Open Access Article
Open Access ArticleIdentification of critical growth factors for peripheral nerve regeneration
Ruirui Zhang†
 a,
Yan Zhang†b and
Sheng Yi*a
a,
Yan Zhang†b and
Sheng Yi*a
aKey Laboratory of Neuroregeneration of Jiangsu and Ministry of Education, Co-innovation Center of Neuroregeneration, Nantong University, Nantong, Jiangsu 226001, China. E-mail: syi@ntu.edu.cn; Fax: +86-513-85511585; Tel: +86-513-85511585
bDepartment of Respiratory and Critical Care Medicine, Affiliated Hospital of Nantong University, Nantong, Jiangsu 226001, China
First published on 5th April 2019
Abstract
Growth factors are essential for the repair and regeneration of tissues and organs, including injured peripheral nerves. However, the expression changes of growth factors during peripheral nerve regeneration have not been fully elucidated. To obtain a global view of alternations of growth factors during the regeneration process, we explored previously achieved sequencing data of rat sciatic nerve stumps at 0 h, 1 d, 4 d, 7 d, and 14 d after nerve crush injury and screened differentially expressed upstream growth factors using Ingenuity Pathway Analysis (IPA) bioinformatic software. Differentially expressed growth factors were then subjected to Gene Ontology (GO) annotation and Kyoto Enrichment of Genes and Genomes (KEGG) pathway analysis. Regulatory networks of the differentially expressed growth factors in axon growth-related biological processes were constructed. Pivotal growth factors involved in axon growth were identified and validated by qRT-PCR. Our current study identified differentially expressed growth factors in the injured nerve stumps after peripheral nerve injury, discovered key growth factors for axon growth and nerve regeneration, and might facilitate the discovery of potential therapeutic targets of peripheral nerve injury.
Introduction
Peripheral nerve injury is a worldwide clinical problem with a quite high incidence.1,2 It affects millions of patients every year, causes severe pain and disability, and induces tremendous social and economical burdens.3 Morphological studies have demonstrated that a complex series of biological changes, including axon and myelin sheath disintegration, wallerian degeneration, debris removal, regenerative tunnel formation, axon extension, axon remyelination, and target reinnervation occur after peripheral nerve injury.4,5 The application of large-scale genetic studies, such as microarray and deep sequencing, have further revealed biological alterations at the cellular, molecular, and genetic levels.6–9 Further elucidation of these genetic changes will help to discover key regulatory factors of peripheral nerve repair and identify potential therapeutic approaches for the treatment of peripheral nerve injury.Growth factors are generally proteins or steroid hormones that are capable of instructing cellular behavior and motivating cellular growth, proliferation, differentiation, and maturation.10 Various growth factors, including epidermal growth factor (Egf), vascular endothelial growth factor (Vegf), platelet derived growth factor (Pdgf), fibroblast growth factor (Fgf), and transforming growth factor-β (Tgf-β) are involved in wound healing and tissue remodeling.11–14 Growth factors are further utilized in regenerative medicine applications to guide and promote tissue regrowth and renewal.15
Emerging studies have shown that many growth factors, especially many neurotrophic factors such as nerve growth factor (Ngf) and brain-derived neurotrophic factor (Bdnf), are very critical for the development and regeneration of the nervous system.16–19 However, there is no systemical investigation of the involvement of growth factors in the biological processes of peripheral nerve injury and repair. In our previous study, by using RNA deep sequencing, we fully determined gene expressions in the sciatic nerve stumps at 0 h, 1 d, 4 d, 7 d, and 14 d after rat sciatic nerve crush injury and profiled differentially expressed genes.20 Here, we further conducted upstream analysis of these differentially expressed genes and screened differentially expressed upstream growth factors using Ingenuity Pathway Analysis Software (IPA). These dysregulated upstream growth factors were functionally analyzed by Database for Annotation, Visualization, and Integrated Discovery (DAVID) to identify critical Gene Ontology (GO) terms and Kyoto Enrichment of Genes and Genomes (KEGG) pathways. Growth factors that were highly related with axon growth, a critical biological process for nerve regeneration and functional recovery, were investigated in detail.
Results and discussion
Differentially expressed upstream growth factors in sciatic nerve stumps after nerve crush injury
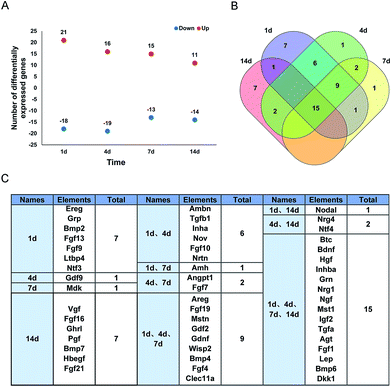
RNA deep sequencing is a very powerful high-throughput analysis tool in examining expression levels of thousands of genes and is now widely applied in biomedical science.21 Previously, by using RNA deep sequencing, we investigated global transcriptional changes in the sciatic nerve stumps after rat sciatic nerve injury.20,22 Differentially expressed genes were then analyzed according to their biological functions. For example, differentially expressed genes in injury-induced robust immune/inflammatory responses,23 actin cytoskeleton organization,24 tight junction signaling,25 and adherens junction signaling and remodeling26 were screened. Besides these critical biological activities, growth factors and growth factors-involved biological processes might also be essential for the repair and regeneration of peripheral nerves. Therefore, in the current study, we systematically characterized the dynamic transcript changes of upstream growth factors of differentially expressed genes.Previously identified differentially expressed genes in the sciatic nerve stumps at 1 d, 4 d, 7 d, and 14 d after rat sciatic nerve crush injury were core analyzed to identify upstream regulators at each time point. Upstream growth factors were selected from these upstream regulators. A total of 66, 71, 72, and 57 upstream growth factors were recognized at 1 d, 4 d, 7 d, and 14 d, respectively. Upstream growth factors with a fold change >2 or <−2 were further screened as differentially expressed upstream growth factors. 21 upstream growth factors were up-regulated and 18 upstream growth factors were down-regulated at 1 d after nerve crush injury (Fig. 1A). Relatively few upstream growth factors were differentially expressed at later time points (Fig. 1A). These differentially expressed upstream growth factors during the time course were further analyzed and illustrated by the Venn diagram (Fig. 1B) and listed (Fig. 1C). A total of 15 genes coding for growth factors, including betacellulin (Btc), Bdnf, hepatocyte growth factor (Hgf), insulin subunit beta A (Inhba), granulin precursor (Grn), neuregulin 1 (Nrg1), Ngf, macrophage stimulating 1 (Mst1), insulin like growth factor 2 (Igf2), transforming growth factor alpha (Tgfa), angiotensinogen (Agt), fibroblast growth factor 1 (Fgf1), leptin (Lep), bone morphogenetic protein 6 (Bmp6), and dickkopf WNT signaling pathway inhibitor 1 (Dkk1) were differentially expressed, indicating that these consistently differentially expressed growth factors might be critical for peripheral nerve repair and regeneration.
Validation of the expression levels of differentially expressed growth factors
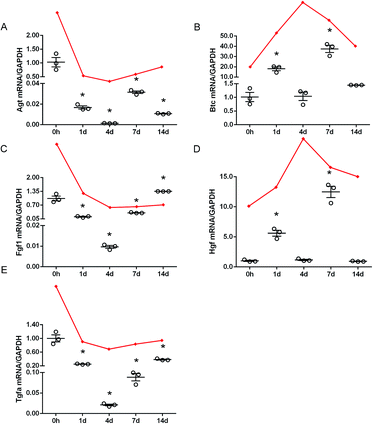
To validate the accuracy of outcomes from RNA deep sequencing, qRT-PCR experiments were conducted to examine the expression levels of Agt, Btc, Fgf1, Hgf, and Tgfa, 5 genes that were consistently differentially expressed after rat sciatic nerve crush injury. qRT-PCR outcomes showed that Agt, Fgf1, and Tgfa were robustly down-regulated in the sciatic nerve stumps after nerve injury while Btc and Hgf were mainly up-regulated after nerve injury (Fig. 2). These results were generally in consistent with RNA deep sequencing outcomes.However, these still existed some inconsistency. RNA deep sequencing outcomes showed that Btc and Hgf were kept up-regulated after sciatic nerve crush injury while qRT-PCR outcomes showed that Btc and Hgf were not significantly up-regulated at 4 d after nerve crush injury. These inconsistent results might be related with many factors such as RNA quality, different batches of samples, different efficiencies of reverse transcriptases, different priming methods, and different data normalization methods, and were previously identified and reported in other studies.27–30
Hgf has been shown to promote survival and axon outgrowth of cultured motor neurons in peripheral nervous system.31 Also, Hgf could induce the activation of Schwann cells to promote re-myelination of injured nerves, and exogenous addition of Hgf could expedite peripheral nerve regeneration.32 A recent study identified Btc as a new regulator of axon-Schwann cell interactions in the mammalian peripheral nerve, and regulates myelin formation and peripheral nerve repair.33
Enriched biological activities of differentially expressed growth factors
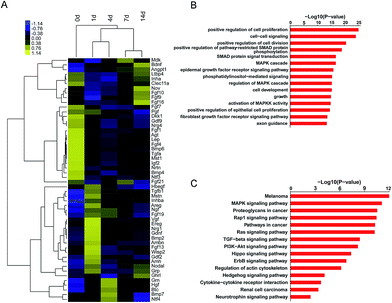
To obtain an overview of the temporal expression patterns of differentially expressed growth factors after sciatic nerve crush injury, the expression levels of these differentially expressed growth factors were displayed in a heatmap. It was demonstrated that about 1/2 of these differentially expressed growth factors were highly expressed at early time points after sciatic nerve crush injury (1 d and 4 d), about 1/5 of differentially expressed growth factors were highly expressed at later time points after sciatic nerve crush injury (14 d), and about 1/3 of differentially expressed growth factors were kept down-regulated after sciatic nerve crush injury (Fig. 3A).Differentially expressed growth factors were then categorized to GO terms and KEGG pathways to discover possible biological activities involved in peripheral nerve injury. Top GO terms showed that cellular behaviors-related GO terms, including positive regulation of cell proliferation, cell–cell signaling, positive regulation of cell division, cell development, growth, and positive regulation of epithelial cell proliferation were highly enriched (Fig. 3B). Top KEGG pathways showed that differentially expressed growth factors were enriched in tumor and cancer-related pathways (melanoma, proteoglycans in cancer, pathways in cancer, and renal cell carcinoma), cellular cytoskeleton-related pathways (Ras signaling pathway, and regulation of actin cytoskeleton), and cellular behaviors-related pathways (MAPK signaling pathway, TGF-beta signaling pathway, and PI3K-Akt signaling pathway) (Fig. 3C).
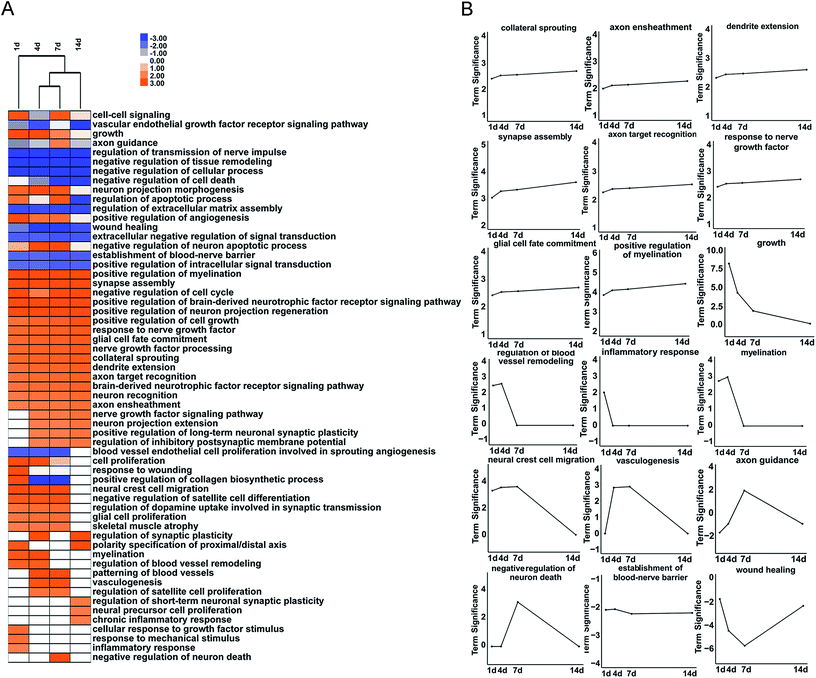
To further reveal differentially expressed growth factors-involved biological processes after peripheral nerve injury, GO biological process terms, especially GO biological process terms related with wounding healing and tissue remodeling, were investigated in detail. The majority of growth factors-involved GO biological process terms, such as positive regulation of myelination, synapse assembly, and positive regulation of cell growth, were kept up-regulated after nerve crush injury (Fig. 4A).
The expression changes of differentially expressed growth factors involved in key categories in nerve regeneration were further demonstrated in curves (Fig. 4B). Many neurite growth and neurite function-related GO biological process terms, including collateral sprouting, dendrite extension, axon ensheathment, synapse assembly, axon target recognition, response to nerve growth factor, glial cell fate commitment, and positive regulation of myelination were kept activated at all time points after nerve crush injury. Some GO biological processes, including growth, regulation of blood vessel remodeling, neural crest cell migration, myelination, and inflammatory response were highly activated at early time points but less significance at later time points. And some GO biological processes were robustly activated at a single time point. For instance, growth factors involved in axon guidance was robustly activated at 7 d after nerve crush injury, suggested that 7 d might be critical for the directional guidance and pathfinding of axons.
Involvement of differentially expressed growth factors in axon growth-related biological activities
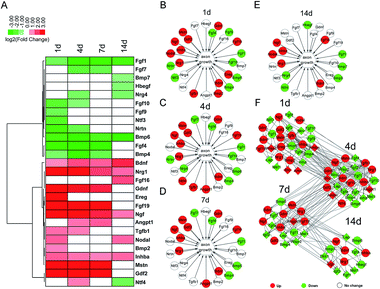
Exploring effective approaches to promote the growth of axons and guide axons to their targets are critical for the functional recovery of injured nerves. Therefore, we specifically investigated the involvement of differentially expressed growth factors in axon growth-related biological activities, including regulation of axonogenesis, axon extension, growth, and axon guidance. The temporal expression patterns of growth factors involved in these GO terms were shown in a heatmap (Fig. 5A). Moreover, functional networks at different time points after nerve crush injury were constructed to display the importance of these differentially expressed growth factors (Fig. 5B–E). A regulatory network of differentially expressed growth factors in axon growth-related biological activities to demonstrate the cascade relationship of these growth factors (Fig. 5F).Ngf and Bdnf, not surprisingly, were kept up-regulated after nerve injury. Our outcomes suggested that besides these two well-known growth factors, the expression levels of Nrg1 and Inhba were also kept increased in the injured sciatic nerve stumps. Nrg1 has been implicated in the maturation, survival, and motility of Schwann cells, the main glial cells in the peripheral nervous system.34 Moreover, Nrg1 has also been involved in the myelination of oligodendrocytes, the glial cells in the central nervous system.35 Therefore, it is likely that up-regulated Nrg1 might also regulate the myelination of Schwann cells and participate in constructing a permissive regenerative microenvironment and promoting axon growth by regulating the phenotype of Schwann cells. The expressions and functions of Inhba in the nervous system have not been fully investigated. A previous study showed that the serum level of activin A, a protein subunit encoded by Inhba, was increased after ischemia and reperfusion injury of rat sciatic nerves.36 The biological functions of Inhba needed to be further explored.
Meanwhile, it is worth noting that our observation showed that neurotrophin-4 (Ntf4), a distinguished neurotrophic factor that is critical for neuron survival,37,38 was first found to be first up-regulated and then down-regulated in the injured sciatic nerve stumps. Ntf4 was indicated to participate in the myelination of Schwann cells39,40 and the successful early growth of regenerating axons.41 Therefore, up-regulated Ntf4 at 4 d after nerve crush injury might contribute to nerve regeneration. However, the biological roles of down-regulated Ntf4 at later time points (14 d) remain unclear and needs to be investigated. Previously, Ntf4 was delivered as a supporting factor by fibrin glue to the injured nerve stumps to promote peripheral nerve regeneration.42 However, our current study showed that under pathological conditions, the expression levels of Ntf4 were not consistent up-regulated but first up-regulated and then down-regulated. Therefore, the concentrations of growth factors might need to be determined when constructing Ntf4-based tissue engineered nerve grafts or other growth factor-based tissue engineered nerve grafts to achieve good functional recovery.
Experimental
Animal surgery
A total of 30 Adult Sprague-Dawley (SD) rats, weighting 180–220 g, were purchased from the Experimental Animal Center of Nantong University were subjected to sciatic nerve crush injury as previously mentioned.20 Briefly, SD rats were anesthetized by the injection of mixed narcotics (85 mg kg−1 trichloroacetaldehyde monohydrate, 42 mg kg−1 magnesium sulfate, and 17 mg kg−1 sodium pentobarbital) and rat left sciatic nerves at 10 mm above the bifurcation into the tibial and common fibular nerves were crushed with a forceps for 3 times with 10 s for each time. Rats were randomly divided into five group with 6 rats in each group and were sacrificed by decapitation at 0 h, 1 d, 4 d, 7 d, and 14 d after nerve crush injury. Rats in the 0 h group were subjected to sham-surgery and considered as control animals. Animal procedures were ethically approved by the Administration Committee of Experimental Animals and conducted in accordance with Institutional Animal Care guideline of Nantong University.Bioinformatic analysis
Previously obtained RNA sequencing data20 of rat sciatic nerve stumps at 0 h, 1 d, 4 d, 7 d, and 14 d after nerve crush injury (National Center for Biotechnology Information database: accession number PRJNA394957, SRP113121) were uploaded to IPA software (Ingenuity Systems Inc., Redwood City, CA, USA) for core analysis. The expression levels of genes at 1 d, 4 d, 7 d, and 14 d after crush injury were compared with their expression levels at 0 h. Genes with a fold change >2 or <−2 and a false discovery rate <0.05 were considered as differentially expressed genes. Differentially expressed genes were then subjected to IPA upstream analysis to screen upstream regulators and select differentially expressed upstream growth factors. Venn diagram analysis (Venny 2.1.0 online tool; http://bioinfogp.cnb.csic.es/tools/venny/index.html) was used to discover intersecting differentially expressed growth factors. DAVID was used to enrich significant GO terms and KEGG pathways.43,44Tissue collection and RNA extraction
After sacrificing rats at different time points after nerve crush injury, 5 mm long sciatic nerve stumps at the crush sites were collected. Total RNA was isolated from sciatic nerve stumps using TRIzol reagent (Life Technologies, Carlsbad, CA, USA). RNase-free DNase I (TaKaRa, Dalian, China) was added to remove contaminating DNA. Purified RNA was quantified using a NanoDrop ND-1000 spectrophotometer (Infinigen Biotechnology Inc., City of Industry, CA, USA), reverse transcribed into cDNA with the Prime-Script reagent Kit (TaKaRa, Dalian, China), and subjected to subsequent RT-PCR experiments.Quantitative real time RT-PCR (qRT-PCR)
qRT-PCR was performed for quantification of gene expression with SYBR Premix Ex Taq (TaKaRa) on an Applied Biosystems Stepone real-time (Applied Biosystems, Foster City, CA, USA). Relative gene expression levels were quantified using the comparative 2−ΔΔCt method with GAPDH as the reference gene. Primer pair sequences were listed in Table 1.| Gene | Primers | Sequences |
|---|---|---|
| a Agt: angiotensinogen; Btc: betacellulin; Fgf1: fibroblast growth factor 1; Hgf: hepatocyte growth factor; Tgfa: transforming growth factor alpha; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; qRT-PCR: quantitative real time RT-PCR. | ||
| Agt | Agt-forward | 5′-GGCAAGATGGGTGACACCA-3′ |
| Agt-reverse | 5′-CTGCTTGGAGTTCAAGGAGGAT-3′ | |
| Btc | Btc-forward | 5′-TCTCCAGTGCGTGGTGG-3′ |
| Btc-reverse | 5′-CGAGAGAAGTGGGTTTTCGATT-3′ | |
| Fgf1 | Fgf1-forward | 5′-ATGGGACCAGGGACAGGAG-3′ |
| Fgf1-reverse | 5′-TGGTGTCTGCGAGCCGTAT-3′ | |
| Hgf | Hgf-forward | 5′-GCAAGACATGTCAGCGCTGG-3′ |
| Hgf-reverse | 5′-CCAAGGGGTGTCAGGGTCAA-3′ | |
| Tgfa | Tgfa-forward | 5′-CTGCAGGTTTTTGGTGCAGG-3′ |
| Tgfa-reverse | 5′-TGATGGCCTGCTTCTTCTGG-3′ | |
| GAPDH | GAPDH-forward | 5′-ACAGCAACAGGGTGGTGGAC-3′ |
| GAPDH-reverse | 5′-TTTGAGGGTGCAGCGAACTT-3′ | |
Statistical analysis
Statistical analyses were performed one-way analysis of variance using GraphPad Prism 6.0 (GraphPad Software, Inc., San Diego, CA, USA). Data were summarized from 3 paired experiments and expressed as the mean ± SEM. A p-value < 0.05 was considered as statistically significant.Conclusions
The current study provided valuable information regarding the dynamic changes of growth factors in the sciatic nerve stumps at different time points after rat nerve crush injury, elucidated the biological processes of differentially expressed growth factors, identified critical growth factors for peripheral nerve repair and regeneration, and thus might contribute to identification of potential treatments for peripheral nerve repair and regeneration.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This study was supported by a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions [PAPD].References
- R. Li, Z. Liu, Y. Pan, L. Chen, Z. Zhang and L. Lu, Cell Biochem. Biophys., 2014, 68, 449–454 CrossRef CAS PubMed.
- L. Tian, M. P. Prabhakaran and S. Ramakrishna, Regener. Biomater., 2015, 2, 31–45 CrossRef CAS PubMed.
- X. Gu, F. Ding, Y. Yang and J. Liu, Prog. Neurobiol., 2011, 93, 204–230 CrossRef CAS PubMed.
- A. Faroni, S. A. Mobasseri, P. J. Kingham and A. J. Reid, Adv. Drug Delivery Rev., 2015, 82–83, 160–167 CrossRef CAS PubMed.
- A. D. Gaudet, P. G. Popovich and M. S. Ramer, J. Neuroinflammation, 2011, 8, 110 CrossRef PubMed.
- S. Li, Q. Liu, Y. Wang, Y. Gu, D. Liu, C. Wang, G. Ding, J. Chen, J. Liu and X. Gu, PLoS One, 2013, 8, e57000 CrossRef CAS PubMed.
- B. Pan, Y. Liu, J. Y. Yan, Y. Wang, X. Yao, H. X. Zhou, L. Lu, X. H. Kong and S. Q. Feng, Muscle Nerve, 2017, 55, 373–383 CrossRef CAS PubMed.
- T. Qian, C. Fan, Q. Liu and S. Yi, Mol. Brain, 2018, 11, 73 CrossRef PubMed.
- C. Yao, Y. Wang, H. Zhang, W. Feng, Q. Wang, D. Shen, T. Qian, F. Liu, S. Mao, X. Gu and B. Yu, J. Neurosci., 2018, 38, 6574–6585 CrossRef CAS PubMed.
- S. A. Aaronson, Science, 1991, 254, 1146–1153 CrossRef CAS.
- D. Bose, F. Meric-Bernstam, W. Hofstetter, D. A. Reardon, K. T. Flaherty and L. M. Ellis, Lancet Oncol., 2010, 11, 373–382 CrossRef CAS.
- M. K. Lichtman, M. Otero-Vinas and V. Falanga, Wound Repair Regen., 2016, 24, 215–222 CrossRef PubMed.
- M. Rodrigues, N. Kosaric, C. A. Bonham and G. C. Gurtner, Physiol. Rev., 2019, 99, 665–706 CrossRef PubMed.
- S. Werner and R. Grose, Physiol. Rev., 2003, 83, 835–870 CrossRef CAS PubMed.
- A. C. Mitchell, P. S. Briquez, J. A. Hubbell and J. R. Cochran, Acta Biomater., 2016, 30, 1–12 CrossRef CAS PubMed.
- S. P. Frostick, Q. Yin and G. J. Kemp, Microsurgery, 1998, 18, 397–405 CrossRef CAS PubMed.
- S. Madduri and B. Gander, J. Peripher. Nerv. Syst., 2010, 15, 93–103 CrossRef CAS PubMed.
- A. D. Sebben, M. Lichtenfels and J. L. da Silva, Rev. Bras. Ortop., 2011, 46, 643–649 CrossRef PubMed.
- A. D. Widgerow, A. A. Salibian, S. Lalezari and G. R. Evans, J. Neurosci. Res., 2013, 91, 1517–1524 CrossRef CAS PubMed.
- S. Yi, H. Zhang, L. Gong, J. Wu, G. Zha, S. Zhou, X. Gu and B. Yu, PLoS One, 2015, 10, e0143491 CrossRef PubMed.
- Z. Wang, M. Gerstein and M. Snyder, Nat. Rev. Genet., 2009, 10, 57–63 CrossRef CAS PubMed.
- L. Zhao and S. Yi, J. Cell. Physiol., 2019, 234, 6876–6885 CrossRef CAS PubMed.
- L. Xing, Q. Cheng, G. Zha and S. Yi, Mediators Inflammation, 2017, 2017, 3827841 Search PubMed.
- Y. Wang, Q. Shan, J. Pan and S. Yi, Front. Physiol., 2018, 9, 23 CrossRef PubMed.
- X. Wang, Y. Miao, J. Ni, Y. Wang, T. Qian, J. Yu, Q. Liu, P. Wang and S. Yi, Front. Physiol., 2018, 9, 1519 CrossRef PubMed.
- S. Yi, X. H. Wang and L. Y. Xing, Neural Regener. Res., 2018, 13, 1804–1810 CrossRef PubMed.
- J. S. Morey, J. C. Ryan and F. M. Van Dolah, Biol. Proced. Online, 2006, 8, 175–193 CrossRef CAS PubMed.
- K. B. Beckman, K. Y. Lee, T. Golden and S. Melov, Mitochondrion, 2004, 4, 453–470 CrossRef CAS PubMed.
- W. Etienne, M. H. Meyer, J. Peppers and R. A. Meyer Jr, BioTechniques, 2004, 36, 618–620 CrossRef CAS PubMed.
- T. E. White, M. C. Surles-Zeigler, G. D. Ford, A. S. Gates, B. Davids, T. Distel, M. C. LaPlaca and B. D. Ford, BMC Genomics, 2016, 17, 130 CrossRef PubMed.
- Q. Yin, G. J. Kemp, L. G. Yu, S. C. Wagstaff and S. P. Frostick, Neuroscience, 2001, 105, 779–783 CrossRef CAS PubMed.
- A. W. English, W. Meador and D. I. Carrasco, Eur. J. Neurosci., 2005, 21, 2624–2634 CrossRef PubMed.
- Q. Yin, G. J. Kemp, L. G. Yu, S. C. Wagstaff and S. P. Frostick, Muscle Nerve, 2001, 24, 345–351 CrossRef CAS PubMed.
- M. Freidin, S. Asche, T. A. Bargiello, M. V. Bennett and C. K. Abrams, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 3567–3572 CrossRef CAS PubMed.
- I. Lundgaard, A. Luzhynskaya, J. H. Stockley, Z. Wang, K. A. Evans, M. Swire, K. Volbracht, H. O. Gautier, R. J. Franklin, F.-C. Charles, D. Attwell and R. T. Karadottir, PLoS Biol., 2013, 11, e1001743 CrossRef PubMed.
- O. T. Bagdatoglu, G. Polat, C. Bagdatoglu and U. Atik, Turk. Neurosurg., 2008, 18, 149–156 Search PubMed.
- A. Ebens, K. Brose, E. D. Leonardo, M. G. Hanson Jr, F. Bladt, C. Birchmeier, B. A. Barres and M. Tessier-Lavigne, Neuron, 1996, 17, 1157–1172 CrossRef CAS PubMed.
- K. R. Ko, J. Lee, D. Lee, B. Nho and S. Kim, Sci. Rep., 2018, 8, 8316 CrossRef PubMed.
- G. Fan, C. Egles, Y. Sun, L. Minichiello, J. J. Renger, R. Klein, G. Liu and R. Jaenisch, Nat. Neurosci., 2000, 3, 350–357 CrossRef CAS PubMed.
- N. Vallieres, B. Barrette, L. X. Wang, E. Belanger, L. Thiry, M. R. Schneider, M. Filali, D. Cote, F. Bretzner and S. Lacroix, Glia, 2017, 65, 657–669 CrossRef PubMed.
- L. D'Angelo, L. Avallone, A. Cellerino, P. de Girolamo, M. Paolucci, E. Varricchio and C. Lucini, Ann. Anat., 2016, 207, 47–54 CrossRef PubMed.
- W. Guo, Y. Li, C. Sun, H. Q. Duan, S. Liu, Y. Q. Xu and S. Q. Feng, Anim. Cells Syst., 2017, 21, 84–92 CrossRef CAS PubMed.
- W. Huang da, B. T. Sherman and R. A. Lempicki, Nucleic Acids Res., 2009, 37, 1–13 CrossRef PubMed.
- W. Huang da, B. T. Sherman and R. A. Lempicki, Nat. Protoc., 2009, 4, 44–57 CrossRef PubMed.
Footnote |
| † These authors provided equal contribution to this work. |
| This journal is © The Royal Society of Chemistry 2019 |