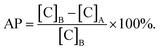Open Access Article
Open Access ArticleLow density lipoprotein adsorption on a titanium surface and its effect on osteoblast behaviors
Li-na Xuab,
Shui-yi Shaoab,
Wen-qing Zhuab,
Chao Chenc,
Song-mei Zhangd and
Jing Qiu *ab
*ab
aDepartment of Oral Implantology, Affiliated Hospital of Stomatology, Nanjing Medical University, Nanjing, PR China
bJiangsu Key Laboratory of Oral Disease, Nanjing Medical University, Nanjing, 210029, PR China. E-mail: qiujing@njmu.edu.cn; Tel: +86 25 85031834
cDepartment of Oral Implantology, Huaxia Hospital of Stomatology, Suzhou, PR China
dDepartment of General Dentistry, Eastman Institute for Oral Health, University of Rochester, Rochester, NY, USA
First published on 12th June 2019
Abstract
Objective: This study aims to investigate the adsorption of low density lipoprotein (LDL) on a titanium surface and to explore its effect on osteoblast behaviors. Materials and methods: LDL adsorption on a titanium surface was analyzed using LDL assay and X-ray photoelectron spectroscopy (XPS). Physical properties, including topography, surface roughness and wettability of a control smooth titanium surface and a LDL pre-adsorbed titanium surface, were assessed. Subsequently, the adhesion, proliferation and differentiation abilities of MC3T3-E1 cells (an osteoblast-like cell line) on the surfaces of control titanium and LDL pre-adsorbed titanium were investigated. Results: LDL assay and XPS confirmed LDL adsorption on the titanium surface. The maximum adsorption of LDL on the titanium surfaces was observed after 150 minutes of incubation. In comparison with the control smooth titanium surface, the roughness and hydrophilicity of the LDL pre-adsorbed titanium surface were significantly altered. Furthermore, in vitro studies demonstrated that LDL adsorption obviously attenuated the adhesion, proliferation and differentiation of MC3T3-E1 cells on the titanium surface. Conclusion: LDL could adsorb on a titanium surface. Meanwhile, LDL adsorption changed the characteristics of the titanium surface, which, in turn, negatively regulated osteoblast behaviors.
1. Introduction
Titanium and its alloys are extensively applied in dental implants owing to their advantages of excellent biocompatibility, great tensile strength, and recognizable corrosion resistance.1 The surface properties of titanium indicate the existence of amorphous TiO2, which is known as a stable oxide layer with a thickness between 3 and 7 nm.2 Blood (plasma) infiltration occurs during the first process of contact with an implant surface once a dental implantation is finished.3 It is well known that, immediately after plasma contact, rapid adsorption of plasma proteins onto the biomedical material takes place.4 In bone-to-implant binding, adsorption of blood-derived proteins onto the implant surface can influence subsequent cell attachment, spreading, proliferation, differentiation and regulation of material–cell interactions.5,6 Thus, the exploration of protein adsorption on a titanium surface is necessary to elucidate the factors influencing dental implant success.Protein adsorption, such as human serum protein on TiO2, is of great importance due to their presence in plasma and potential functions in wound healing and inflammatory response.7 Previous reports have investigated several blood-derived proteins existed in plasma, such as fibronectin, serum albumin and collagen. The potential effects of their adsorption on titanium surface and osteoblasts in vitro have been identified.8–10 These proteins can concentrate on the interface between implant surfaces and the surrounding matrix. In addition, the adsorption of fibronectin is considered to be affected by the wettability of implant surface, with greater binding observed on hydrophobic surfaces.11,12
Low density lipoprotein (LDL), one of plasma lipoproteins, is considered as a macro-molecular assembly of proteins and lipids in blood.13 It consists of triglycerides, cholesteryl esters and apolipoprotein B-100 (apoB-100). ApoB-100 is one of the largest monomeric proteins with 4536 residues.14 In hyperlipidemia, plasma concentration of LDL tends to be excessive. Hyperlipidemia induced by high-fat (HF) diet is characterized by elevated lipids in the bloodstream.15 Some studies have indicated that hyperlipidemia can significantly interfere with the process of implant osseointegration, decreasing bone formation and strength of bone-to-implant interface, as well as increasing implant loss. This may eventually lead to adverse consequences in dental implant therapy.16,17 However, recent in vivo studies also pointed out that high LDL is a neglected biologic risk factor in dental implant osseointegration.18,19 Since the effect of high LDL on bone development or implant remains controversial, more studies have been performed to illustrate the relationship between high level of LDL and osteoblast behaviors.20–22 At the cellar level, osteoblasts are able to bind, internalize and degrade LDL by SR-B receptors. Meanwhile, they can oxidize LDL into oxidized-LDL (ox-LDL), which has been proved to inhibit osteoblast differentiation and result in adverse effects on bone formation.23 Furthermore, it has been found that ox-LDL is negatively associated with bone mineral density, leading to osteoporosis in post-menopausal women.24 Consequently, the potential role of LDL in the interaction between the implant surface and surrounding matrix should be clarified, especially in hyperlipidemia patient.
Cellular responses are mediated through proteins and protein ligands. It is generally believed that the initial event in the interaction between material surfaces and blood is protein adsorption onto the surface.25 The relationship between the adsorption of specific proteins and the surface reactivity of materials has not so far been fully explicated. As a risk factor of hyperlipidemia, LDL was speculated to induce adverse effects during the process of dental implant osseointegration. However, to the best of our knowledge, scientific evaluations on the surface characteristics of titanium surface after LDL adsorption have not been reported. Therefore, in the present study, we hypothesized that early osteoblast behaviors could be affected by LDL pre-adsorbed titanium surface. The existence of LDL adsorption on titanium surface in vitro was explored and the osteoblast behaviors on pre-adsorbed titanium surface were investigated. Such information would promote the understanding of potential influence of hyperlipidemia on dental implant treatment.
2. Materials and methods
2.1. Specimen preparation
Commercially pure titanium (99.5 wt% purity, Shanghai, China) was applied in this study. All specimens were polished using waterproof silicon carbide paper from #400, #600, #800, #1000, #1200 to #1500, followed by ultra-sonically cleaning with ethanol and distilled water. Subsequently, samples were dried in an oven at 65 °C for 24 h. All samples were disinfected by disinfecting pan and dried for 24 h.2.2. Evaluation of LDL adsorption capacity
Adsorption percentage was calculated in accordance with the following equation:In this equation, adsorption percentage was represented by AP. [C]B was the concentration before adsorption. [C]A was the concentration after adsorption. LDL was purchased from Yi-Yuan Limited Liability Company (Guangzhou, China). Protein purity was identified using agarose gel electrophoresis according to manufacturer's instructions. LDL was diluted to 100 μg ml−1 by phosphate-buffered saline (PBS), and then transferred onto the surface of titanium specimens in a 96-well plate. Subsequently, they were incubated in a 5% CO2 humidified environment at 37 °C for 180 min. The concentration of LDL was determined in strict accordance with a LDL assay kit (Jian-Cheng Bioengineering Institute, Nanjing, PRC). Measurements were conducted for three times for each time point.
2.3. Analysis of surface characterization
According to the concentration of LDL (0, 50, 100 and 200 μg ml−1), titanium specimens were divided into four groups. Titanium specimens of different groups were immersed in 0, 50, 100 and 200 μg ml−1 LDL solutions respectively, and then incubated in a 5% CO2 humidified atmosphere at 37 °C for an optimal time, which was determined by above adsorption test. After that, samples were washed with deionized water and dried in a vacuum oven at room temperature. The surface chemical composition of each sample was analyzed by X-ray photoelectron spectroscopy (XPS, Thermo Scientific Escalab 250Xi, USA) utilizing a monochromatic Al Kα electrode at 15 kV and 150 W with a 45° take-off angle. Survey and high-resolution spectra were obtained using pass energies of 160 and 40 eV, respectively. Reference binding energy of each element was obtained from the National Institute of Standards and Technology XPS Online Database (http://srdata.nist.gov/xps/). Spectra were calibrated by adjusting the binding energy of C 1s to 284.8 eV.The surface roughness of each sample was measured using an optical profilometer (MicroXamTM, Phase-Shift, UP, Rtec co, USA). The scanning area on each sample was 50 μm × 50 μm. The surface wettability of each sample was evaluated from contact angle by measuring the droplet of pure water on the substrate using Automatic Contact Angle Meter Model SL200B (Kenuo, USA) in an ambient environment. All measurements were performed in triplicate.
2.4. Cell culture
MC3T3-E1 cells, an osteoblast-like cell line, were chosen in this study. The cells were purchased from Chinese Academy of Sciences Cell Bank (Shanghai, China). The cells were cultured in α-Minimum Essential Medium (α-MEM, Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin, and maintained in a humidified atmosphere of 5% CO2 and 95% air at 37 °C. The medium was changed every two days. Cells were passaged on reaching 80% confluence for subsequent use.2.5. Cell adhesion assay
To observe the morphology of cells growing on titanium specimens, MC3T3-E1 cells (5 × 103 cells per well) were seeded on different samples pre-adsorbed with 0, 50, 100 and 200 μg ml−1 LDL in 96-well plates. After culturing for 4 h, each sample was rinsed with PBS and fixed with 4% paraformaldehyde in PBS for 10 minutes at room temperature. Subsequently, each sample was stained with rhodamine phalloidin (Cytoskeleton, USA) at room temperature in the dark for 30 minutes. Afterwards, each sample was stained with 4′,6′-diamidino-2-phenylindole (DAPI) (Beyotime, Shanghai, China) in the dark for 30 seconds. The morphology of adherent MC3T3-E1 cells in five random fields on each sample were observed under a laser scanning confocal microscope (LSM710, Zeiss, Germany) at 400× magnification.2.6. Cell proliferation assay
Cell proliferation assay was performed to assess cell viability on different titanium specimens pre-adsorbed with 0, 50, 100 and 200 μg ml−1 LDL using CCK-8 assay. MC3T3-E1 cells (2 × 103 cells per well) were seeded on samples in 96-well plates. After culturing for 1, 2 and 3 days, 100 μL of culture medium containing 10 μL of CCK-8 solution (Beyotime, Shanghai, China) was added to each well and then incubated for 2 h. The absorbance at 450 nm was measured by a microplate reader (Spectramax190, MD, USA). Each experiment was performed in triplicate.2.7. Alkaline phosphatase (ALP) activity assay
MC3T3-E1 cells (2 × 105 cells per well) were seeded on different samples pre-adsorbed with 0, 50, 100 and 200 μg ml−1 LDL in 6-well plates for ALP activity assay. After 7 and 14 days of incubation, samples was washed with PBS and the cells on samples were lysed with radio-immunoprecipitation assay buffer (Beyotime, Shanghai, China) at 4 °C for 30 minutes. Subsequently, the lysates were centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C for 10 minutes to remove cellular debris and the liquid supernatants were then collected. ALP activity was obtained using an ALP Assay Kit (Jian-Cheng Bioengineering Institute, China). Total protein content was determined by a BCA protein assay kit (KeyGEN BioTECH, Nanjing, China). After protein content normalization, ALP activity relative to control was calculated. Each experiment was performed in triplicate.
000 rpm at 4 °C for 10 minutes to remove cellular debris and the liquid supernatants were then collected. ALP activity was obtained using an ALP Assay Kit (Jian-Cheng Bioengineering Institute, China). Total protein content was determined by a BCA protein assay kit (KeyGEN BioTECH, Nanjing, China). After protein content normalization, ALP activity relative to control was calculated. Each experiment was performed in triplicate.
2.8. Real-time quantitative PCR
Expression levels of runt-related transcription factor 2 (Runx2) and osteocalcin (OCN) genes of cells on different samples pre-adsorbed with 0, 50, 100 and 200 μg ml−1 LDL were measured using real-time quantitative PCR. MC3T3-E1 cells at a density of 2 × 105 cells per well were seeded on samples in 6-well plates. After culturing for 7 and 14 days, total RNA in MC3T3-E1 cells was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Complementary DNA (cDNA) was synthesized according to the instruction of PrimeScript RT Master mix (Takara, Kusatsu, Japan). For real-time PCR assay, each cDNA was amplified using SYBR Premix Ex Taq (Takara, Kusatsu, Japan) via an ABI real-time PCR 7300 system (Applied Biosystems, Foster City, CA, USA). All primer sequences used for target genes were shown in Table 1. Expression levels of all target genes were normalized to that of β-actin. Real-time PCR was performed in triplicate for each group.| Gene | Primer sequence (F: forward; R: reverse) | Product size (bp) |
|---|---|---|
| Runx2 | F: CCGAAATGCCTCCGCTGTTATG | 194 |
| R: TCTGTCTGTGCCTTCTTGGTTCC | ||
| OCN | F: CACCTAGCAGACACCATGAGGAC | 180 |
| R: GACTGAGGCTCCAAGGTAGCG | ||
| β-actin | F: GTGCTATGTTGCTCTAGACTTCG | 174 |
| R: ATGCCACAGGATTCCATACC |
2.9. Western blotting
Protein expressions of Runx2 and OCN of cells on titanium surfaces were examined by Western blotting. After culturing in 6-well plates on different samples pre-adsorbed with 0, 50, 100 and 200 μg ml−1 LDL for 3, 7 and 14 days, MC3T3-E1 cells (2 × 105 cells per well) were rinsed with cold PBS and protein samples were harvested by lysis in radio-immunoprecipitation (RIPA) buffer. The concentration of extracted total proteins was determined using the BCA protein assay kit (Key-GEN BioTECH, Nanjing, China). Protein samples (20 μg) were separated by electrophoresis and then transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). After blocking with 5% non-fat milk for 1 h at room temperature, the membranes were incubated with primary antibodies against Runx2 (12556; CST, Beverly, MA, USA), OCN (ab93876; Abcam, Cambridge, Ma, USA), and β-actin (BM0627, Boster, Wuhan, China) at 4 °C overnight. On the next day, the membranes were incubated with corresponding secondary antibodies (ZB-2301, Goat anti-Rabbit IgG, ZSGB-BIO, Beijing, China; AP124P, Goat anti-Mouse IgG, Millipore, USA) at room temperature for 2 h. After that, the membranes were visualized using the ECL Western Blot Kit (Millipore, USA). The protein expressions were determined relative to that of β-actin. The gray values of protein levels were quantified using the Photoshop software.2.10. Statistical analysis
Experimental results were expressed as means ± standard error. SPSS 22.0 software (IBM, Armonk, NY, USA) was used for the statistical analysis. Data were analyzed by one way analysis of variance (ANOVA) with Student–Newman–Keuls method for multiple comparisons. The homogeneity of variance was tested before performing ANOVA. P < 0.05 was considered statistically significant.3. Results and discussion
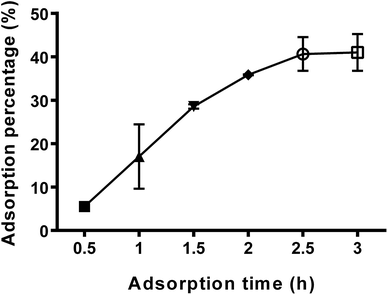
3.1. Effect of time on LDL adsorption percentage
Fig. 1 illustrated the percentage of LDL adsorption on titanium surface at different adsorption intervals during 180 min incubation. The adsorption percentage increased with the prolongation of adsorption time. At 2.5 h (150 min), it reached an optimal condition and the adsorption capacity of LDL was about 41%. For this reason, in the following experiments of this study, 150 min had been selected as the optimal time for LDL pre-adsorption on titanium surface. Similar with other studies, it was reported that the optimal time for LDL adsorption was within the range of 1.5–3 h.26,27 Several metal materials, including alloys (stainless steel and Ti6Al4V) and pure metals (Ti, Al, Mo, Co, Ni and Ta), adsorbed only very low amounts of protein at the initial stage of exposure to the protein solution, and then the amount adsorbed rising gradually to reach a steady value.28 LDL is a principal transporter for cholesterol in the blood. LDL adsorption kinetics suggested that LDL adsorption on titanium surface was restricted to the availability of binding sites.28 The adsorption capacity reached maximum once binding sites were occupied. ApoB-100 is the only protein component of LDL.29 Thus, it can be reasonably supposed that LDL adsorbed on titanium surface via its protein moiety apoB-100.3.2. Characterization of titanium surface
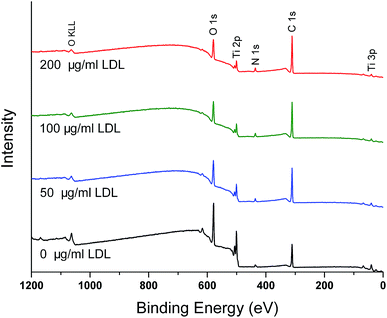 | ||
| Fig. 2 XPS survey spectra of different titanium surfaces pre-adsorbed with 0, 50, 100 and 200 μg ml−1 LDL. | ||
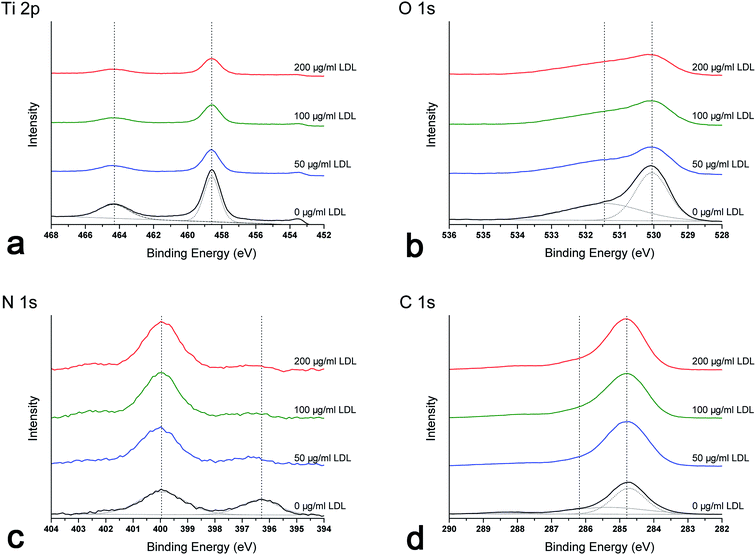 | ||
| Fig. 3 XPS high-resolution spectra of different titanium surfaces pre-adsorbed with 0, 50, 100 and 200 μg ml−1 LDL: (a) Ti 2p; (b) O 1s; (c) N 1s; (d) C 1s. | ||
As shown in XPS survey and high-resolution spectra (Fig. 2 and 3), the results indicated that LDL adsorbed to titanium surface and the titanium oxide was covered after LDL adsorption. Previous studies have reported that titanium oxide played an implant role in the protein adsorption on titanium implants.2,6 There was a clear trend for enhanced LDL adsorption along with the increase of LDL concentration in this study. N 1s signal is specific to the protein. Generally, C 1s spectra consist of different peaks because of many functional groups existing in protein molecules.4 Such groups are carbon singly bonded with oxygen or nitrogen, or carbon bonded with one or two (NH2–), etc. The adsorbed protein amount can be monitored by the experimental area ratio of N 1s and Ti 2p peaks, N 1s/Ti 2p.30 Higher value of this ratio indicated more adsorbed protein.
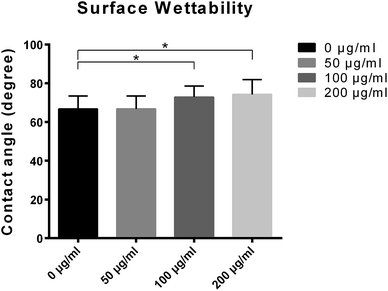 | ||
| Fig. 4 Contact angles of different titanium surfaces pre-adsorbed with 0, 50, 100 and 200 μg ml−1 LDL. | ||
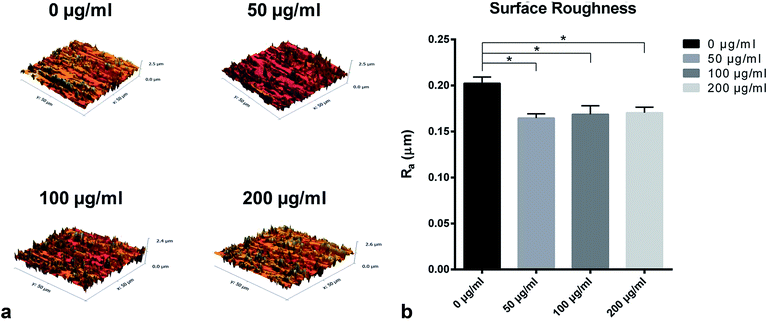
Fig. 5 presented the three-dimensional surface topography and surface roughness values of different samples with or without LDL adsorption. Three-dimensional images revealed that the control titanium surface was obviously rougher than LDL pre-adsorbed titanium surfaces. Correspondingly, significant changes of roughness values were found between LDL pre-adsorbed titanium surfaces and control titanium surface. It is well known that surface roughness has a significant impact on cell response.34 The decline of surface roughness may have impacts on cell response. So we further investigated the cell adhesion, proliferation and differentiation on LDL pre-adsorbed titanium surfaces.
3.3. Cell adhesion and spreading
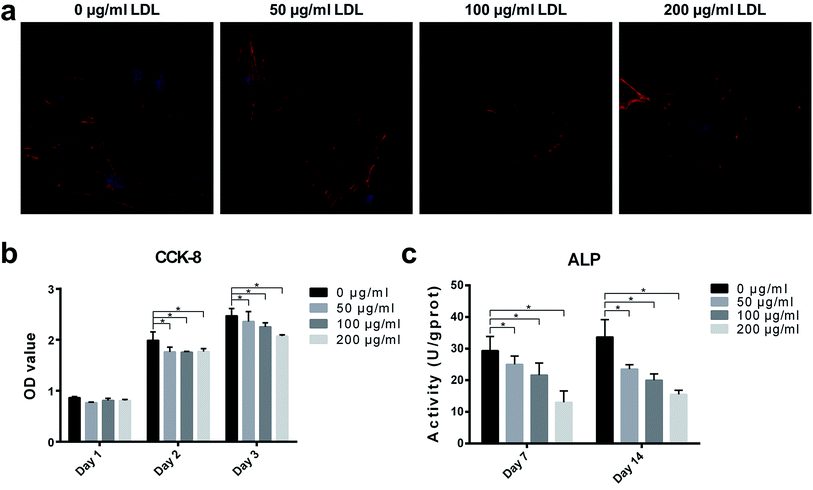
After 4 h cell culture, the morphology of MC3T3-E1 cells varied on different samples with or without LDL adsorption (Fig. 6(a)). As shown in Fig. 6(a), the synapse of MC3T3-E1 cells on LDL pre-adsorbed substrates was obviously shorter than those on the control substrate. It revealed that LDL pre-adsorbed titanium surfaces suppressed the attachment of osteoblasts. Other studies have found that the effects of different proteins on cell attachment were observed to be significantly different.11,35,36 It was observed that titanium surfaces adsorbed with fibronectin significantly enhanced cell attachment.35 However, titanium surfaces adsorbed with albumin had no obvious effect on cell attachment.11 As is well known, protein adsorption on the biomaterial surface is the initial biological response, which can elicit a variety of cell responses.36 In this work, attenuation of cell spreading on LDL pre-adsorbed titanium surfaces might influence the following cell proliferation.3.4. Cell proliferation
Cell proliferation was measured by CCK-8 assay, and the results were presented in Fig. 6(b). With the prolongation of incubation time, MC3T3-E1 cells proliferated on all samples. After 2 and 3 days of culture, compared with the control titanium surface, proliferation of MC3T3-E1 cells exhibited significant reductions on LDL pre-adsorbed titanium surfaces. Hamel et al.37 found that native LDL particles may not alter the proliferation of MG-63 cells. But another study of Kumagai et al.38 illustrated that LDL had an important effect on cytotoxic activity of peritoneal macrophages in mice. In the present study, LDL pre-adsorption titanium surface greatly influenced proliferation of MC3T3-E1 cells. Interaction between LDL and titanium may be the cause of changes in cell proliferation. It has been reported that titanium exhibited oxidant activity, which could increase the oxidation of LDL.39 Ox-LDL, as the oxidation product of LDL, is known to be able to decrease the proliferation abilities of BMSC, MG-63, MC3T3-E1 cells.40 Learning from the results, it was found that LDL pre-adsorbed titanium surface played a negative role on the proliferation of osteoblasts. Furthermore, cell spreading, at the first phase of cell/biomaterial interaction, acts a key role in regulating cell proliferation.413.5. ALP activity
The consequences of ALP activity were shown in Fig. 6(c). After culturing for 7 days, ALP activities of MC3T3-E1 cells on LDL pre-adsorbed titanium surfaces significantly decreased compared with that of MC3T3-E1 cells on control titanium surface. After culturing for 14 days, MC3T3-E1 cells on LDL pre-adsorbed titanium surfaces exhibited more significant reductions of ALP activities compared to the control group. ALP, which is expressed in the early stage of osteogenic differentiation, is an osteogenic marker of osteoblast differentiation. The results indicated that osteoblastic functions of MC3T3-E1 cells were significantly altered after LDL adsorption on titanium surface. Protein adsorption is another critical determinant of cell responses to biomaterials. Various cell behaviors are related to the properties of adsorbed proteins on substrates. Previous studies have shown that the interaction of osteoblast cells with extra cellular matrix (ECM) proteins, collagen-1 (coll-1), fibronectin (FN) may influence matrix mineralization, expressions of osteogenic genes and ALP activity of osteoblasts in vitro.11,36,42 For example, FN played an important role in initial cell attachment and spreading on biomaterials since it was able to specially bind to integrin on cell membranes.43 However, having insight into the role of adsorbed proteins on dental implant surfaces is still a challenge.3.6. Real-time quantitative PCR analysis
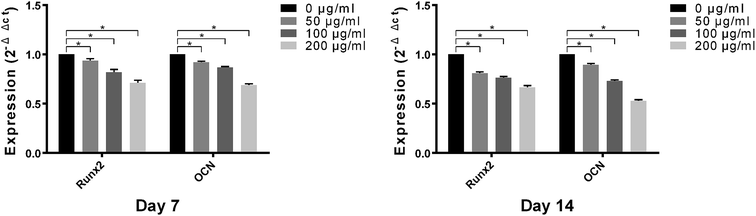
Osteogenic-related genes, including Runx2 and OCN, were determined by the quantitative real-time PCR technique. As shown in Fig. 7, the genes expression levels of Runx2 and OCN in MC3T3-E1 cells on control titanium surface were significantly higher than those on LDL pre-adsorbed titanium surface after culturing for 7 and 14 days. With the increase of LDL concentration, the gene expression levels exhibited gradual declining trend. It demonstrated that LDL played a negative role on expressions of bone formation-related genes after its adsorption on titanium surface.3.7. Western blotting analysis
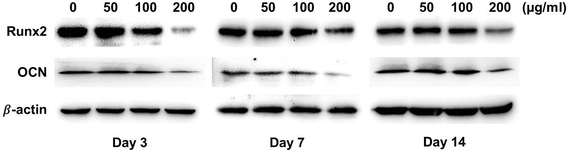
Western blotting analysis was performed to verify the expression levels of osteogenic-related proteins on titanium surfaces with or without LDL adsorption. As shown in Fig. 8, MC3T3-E1 cells on LDL pre-adsorbed titanium surfaces expressed lower protein levels of Runx2 and OCN than those on control titanium surfaces after culturing for 3, 7 and 14 days. The significant differences in the protein levels between different groups can be found from 3 days of incubation. Moreover, as the concentration of LDL rising, the protein expression levels of Runx2 and OCN tended to decrease. Fig. 9 displayed the gray values of protein levels quantified using the Photoshop software. As shown in Fig. 9, significant differences were found between the protein levels of Runx2 and OCN on titanium surfaces pre-adsorbed with 200 μg ml−1 LDL and control titanium surface for 3, 7 and 14 days.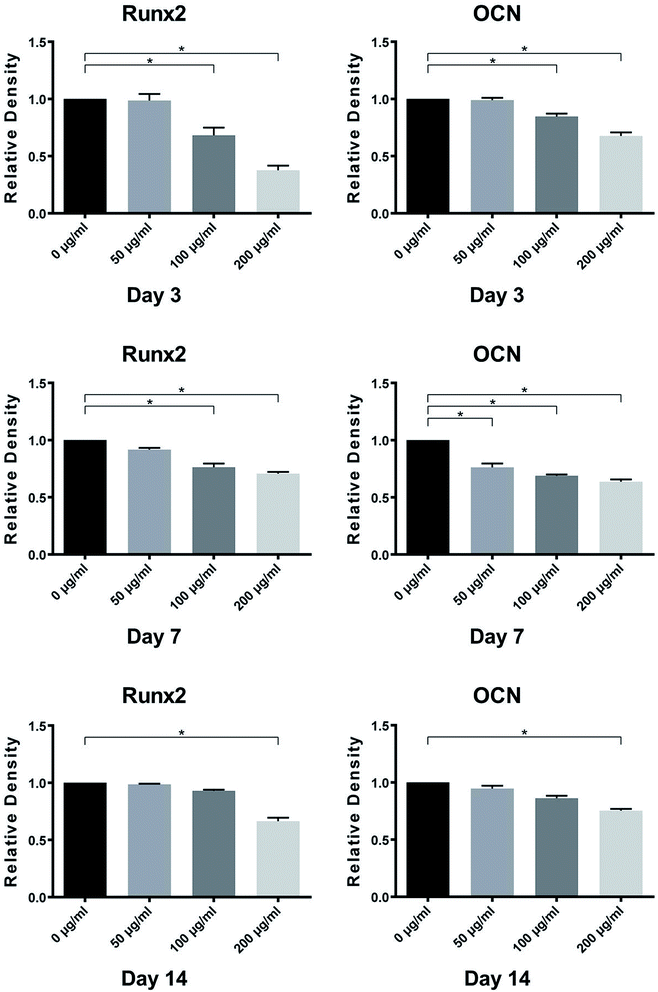 | ||
| Fig. 9 Quantification of protein levels of Runx2 and OCN using the Photoshop software. * indicated significant differences between different groups (P < 0.05). | ||
Both quantitative PCR and Western blotting analysis demonstrated that LDL pre-adsorbed titanium surface significantly down-regulated the expression levels of Runx2 and OCN in MC3T3-E1 cells in comparison with those on control titanium surface. Runx2 and OCN, as prominent osteogenesis markers, were selected to analyze osteoblastic differentiation activities in our work. Runx2 is expressed in the early stage of osteogenic differentiation. OCN is recognized as a marker of late-stage osteogenic differentiation and directly participates in the mineralization process.44 From the results of Fig. 7 and 8, it was believed that LDL down-regulated gene and protein expressions of bone formation after its adsorption on titanium surface. Correspondingly, ALP activity of cells cultured on LDL pre-adsorbed titanium surface was remarkably down-regulated. This was in agreement with the adverse effect of LDL adsorption on cell spreading and proliferation abilities. Furthermore, it has been noticed that osteoblasts could bind, internalize and degrade LDL while LDL adsorbed on titanium surface. This might eventually lead to osteoblasts shedding, which was adverse to bone formation.45
Talking about the results above, the LDL adsorption on titanium surface was confirmed by LDL assay and XPS analysis, and LDL pre-adsorbed titanium surface exhibited significantly worse in vitro bioactivity than that of control titanium surface. Previous studies have reported that hyperlipidemia can significantly compromise implant osseointegration and cause poor outcome in dental implant therapy.16,17 And that, it was found that hyperlipidemia may negatively affect the implant stability and decrease peri-implant bone regeneration.46 From our study, it could be speculated that LDL adsorption on titanium surface might be the possible original cause of hyperlipidemia-induced poor implant osseointegration. Given this viewpoint, it is necessary to control serum LDL level of hyperlipidemia patients before dental implant treatment. However, the underlying mechanism of LDL adsorption regulating osteoblast behaviors and bone formation process required further investigations.
4. Conclusions
In the present study, we explored LDL adsorption on titanium surface and confirmed that the maximum adsorption of LDL on titanium surfaces occurred after 150 min incubation. LDL adsorption could significantly change the roughness and hydrophilicity of titanium surface. Furthermore, it was found that LDL pre-adsorbed titanium surface exhibited deleterious effects on the spreading, proliferation and differentiation of osteoblasts.Author contributions
LNX contributed to design, data acquisition and analysis, and drafted the manuscript. SYS, WQZ contributed to design, data acquisition and analysis. CC, SMZ contributed to data acquisition. The corresponding author JQ contributed to conception, design, data interpretation, and critically revised the manuscript. All authors give final approval and agree to be accountable for all aspects of the work.Conflicts of interest
All authors declare that they have no conflicts of interest with the contents of this article.Acknowledgements
This study was supported by the National Natural Science Foundation of China (Project Number: 81472928, 81870799), the Medical Science Foundation of Jiangsu Provincial Health Department (Project Number: H201641), the Jiangsu Provincial Medical Youth Talent (Project Number: QNRC2016850), the Southeast University-Nanjing Medical University Cooperative Research Project (Project Number: 2242017K3DN14), the Nanjing Medical University-SUYAN Group Intelligent Innovation Research and Development Project (Project Number: NMU-SY201806), and the Science and Technology Development Foundation of Nanjing Medical University (Project Number: 2017NJMU101).References
- M. Kulkarni, A. Flasker, M. Lokar, K. Mrak-Poljsak, A. Mazare, A. Artenjak, S. Cucnik, S. Kralj, A. Velikonja, P. Schmuki, V. Kralj-Iglic, S. Sodin-Semrl and A. Iglic, Int. J. Nanomed., 2015, 10, 1359–1373 CAS
.
- M. Lehnert, M. Gorbahn, C. Rosin, M. Klein, I. Koper, B. Al-Nawas, W. Knoll and M. Veith, Langmuir, 2011, 27, 7743–7751 CrossRef CAS PubMed
.
- J. Park and J. Davies, Clin. Oral Implants Res., 2000, 11, 530–539 CrossRef CAS PubMed
.
- Y. Yang, R. Cavin and J. L. Ong, J. Biomed. Mater. Res., 2003, 67, 344–349 CrossRef PubMed
.
- R. Vaz, G. G. Martins, S. Thorsteinsdottir and G. Rodrigues, Cell Tissue Res., 2012, 348, 569–578 CrossRef CAS PubMed
.
- J. Pei, H. Hall and N. D. Spencer, Biomaterials, 2011, 32, 8968–8978 CrossRef CAS PubMed
.
- Y. Yang, R. Cavin and J. L. Ong, J. Biomed. Mater. Res., 2003, 67, 344–349 CrossRef PubMed
.
- S. Bauer, J. Park, A. Pittrof, Y. Y. Song, K. von der Mark and P. Schmuki, Integr. Biol., 2011, 3, 927–936 CrossRef CAS PubMed
.
- J. Park, S. Bauer, K. A. Schlegel, F. W. Neukam, K. von der Mark and P. Schmuki, Small, 2009, 5, 666–671 CrossRef CAS
.
- W. Yang, X. Xi, Q. Ran, P. Liu, Y. Hu and K. Cai, Mater. Sci. Eng., C, 2014, 34, 410–416 CrossRef CAS
.
- F. Grinnell and M. K. Feld, J. Biol. Chem., 1982, 257, 4888–4893 CAS
.
- G. Altankov and F. Groth, J. Biomed. Mater. Res., 1996, 30, 385–391 CrossRef CAS PubMed
.
- J. S. Samra, L. K. Summers and K. N. Frayn, Br. J. Surg., 1996, 83, 1186–1196 CrossRef CAS PubMed
.
- R. Prassl and P. Laggner, Eur. Biophys. J., 2009, 38, 145–158 CrossRef CAS PubMed
.
- S. Dundar, F. Yaman, M. F. Ozupek, A. Saybak, M. Gul, F. Asutay, M. Kirtay and I. H. Ozercan, J. Korean Assoc. Oral Maxillofac. Surg., 2016, 42, 187–192 CrossRef PubMed
.
- F. Pirih, J. Lu, F. Ye, O. Bezouglaia, E. Atti, M. G. Ascenzi, S. Tetradis, L. Demer, T. Aghaloo and Y. Tintut, J. Bone Miner. Res., 2012, 27, 309–318 CrossRef CAS PubMed
.
- A. Keuroghlian, A. D. Barroso, G. Kirikian, O. Bezouglaia, Y. Tintut, S. Tetradis, P. Moy, F. Pirih and T. Aghaloo, J. Oral Implantol., 2015, 41, e7–e11 CrossRef PubMed
.
- J. Choukroun, G. Khoury, F. Khoury, P. Russe, T. Testori, Y. Komiyama, G. Sammartino, P. Palacci, M. Tunali and E. Choukroun, J. Oral Implantol., 2014, 40, 110–114 CrossRef
.
- M. Muluke, T. Gold, K. Kiefhaber, A. Al-Sahli, R. Celenti, H. Jiang, S. Cremers, T. Van Dyke and U. Schulze-Spate, J. Dent. Res., 2016, 95, 223–229 CrossRef CAS
.
- J. Makovey, J. S. Chen, C. Hayward, F. M. Williams and P. N. Sambrook, Bone, 2009, 44, 208–213 CrossRef CAS PubMed
.
- J. Ryo, I. Mikael, S. Young-Taeg and C. B. Johansson, J. Oral Maxillofac. Res., 2010, 1, 3–9 Search PubMed
.
- E. Luegmayr, H. Glantschnig, G. A. Wesolowski, M. A. Gentile, J. E. Fisher, G. A. Rodan and A. A. Reszka, Cell Death Differ., 2004, 11(suppl. 1), S108–S118 CrossRef CAS PubMed
.
- M. R. Brodeur, L. Brissette, L. Falstrault, V. Luangrath and R. Moreau, J. Bone Miner. Res., 2008, 23, 326–337 CrossRef CAS PubMed
.
- T. Yamaguchi, T. Sugimoto, S. Yano, M. Yamauchi, H. Sowa, Q. Chen and K. Chihara, Endocr. J., 2002, 49, 211–217 CrossRef CAS PubMed
.
- S. Runa, M. Lakadamyali, M. L. Kemp and C. K. Payne, J. Phys. Chem. B, 2017, 121, 8619–8625 CrossRef CAS PubMed
.
- S. Q. Wang, Y. T. Yu, T. Cui and Y. Cheng, Biomaterials, 2003, 24, 2799–2802 CrossRef CAS PubMed
.
- S. Zhao, F. Wen, F. He, L. Liu and G. Yang, Int. J. Oral Maxillofac. Implant., 2014, 29, 211–220 CrossRef PubMed
.
- R. L. Williams and D. F. Williams, Biomaterials, 1988, 9, 206–212 CrossRef CAS
.
- D. D. Deligianni, N. Katsala, S. Ladas, D. Sotiropoulou, J. Amedee and Y. F. Missirlis, Biomaterials, 2001, 22, 1241–1251 CrossRef CAS
.
- M. D. Carroll and D. T. Nguyen, NCHS Data Brief., 2017, 290, 1–8 Search PubMed
.
- American Society for Testing and Materials (ASTM), D7334–08, Standard Practicefor Surface Wettability of Coatings, Substrates and Pigments by Advancing Contact Angle Measurement, ASTM International, West Conshohocken, PA, USA, 2013 Search PubMed
.
- D. Kohavi, L. B. Hauslich, G. Rosen, D. Steinberg and M. N. Sela, Clin. Oral Implant. Res., 2013, 24, 1002–1008 Search PubMed
.
- T. Murtola, T. A. Vuorela, M. T. Hyvonen, S. J. Marrink, M. Karttunen and I. Vattulainen, Soft Matter, 2011, 7, 8135–8141 RSC
.
- E. M. Lee, K. Smith, K. Gall, B. D. Boyan and Z. Schwartz, Biomaterials, 2016, 110, 34–44 CrossRef CAS PubMed
.
- T. Hayakawa, M. Yoshinari and K. Nemoto, Biomaterials, 2004, 25, 119–127 CrossRef CAS
.
- N. R. Gandavarapu, P. D. Mariner, M. P. Schwartz and K. S. Anseth, Acta Biomater., 2013, 9, 4525–4534 CrossRef CAS PubMed
.
- P. Hamel, E. Abed, L. Brissette and R. Moreau, Am. J. Physiol. Cell Physiol., 2008, 294, C1021–C1033 CrossRef CAS PubMed
.
- T. Kumagai, T. Nagata, Y. Kudo, Y. Fukuchi, K. Ebina and K. Yokota, Yakugaku Zasshi, 2001, 121, 277–281 CrossRef CAS
.
- M. Ani, A. A. Moshtaghie and H. Ahmadvand, Iran. Biomed. J., 2007, 11, 113–118 CAS
.
- M. R. Brodeur, L. Brissette, L. Falstrault, P. Ouellet and R. Moreau, Free Radic. Biol. Med., 2008, 44, 506–517 CrossRef CAS PubMed
.
- F. Liu, Z. Wang, F. Liu, J. Xu, Q. Liu, K. Yin and J. Lan, Cell Biosci., 2018, 8, 55 CrossRef PubMed
.
- Q. Q. Wang, W. Li and B. C. Yang, J. Biomed. Mater. Res. A, 2011, 99, 125–134 CrossRef CAS PubMed
.
- A. M. Pereira, R. Machado, A. da Costa, A. Ribeiro, T. Collins, A. C. Gomes, I. B. Leonor, D. L. Kaplan, R. L. Reis and M. Casal, Acta Biomater., 2017, 47, 50–59 CrossRef CAS PubMed
.
- P. P. Ming, S. Y. Shao, J. Qiu, J. Yang, Y. J. Yu, J. X. Chen, W. Q. Zhu and C. B. Tang, Appl. Surf. Sci., 2017, 416, 790–797 CrossRef CAS
.
- M. Okayasu, M. Nakayachi, C. Hayashida, J. Ito, T. Kaneda, M. Masuhara, N. Suda, T. Sato and Y. Hakeda, J. Biol. Chem., 2012, 287, 19229–19241 CrossRef CAS PubMed
.
- M. B. Tekin and H. Toker, Int. J. Implant Dent., 2019, 15, 18–25 CrossRef PubMed
.
| This journal is © The Royal Society of Chemistry 2019 |