Open Access Article
Open Access ArticlePoly(adenine)-mediated DNA-functionalized gold nanoparticles for sensitive detection of mercury ions in aqueous media
Jinjin Yin,
Jiuchao Wang,
Xiyue Yang,
Tao Wu *,
Huashan Wang
*,
Huashan Wang * and
Xiaoming Zhou
* and
Xiaoming Zhou
College of Chemical Engineering and Materials Science, State Key Laboratory of Food Nutrition and Safety, Key Laboratory of Food Nutrition and Safety, Tianjin University of Science and Technology, Tianjin 300457, China. E-mail: wutaoxx@gmail.com; whs@tust.edu.cn
First published on 14th June 2019
Abstract
In this work, a facile and sensitive colorimetric sensor for Hg2+ ions based on poly (adenine)-mediated DNA-functionalized gold nanoparticles (Au NPs) is reported. One DNA sequence consisting of poly-A and T-rich DNA was designed rationally. Poly-A was used as an anchoring block to bind tightly to Au NPs, and T-rich DNA was utilized for specific recognition of Hg2+ ions. With the assistance of poly-A, T-rich DNA was easily introduced onto the surface of Au NPs and kept an upright orientation. In the presence of Hg2+ ions, T base binding with Hg2+ ions results in the formation of “T–Hg2+–T” among the Au NPs, which caused aggregation of the Au NPs and a subsequent change in the color of the solution, from wine red to grayish blue. On this occasion, the limit of detection (LOD) was 3.75 nM Hg2+ ions with a linear range from 5 nM to 200 nM, as measured by UV-Vis spectroscopy. Moreover, successful application of this method for the detection of Hg2+ ions in real samples was demonstrated.
Introduction
It is generally acknowledged that mercury is one of the severest pollutants with different forms in the natural environment.1 Mercury ions (Hg2+) are the main and most widely distributed form that could be converted to organic mercury with higher toxicity by microorganisms. Organic mercury could be accumulated in organisms in the food chain and result in acute damage to human health, such as permanent damage to the central nervous system, brain and liver.2–5 These serious health problems have encouraged researchers to develop efficient, facile and fast methods for detection of Hg2+ in the environment with high selectivity and sensitivity to reduce the conversion of organic mercury.6–11 The World Health Organization has established a maximum permissible Hg2+ level of 5 nM in drinking water.12 At present, the conventional analysis methods for Hg2+ ions with responsive sensitivity include absorption spectrometry, inductively coupled plasma-mass spectrometry and so on.13–15 However, these methods suffer from their own disadvantages, such as complicated sample treatment, and requiring trained personnel, expensive equipment and off-site analysis.Oligonucleotides are not only genetic material in biology, but also useful tools in chemistry, such as used as a template to prepare nano-materials16–18 used as an element to engineer nano-probe and used as the “chemistry antibody” to recognize targets (aptamer) and so on.19–21 The thymine (T) base of oligonucleotide specific binding with Hg2+ to form “T–Hg2+–T” complex, and since it was reported,22 many sensors for detection of Hg2+ have been developed based on it.23,24 Liu developed a Hg2+ detection method with a detection limit of 0.5 nM by using Hg2+ specific probe (18T) and the intercalation dye SYBR Green I.25 In order to expand the application of detection methods, nanomaterials have been employed to construct sensitive sensors for Hg2+. Gold nanoparticles (Au NPs) are widely used to engineer nano-sensors in the fields of analytical chemistry and bio-sense because of their excellent physicochemical properties.26–28 In 2007, Mirkin developed a colorimetric detection of Hg2+ in aqueous media using DNA-functionalized Au NPs for the first time.29 Furthermore, Liu reported a room temperature colorimetric method to detect Hg2+ based on DNA conjugated Au NPs.30 Recently, Chen and co-workers developed a colorimetric sensor for Hg2+ based on “T–Hg2+–T” coordination to grow Au NPs furtherly.10
Up to now, most DNA-functionalized Au NPs are usually thiolate-modified DNA probes on the surface of Au NPs, and it is a multi-step and time-consuming chemical process to construct them.31,32 Fortunately, it has been reported that poly-A as an effective anchoring block could bind tightly to the surface of Au NPs, which is a facile and label-free process. Moreover, the DNA probe connected with poly-A could maintain an upright orientation, which favours the binding of the target to the DNA probe.33,34 Based on this principle, we employed a single-stranded DNA (ssDNA) containing thymine (T)-rich DNA and poly-adenine to engineer a versatile and sensitive colorimetric detection method specific to Hg2+ ions based on Au NPs. Poly-A brings the T-rich DNA to Au NPs, and T-rich DNA strands can be hybridized with each other among the Au NPs because of Hg2+ ions lead to form the structure of “T–Hg2+–T”. Then the colour of solution of Au NPs changes when the Au NPs aggregate. The limit of detection (LOD) was measured at 3.75 nM Hg2+ ions using UV-Vis spectroscopy. And this strategy specific to Hg2+ ions cannot be affected by other metal ions. It is a facile and fast detection sensor for Hg2+ ions with high sensitivity and selectivity.
Experimental
Materials and chemicals
Chloroauric acid (HAuCl4) was purchased from Sigma-Aldrich Chemicals. A stock solution of HAuCl4 was prepared by dissolving 1.0 g HAuCl4 in 100 mL ultrapure water (18.2 MΩ cm−1) from a Millipore water purification system. Sodium citrate and hydrochloric acid were purchased from Sinopharm Chemical Reagent Co. Ltd. Purified DNA sequences were purchased from Sangon Biotechnology Co. Ltd (Shanghai, China) and schematically shown in Table 1. Standard liquid elemental mercury (1 mg mL−1) was purchased from Aladdin Industrial Corporation. Unless otherwise noted, all reagent grade chemicals were used directly without further purification.| DNA name | DNA sequences (5′–3′) |
|---|---|
| T15 | TTTTTTTTTTTTTTT |
| A5R | AAAAA-random sequence (10 bases) |
| A5T15 | AAAAATTTTTTTTTTTTTTTTTTTT |
| A5T10 | AAAAATTTTTTTTTT |
| A5T5 | AAAAATTTTT |
| A10T10 | AAAAAAAAAATTTTTTTTTT |
| A15T10 | AAAAAAAAAAAAAAATTTTTTTTTT |
| A20T10 | AAAAAAAAAAAAAAAAAAAATTTTTTTTTT |
| A5A | AAAAATTCTTCTTTCTTCCCCCCTTGTTTGTTGTTT |
Preparation and characterization of Au NPs
All glassware was washed and then thorough rinsed with chloroazotic acid before use in this work. Au NPs (an average diameter of 13 nm) were prepared by the reduction of HAuCl4 solution with sodium citrate.35 Briefly, 50 mL of 1% HAuCl4 solution was heated to 100 °C. Then, 10 mL sodium citrate solution (38.8 mM) was freshly prepared and added rapidly to the HAuCl4 solution. The solution was mixed by stirring and heating until the colour of the solution turned to wine red. The solution was cooled naturally to room temperature and then stored at 4 °C in the dark before using. Ultraviolet-visible (UV-Vis) spectra of the Au NPs solution were characterized at 520 nm using a Cary 50-Bio UV spectrometer (Victoria, Australia). Transmission electron microscopy (TEM) images of the Au NPs were obtained with a 2010FEF microscope (JEOL, Japan).Preparation of DNA–Au NPs conjugates
The preparation of DNA–Au NP conjugates based on poly-adenine-mediation was performed according to a procedure by Opdahl et al.33 Briefly, Au NPs were incubated with A-rich DNA with a molar ratio of 200 (DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Au NPs) for 12 h on ice. The buffer used in this process was composed of 10 mM sodium phosphate and 0.1 M NaCl at pH 7.4. And then the conjugates were washed three times with this buffer at centrifugation with 12
Au NPs) for 12 h on ice. The buffer used in this process was composed of 10 mM sodium phosphate and 0.1 M NaCl at pH 7.4. And then the conjugates were washed three times with this buffer at centrifugation with 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 20 min, 4 °C.
000 rpm, 20 min, 4 °C.
Detection of Hg2+ by our strategy in buffer
For Hg2+ detection, 5 μL solution containing different concentrations of Hg2+ ions were incubated with 200 μL DNA–Au NPs conjugates for 10 min. And then the absorption at 520 nm was recorded by Cary 50-Bio UV spectrometer. For each sample, the experiment was repeated three times. The control experiments using 5 μL of solution without Hg2+ ions were carried out under same conditions.Detection of Hg2+ in real samples
To investigate the capability of our strategy for practical application, tap water and lake water were used as real samples in this work. The tap water was used directly without further measurement, and the lake water samples were collected from a lake located on the campus of the Tianjin University of Science and Technology, was first filtered by using a 0.22 μm membrane to remove the sand, particles and microorganisms. All the water samples were treated with different concentrations of Hg2+ ions on the basis of the presence of possible metal ions in the water. The detection procedure method used was mentioned above.Results and discussion
Sensor operation principle
The colorimetric detection of mercury ions based on poly-adenine-mediated DNA-functionalized gold nanoparticles is shown in Scheme 1. This strategy relies on a well-designed ssDNA containing thymine (T)-rich DNA (in green) and poly-adenine (A) (in blue) to engineer a colorimetric strategy specific to Hg2+ based on Au NPs. In this design, the T-rich portion is used to discern Hg2+, and poly-A portion is used as an anchoring block to bind with Au NPs. With the assistance of poly-A, T-rich DNA could be connected to Au NPs and maintain an upright orientation. In the presence of Hg2+ ions, the precise structure of “T–Hg2+–T” appears in the T-rich DNA portion, which will result in aggregation of the Au NPs. In contrast, in the absence of Hg2+ ions, T-rich DNA portion cannot hybridize between each other and not form aggregated Au NPs. The different colored solutions of Au NPs (from wine red to grayish blue) are obtained through the generation of morphologically varied Au NPs.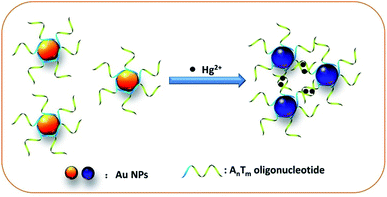 | ||
| Scheme 1 The principle of poly-A mediated DNA-functionalized Au NPs for sensitive detection of Hg2+ ions. | ||
Feasibility study experiment
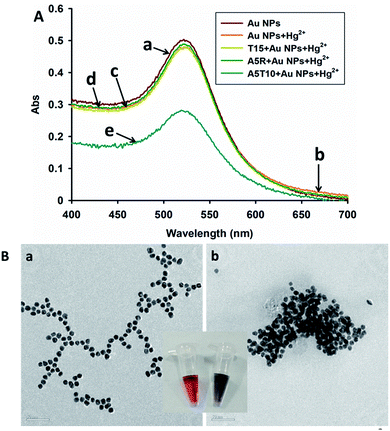
As a proof of feasibility experiment, several DNA sequences were used with Au NPs. As shown in Fig. 1A, the curve a is the absorption spectrum of the Au NPs, curve b is the resolution of Au NPs with 1 μM Hg2+ ions, and curves c–e are the resolution of T15, A5R and A5T10–Au NPs with 1 μM Hg2+ ions, respectively. From Fig. 1A, we can find that the obvious absorption peak change appear in curve e, A5T10–Au NPs with 1 μM Hg2+ ions. The oligonucleotide of A5T10 is consist of poly-A and T-rich base, whereas T15 is only T-rich sequence, and A5R includes poly-A with a random base sequence. The results proved that our designed strategy specific to Hg2+ ions works well. Moreover, TEM images of the solution of A5T10–Au NPs with and without 1 μM Hg2+ ions are shown in Fig. 1B, respectively. An aggregated Au NPs were found after adding 1 μM Hg2+ ions in A5T10–Au NPs, which further confirmed the performance of our design.Optimization of experimental conditions
To achieve a sensitive and selective response of the assay, several analytical parameters were investigated, including the number of adenine and thymine bases, recognition sequence, the concentration of DNA and the pH of solution. Firstly, the number of adenine and thymine bases was studied, as shown in Fig. 2. Different DNA sequences were used to anchor the Au NPs and then 5 μM Hg2+ ions was added. The change in the absorption peak at approximately 520 nm was recorded by UV-Vis spectroscopy. Fig. 2B shows that, the A5T10 anchored Au NPs exhibited the best response to Hg2+ ions in these designed DNA sequences. It should be noted that the aptamer of Hg2+ ions was connected with 5 adenine bases (A5A), which could change the absorption peak at approximately 520 nm of the Au NPs in the presence of Hg2+ ions. However, the change was not as obvious as that of A5T10 (see in Fig. 2). Thus, A5T10 was selected to engineer the strategy in this work.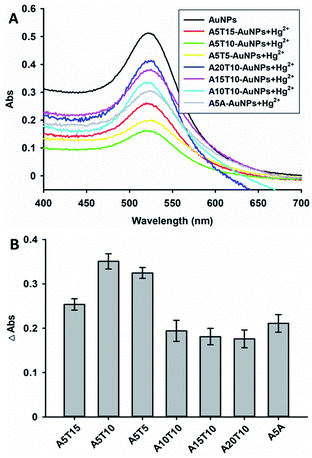 | ||
| Fig. 2 The number of adenine and thymine was optimized with adding 5 μM Hg2+ ions. (A) The UV-vis spectroscopy and (B) corresponding histogram. | ||
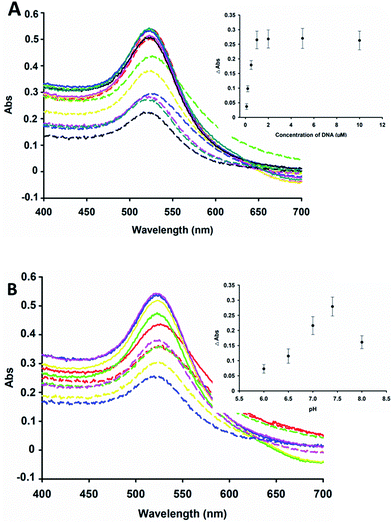
In this design, because the concentration of DNA and the pH of the solution have impacts on the sensitivity of this detection strategy, we investigated the effects of different dosages of DNA and pH of the buffer. The results are shown in Fig. 3A, and indicate that when the concentration of DNA increased, the change in the absorbance at 520 nm did not increase greatly when the concentration of DNA was much more than 1 μM with 500 nM Hg2+ ions. We investigated the pH effects from 6.0 to 8.0, as shown in Fig. 3B. The biggest change in absorbance at 520 nm was obtained at pH 7.4. In summary, the optimum condition was 1 μM A5T10 DNA in pH 7.4 buffer.
Sensitivity and selectivity
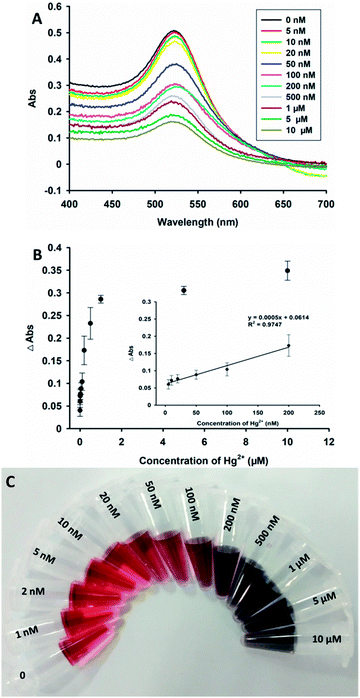
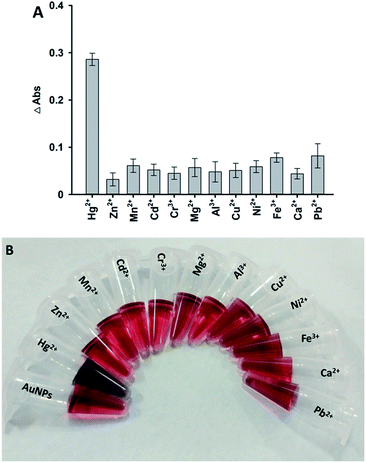
With optimized detection conditions, a series of samples with different concentrations (0, 5 nM, 10 nM, 20 nM, 50 nM, 100 nM, 200 nM, 500 nM, 1 μM, 5 μM and 10 μM) of Hg2+ ions were measured by this method. The absorbance at 520 nm gradually decreased when the amount of Hg2+ ions increased (Fig. 4A). For each sample, the change in the absorbance at 520 nm, Y, was plotted versus the concentration of Hg2+ ions, X, as shown in Fig. 4B. The regression equation of Y = 0.0005X + 0.0614 with a linear range was observed in the presence of 5–200 nM Hg2+ ions (R2 = 0.9747). The detection limit of this method was found to be 3.75 nM, and was estimated based on the 3δ/slope rule. Compared to other colorimetric methods for the assay of Hg2+ ions,10,36,37 this proposed strategy was shown to have a comfortable limit of detection based on a very facile experimental process.In addition, the selectivity of the designed assay was also evaluated for the determination of other metal ions, including Zn2+, Mn2+, Cd2+, Cr3+, Mg2+, Al3+, Cu2+, Ni2+, Fe3+, Ca2+, and Pb2+ ions instead of Hg2+ ions in the solution. Fig. 5A shows the changes in absorbance at 520 nm of this system when it was treated with different metal ions. An obvious change was observed from the solution in the presence of 1 μM Hg2+ ions, while very low changes were observed in the presence of other metal ions (10 μM). Thus, the results proved that the detection method has high selectivity for Hg2+ ions.
Detection of Hg2+ ions in real samples
Furthermore, the developed colorimetric assay was applied to the determination of Hg2+ ions in real samples, the tap water and the lake water (treated by the method mentioned in Experimental section) to realize its applicability for real sample analysis. The results are shown in Table 2, and the recovery values ranged from 92 to 105.2% using our designed method. The excellent accuracy of this assay indicates that its application to real samples would be ideal.| Added (nM) | ICP-MS (nM) | Mean found (nM) | Recovery (%) |
|---|---|---|---|
| 50 | 49 | 46 | 92 |
| 200 | 204 | 195 | 97.5 |
| 500 | 508 | 526 | 105.2 |
Conclusions
In this work, we have developed a facile and sensitive colorimetric assay for the specific detection of Hg2+ ions based on poly-A mediated DNA-functionalized Au NPs. A poly-A and T-rich DNA sequence was selected to engineer the strategy for determination of Hg2+ ions. Poly-A was used to bring the T-rich portion onto the surface of Au NPs, and then binding specifically occurred between the T-rich portion of the oligonucleotide and the Hg2+ ions to form the complex of “T–Hg2+–T” among the Au NPs. The color of the solution then changed from wine red to grayish blue. Under optimized detection conditions, this method can detect Hg2+ ions with a LOD of 3.75 nM Hg2+ ions in a linear range from 5 nM to 200 nM by using UV-Vis spectroscopy. Furthermore, this method had good accuracy that can be applied to real samples. Thus, a simple and low-cost colorimetric assay for Hg2+ ions with high sensitivity and selectivity was developed that has great potential for monitoring Hg2+ ions in the environment.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is supported by the Natural Science Foundation of Tianjin [No. 18JCQNJC0600] and the Youth Innovation Foundation of Tianjin University of Science and Technology [No. 2016LG16].Notes and references
- T. W. Clarkson and L. Magos, Crit. Rev. Toxicol., 2006, 36, 609–662 CrossRef CAS PubMed.
- H. H. Harris, I. J. Pickering and G. N. George, Science, 2003, 301, 1203 CrossRef CAS PubMed.
- Z. Gu, M. X. Zhao, Y. W. Sheng, L. A. Bentolila and Y. Tang, Anal. Chem., 2011, 83, 2324–2329 CrossRef CAS PubMed.
- Y. Choi, Y. Park, T. Kang and L. P. Lee, Nat. Nanotechnol., 2009, 4, 742–746 CrossRef CAS PubMed.
- E. S. Cho, J. Kim, B. Tejerina, T. M. Hermans, H. Jiang, H. Nakanishi, M. Yu, A. Z. Patashinski, S. C. Glotzer and F. Stellacci, Nat. Mater., 2012, 11, 978–985 CrossRef CAS PubMed.
- Z. B. Chen, C. M. Zhang, Q. G. Gao, G. Wang, L. L. Tan and Q. Liao, Anal. Chem., 2015, 87, 10963–10968 CrossRef CAS PubMed.
- C. Yuan, B. Liu, F. Liu, M. Y. Han and Z. Zhang, Anal. Chem., 2014, 86, 1123–1130 CrossRef CAS PubMed.
- J. Du, B. Zhu and X. Chen, Small, 2013, 9, 4104–4111 CrossRef CAS PubMed.
- X. Li, J. Q. Xie, B. Y. Jiang, R. Yuan and Y. Xiang, ACS Appl. Mater. Interfaces, 2017, 9, 5733–5738 CrossRef CAS PubMed.
- L. L. Tan, Z. B. Chen, C. Zhang, X. C. Wei, T. H. Lou and Y. Zhao, Small, 2017, 13, 1603370–1603377 CrossRef PubMed.
- Q. F. Zhai, H. H. Xing, X. W. Zhang, J. Li and E. K. Wang, Anal. Chem., 2017, 89, 7788–7794 CrossRef CAS PubMed.
- G. Aragay, J. Pons and A. Merkoçi, Chem. Rev., 2011, 111, 3433–3458 CrossRef CAS PubMed.
- Z. H. Fernández, L. A. V. Rojas, A. M. Álvarez, J. R. E. Álvarez, J. A. dos Santos, I. P. González, M. R. Gonzalez, N. A. Macias, D. L. Sanchez and D. H. Torres, Food Control, 2015, 48, 37–42 CrossRef.
- M. Noël, J. R. Christensen, J. Spence and C. T. Robbins, Sci. Total Environ., 2015, 529, 1–9 CrossRef PubMed.
- W. N. Chen, S. J. Jiang, Y. L. Chen and A. C. Sahayam, Anal. Chim. Acta, 2015, 860, 8–14 CrossRef CAS PubMed.
- J. T. Petty, J. Zheng, N. V. Hud and R. M. Dickson, J. Am. Chem. Soc., 2004, 126, 5207–5212 CrossRef CAS PubMed.
- N. Ma, E. H. Sargent and S. O. Kelley, Nat. Nanotechnol., 2009, 4, 121–125 CrossRef CAS PubMed.
- A. Lopez and J. W. Liu, Can. J. Chem., 2015, 93, 615–620 CrossRef CAS.
- S. J. Reinholt and H. G. Craighead, Anal. Chem., 2018, 90, 2601–2608 CrossRef CAS PubMed.
- P. F. Zhang, Z. Zhao, C. S. Li, H. F. Su, Y. Y. Wu, R. T. K. Kwok, J. W. Y. Lam, P. Gong, L. T. Cai and B. Z. Tang, Anal. Chem., 2018, 90, 1063–1067 CrossRef CAS PubMed.
- J. J. Yin, Y. Q. Liu, S. Wang, J. K. Deng, X. D. Lin and J. T. Gao, Sens. Actuators, B, 2018, 256, 573–579 CrossRef CAS.
- Y. Miyake, H. Togashi, M. Tashiro, H. Yamaguchi, S. Oda, M. Kudo, Y. Tanaka, Y. Kondo, R. Sawa, T. Fujimoto, T. Machinami and A. Ono, J. Am. Chem. Soc., 2006, 128, 2172–2173 CrossRef CAS PubMed.
- X. Y. Jiang, H. J. Wang, H. J. Wang, R. Yuan and Y. Q. Chai, Anal. Chem., 2016, 88, 9243–9250 CrossRef CAS PubMed.
- X. J. Liu, Z. J. Wu, Q. Q. Zhang, W. F. Zhao, C. H. Zong and H. W. Gai, Anal. Chem., 2016, 88, 2119–2124 CrossRef CAS PubMed.
- B. Liu, Biosens. Bioelectron., 2008, 24, 756–760 CrossRef CAS PubMed.
- M. Annadhasan, T. Muthukumarasamyvel, V. R. Sankar Babu and N. Rajendiran, ACS Sustainable Chem. Eng., 2014, 2, 887–896 CrossRef CAS.
- Y. Zhao, L. L. Gui and Z. B. Chen, Sens. Actuators, B, 2017, 241, 262–267 CrossRef CAS.
- X. F. Ding, L. T. Kong, J. Wang, F. Fang, D. D. Li and J. H. Liu, ACS Appl. Mater. Interfaces, 2013, 5, 7072–7078 CrossRef CAS PubMed.
- J. S. Lee, M. S. Han and C. A. Mirkin, Angew. Chem., Int. Ed., 2007, 46, 4093–4096 CrossRef CAS PubMed.
- X. J. Xue, F. Wang and X. G. Liu, J. Am. Chem. Soc., 2008, 130, 3244–3245 CrossRef CAS PubMed.
- T. T. Lou, Z. P. Chen, Y. Q. Wang and L. X. Chen, ACS Appl. Mater. Interfaces, 2011, 3, 1568–1573 CrossRef CAS PubMed.
- J. W. Liu and Y. Lu, Nat. Protoc., 2006, 1, 246–252 CrossRef CAS PubMed.
- A. Opdahl, D. Y. Petrovykh, H. K. Suda, M. J. Tarlov and L. J. Whitman, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 9–14 CrossRef PubMed.
- D. Zhu, P. Song, J. W. Shen, S. Su, J. Chao, A. Aldalbahi, Z. A. Zhou, S. P. Song, C. H. Fan, X. L. Zuo, Y. Tian, L. H. Wang and H. Pei, Anal. Chem., 2016, 88, 4949–4954 CrossRef CAS PubMed.
- H. D Hill and C. A. Mirkin, Nat. Protoc., 2006, 1, 324–336 CrossRef PubMed.
- Z. H. Qing, X. X. He, K. M. Wang, Z. Z. Zou, X. Yang, J. Huang and G. P. Yan, Anal. Methods, 2012, 4, 3320–3325 RSC.
- W. H. Ni, H. J. Chen, J. Su, Z. H. Sun, J. F. Wang and H. K. Wu, J. Am. Chem. Soc., 2010, 132, 4806–4814 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |