-3 converted (S)-equol more efficiently than (R)-equol in whole-cell reactions, whereas HpaBpl-2 exhibited substrate preference for (R)-equol over (S)-equol.
HPLC analyses revealed that the reaction of HpaBec with (S)-equol generated a small peak with a retention time of 12.8 min in addition to a peak eluting at 13.2 min (corresponding to (S)-3′-hydroxyequol) (Fig. S4†). Notably, the reaction of HpaBpl-1 with (S)-equol generated a relatively large peak eluting at 12.8 min, without any detectable by-products (Fig. 1D and S4†). HpaBro-2 also generated a peak at the same retention time, which was smaller than that generated by HpaBpl-1 (Fig. S4†). Although HpaBpl-1 (ca. 50 kDa) is largely insoluble in E. coli cells, solubility increases upon co-expression with the chaperonin GroEL and the cochaperonin GroES (Fig. S9†). In experiments using E. coli cells transformed in this manner, a compound corresponding to the peak eluting at 12.8 min was produced and confirmed to be formed from (S)-equol via one-oxygen addition based on determination of its mass. Furthermore, analysis by NMR spectroscopy (Fig. S10–S13†) identified the product as (S)-6-hydroxyequol (Scheme 1). The C-6 position of (S)-equol was regioselectively hydroxylated by E. coli cells expressing HpaBpl-1.
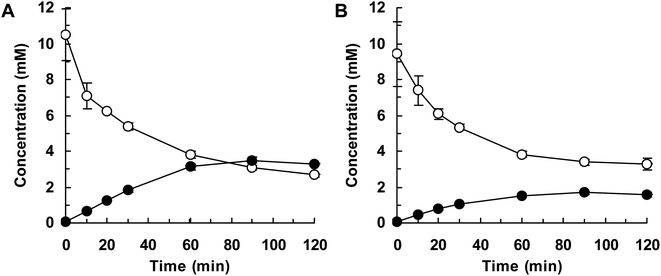
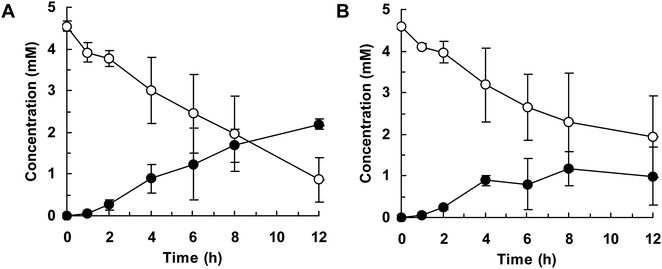
Finally, we attempted flask-scale production of 3′- and 6-hydroxyequols using E. coli cells expressing HpaBro-3 and HpaBpl-1, respectively. Equol was incubated with respective HpaB whole-cell catalyst in a 500 mL flask containing 20 mL of reaction mixture. The time course of equol conversion is shown in Fig. 3 and 4. HpaBro-3-expressing E. coli cells produced 3.5 mM (0.90 g L−1) of (S)-3′-hydroxyequol and 1.7 mM (0.44 g L−1) of (R)-3′-hydroxyequol from 10 mM (S)-equol and (R)-equol, respectively, only in 90 min (Fig. 3). The rate of equol hydroxylation by E. coli cells expressing HpaBpl-1 was low compared with rate of equol hydroxylation by E. coli cells expressing HpaBro-3 (Fig. 4). Nonetheless, HpaBpl-1-expressing E. coli cells produced 2.2 mM (0.57 g L−1) of (S)-6-hydroxyequol and 1.0 mM (0.26 g L−1) of (R)-6-hydroxyequol from 5 mM (S)-equol and (R)-equol, respectively, in 12 h.
Conclusions
In this study, we developed a one-step, product-selective approach for synthesizing 3′- and 6-hydroxyequols from equol using monooxygenases of the HpaBC family. To the best of our knowledge, this is the first report describing enzymes that catalyze the hydroxylation of equol. Previous metabolic studies have suggested that (S)-equol is further metabolized in mammals.11,17 Hydroxyequols synthesized using the approach presented here might be useful in studies evaluating biological activity, including the toxicity of potential metabolites, as well as in investigations of structure–activity relationships. In addition, these hydroxyequols and their derivatives could have applications as nutritional supplements and pharmaceuticals. Although typical HpaB enzymes from P. aeruginosa and E. coli exhibited only low catalytic activity for equol, we identified two novel HpaB homologues exhibiting high activity and differing regioselectivity for equol using a genome mining approach. Although typical HpaB enzymes have been extensively studied, few studies appear to have examined potential exploitation of their homologues. The results of this study suggest that other HpaB homologues exhibiting useful catalytic activities are present in nature. Further investigations will therefore focus on more-intensive screening for enzymes exhibiting greater activity and different selectivity for equol and other flavonoids for the synthesis of valuable polyphenols.Conflicts of interest
There are no conflicts to declare.Notes and references
- J. H. Mitchell, P. T. Gardner, D. B. McPhail, P. C. Morrice, A. R. Collins and G. G. Duthie, Arch. Biochem. Biophys., 1998, 360, 142–148 CrossRef CAS; C. E. Rüfer and S. E. Kulling, J. Agric. Food Chem., 2006, 54, 2926–2931 CrossRef.
- T. D. Lund, D. J. Munson, M. E. Haldy, K. D. Setchell, E. D. Lephart and R. J. Handa, Biol. Reprod., 2004, 70, 1188–1195 CrossRef CAS; R. S. Muthyala, Y. H. Ju, S. Sheng, L. D. Williams, D. R. Doerge, B. S. Katzenellenbogen, W. G. Helferich and J. A. Katzenellenbogen, Bioorg. Med. Chem., 2004, 12, 1559–1567 CrossRef; K. D. Setchell, N. M. Brown and E. Lydeking-Olsen, J. Nutr., 2002, 132, 3577–3584 CrossRef.
- T. Aso, S. Uchiyama, Y. Matsumura, M. Taguchi, M. Nozaki, K. Takamatsu, B. Ishizuka, T. Kubota, H. Mizunuma and H. Ohta, J. Women's Health, 2012, 21, 92–100 CrossRef CAS; A. Oyama, T. Ueno, S. Uchiyama, T. Aihara, A. Miyake, S. Kondo and K. Matsunaga, Menopause, 2012, 19, 202–210 CrossRef; Y. Tousen, J. Ezaki, Y. Fujii, T. Ueno, M. Nishimuta and Y. Ishimi, Menopause, 2011, 18, 563–574 CrossRef; T. Usui, M. Tochiya, Y. Sasaki, K. Muranaka, H. Yamakage, A. Himeno, A. Shimatsu, A. Inaguma, T. Ueno, S. Uchiyama and N. Satoh-Asahara, Clin. Endocrinol., 2013, 78, 365–372 CrossRef.
- F. Rafii, Metabolites, 2015, 5, 56–73 CrossRef CAS; K. D. Setchell and C. Clerici, J. Nutr., 2010, 140, 1355S–1362S CrossRef; J. P. Yuan, J. H. Wang and X. Liu, Mol. Nutr. Food Res., 2007, 51, 765–781 CrossRef.
- A. B. Alvero, M. Kelly, P. Rossi, A. Leiser, D. Brown, T. Rutherford and G. Mor, Future Oncol., 2008, 4, 475–482 CrossRef CAS; S. Mahoney, F. Arfuso, M. Millward and A. Dharmarajan, Cancer Cell Int., 2014, 14, 110 CrossRef.
- M. P. Marques, J. Med. Chem., 2003, 46, 5395–5401 CrossRef CAS; C. Siquet, F. Paiva-Martins, J. L. Lima, S. Reis and F. Borges, Free Radical Res., 2006, 40, 433–442 CrossRef.
- T. Furuya and K. Kino, Tetrahedron Lett., 2014, 55, 2853–2855 CrossRef CAS; T. Furuya, M. Sai and K. Kino, Biosci., Biotechnol., Biochem., 2016, 80, 193–198 CrossRef; T. Furuya, M. Sai and K. Kino, J. Biosci. Bioeng., 2018, 126, 478–481 CrossRef.
- A. Arora, M. G. Nair and G. M. Strasburg, Arch. Biochem. Biophys., 1998, 356, 133–141 CrossRef CAS.
- P. G. Lee, J. Kim, E. J. Kim, S. H. Lee, K. Y. Choi, R. J. Kazlauskas and B. G. Kim, ACS Chem. Biol., 2017, 12, 2883–2890 CrossRef CAS.
- G. Di Nardo and G. Gilardi, Int. J. Mol. Sci., 2012, 13, 15901–15924 CrossRef CAS.
- C. E. Rüfer, H. Glatt and S. E. Kulling, Drug Metab. Dispos., 2006, 34, 51–60 CrossRef.
- S. J. Gharpure, A. M. Sathiyanarayanan and P. Jonnalagadda, Tetrahedron Lett., 2008, 49, 2974–2978 CrossRef CAS.
- T. Furuya and K. Kino, Appl. Microbiol. Biotechnol., 2014, 98, 1145–1154 CrossRef CAS; T. Heine, W. J. H. van Berkel, G. Gassner, K. H. van Pée and D. Tischler, Biology, 2018, 7, E42 CrossRef.
- S. H. Kim, T. Hisano, K. Takeda, W. Iwasaki, A. Ebihara and K. Miki, J. Biol. Chem., 2007, 282, 33107–33117 CrossRef CAS.
- W. Chen, J. Yao, J. Meng, W. Han, Y. Tao, Y. Chen, Y. Guo, G. Shi, Y. He, J. M. Jin and S. Y. Tang, Nat. Commun., 2019, 10, 960 CrossRef CAS PubMed; J. A. Jones, S. M. Collins, V. R. Vernacchio, D. M. Lachance and M. A. Koffas, Biotechnol. Prog., 2016, 32, 21–25 CrossRef; M. A. Prieto and J. L. Garcia, J. Biol. Chem., 1994, 269, 22823–22829 Search PubMed.
- K. D. Setchell and C. Clerici, J. Nutr., 2010, 140, 1363S–1368S CrossRef CAS PubMed.
- R. J. Schwen, L. Nguyen and R. L. Jackson, Food Chem. Toxicol., 2012, 50, 2074–2083 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra03913a |
| This journal is © The Royal Society of Chemistry 2019 |