 Open Access Article
Open Access ArticleGrowing prospects of DNA nanomaterials in novel biomedical applications
Zhiguang Suo†
a,
Jingqi Chen†a,
Xialing Houa,
Ziheng Hua,
Feifei Xingb and
Lingyan Feng *a
*a
aMaterials Genome Institute, Shanghai University, Shanghai 200444, China. E-mail: lingyanfeng@t.shu.edu.cn
bDepartment of Chemistry, College of Science, Shanghai University, Shanghai 200444, China
First published on 28th May 2019
Abstract
As an important genetic material for life, DNA has been investigated widely in recent years, especially in interdisciplinary fields crossing nanomaterials and biomedical applications. It plays an important role because of its extraordinary molecular recognition capability and novel conformational polymorphism. DNA is also a powerful and versatile building block for the fabrication of nanostructures and nanodevices. Such DNA-based nanomaterials have also been successfully applied in various aspects ranging from biosensors to biomedicine and special logic gates, as well as in emerging molecular nanomachines. In this present mini-review, we briefly overview the recent progress in these fields. Furthermore, some challenges are also discussed in the conclusions and perspectives section, which aims to stimulate broader scientific interest in DNA nanotechnology and its biomedical applications.
1 Introduction
Deoxyribonucleic acid (DNA) is a biological molecule that consists of various base pairs for storage and propagation of genetic information. At the same time, DNA has also become a powerful and versatile functional material in nanotechnology and in biomedical applications, which benefit from its exciting physicochemical properties. Apart from the classic right-handed B-form double helix DNA,1 other DNA structures, such as the A-/Z- form and triplex, G-quadruplex and i-motif DNA etc., were developed later and have been widely applied.2 In addition, the more complicated conformational polymorphism DNA structures (e.g. DNA origami) have also been used and assembled in DNA nanotechnology. These functional nanostructures can be considered as templates or scaffolds for biomedical applications with novel functions.3 Furthermore, a great deal of DNA based biocomputing applications have been developed, which cannot be ignored within the rapid growth of DNA nanotechnology.In this mini-review, we mainly focus on the role of DNA in biosensors, disease diagnostics and in therapeutic applications. Furthermore, we summarize the strategies of DNA nanostructures applied in logic gates and biocomputing, as well as in molecular nanomachines. We hope that this review will provide useful information towards broader scientific interests in DNA nano-biomedical/biocomputing applications.
2 DNA biosensors
Biosensors are a class of analytical devices that have biological sensing elements, which convert a biological response into an analytically useful signal via a physicochemical transducer. DNA biosensors have been widely applied in biological fields, environment monitoring and in the food safety industry. DNA plays an important role in the design and application of a biosensor, including as a target,4 a recognition motif,5 a staple strand for linking6,7 and as a signal report probe etc.8,9 DNA-based biosensors have gained significant achievements over the past decades.10 In this section, electrochemical and fluorescent DNA biosensors will be first introduced according to their signal readout, and recent publications of DNA-based biosensors will be mainly summarized, particularly with respect to heavy metal ions, cancer biomedicine, and pathogen detections.A typical electrochemical DNA-based (E-DNA) sensing platform generally involves a redox-labelled single strand DNA (ssDNA) probe immobilized on an electrode surface.11 Then the target-binding event will induce a change in the DNA structure,12 which has been proven to be rapid, sensitive, and reagentless.13 Aptamer, as a new kind of selected DNA/RNA, has been widely employed to construct electrochemical/optical sensors with a highly specific binding ability.14 In addition, some novel nanomaterials, such as CdSe quantum dots, are labelled on the DNA signal strand as probes to realize signal amplification.15 To enhance the performance of electrochemical DNA sensors, Plaxco's group has systematically investigated the influence of different electrode surface modifications for favourable performances.16,17 By adopting two labelled probes (e.g. methylene blue, MB, and anthraquinone, AQ) with distinguishable electroactive potentials, a dual-reporter approach greatly eliminates E-AB baseline drift, even in flowing, undiluted whole blood (Fig. 1A).18 MB, AQ and ferrocene (Fc) are widely used electroactive probes in E-DNA sensors with stable redox signal readouts. Fan et al. developed a series of works using the tetrahedral structure of DNA to construct E-DNA biosensors. In such DNA nanostructures, rigid branched DNA motifs could be obtained through complementary base pairing, and then further assembled into discrete finite objects. In order to investigate the DNA-mediated charge transport (CT) method, they modified the redox probes at different positions on the 3D tetrahedral DNA. CT rate measurements provided unambiguous evidence that the intercalative MB undergoes efficient mediated CT over longer distances along the duplex, whereas the non-intercalative Fc probe tunnels electrons through the space. Furthermore, ultrasensitive microRNA detection has also been realized by combining the tetrahedral DNA nanostructure and hybridization chain reaction amplification (Fig. 1B).19
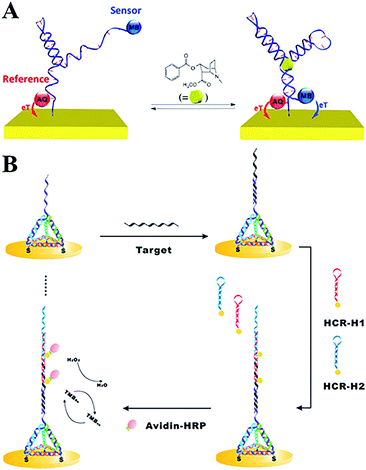 | ||
| Fig. 1 (A) Electrochemical aptamer-based sensors with dual-reporter drift correction. (B) 3D tetrahedral DNA scaffold and hybridization chain reaction for electrochemical miRNA detection. These figures have been adapted from ref. 18 and 19, with permission from the American Chemical Society. | ||
Another typical DNA self-assembled nanostructure, DNA origami, has also been successfully applied in biosensing, smart drug delivery, enzyme cascades and in analysis platforms, etc. Very recently, Hemmig et al. explored the potential applications of DNA origami nanotechnology in novel optical voltage sensing nanodevices. The voltage responsive DNA origami structures were labelled with a single pair of fluorescent dyes and were immobilized on a nanocapillary tip. Upon application of an electric field, controlled structural changes could be obtained. This novel proof-of-concept sensing mechanism opens up a new pathway for determining the mechanical properties of DNA origami, enabling potential application in voltage sensors.20 3D origami structures are stable enough to resist deformation in harsh environments, making them suitable for potential application in in vivo biosensing.
Fluorescence detection demonstrates an immediate response and real time detection with high sensitivity and minimal sample consumption properties. Natural state DNA is usually non-fluorescent, so fluorescent DNA biosensors always employ an external fluorophore.21 With the application of a labelled probe, various fluorescent DNA biosensors have been constructed. Carboxyfluorescein (FAM) and other fluorescent dyes are commonly used to functionalize DNA and are quenched by the complementary strand labelled with a quenching group.22–24
In addition, a very common and effective approach is to introduce novel nanomaterials (gold and silver nanoclusters, carbon dots (CD) and graphene dots) into fluorescent biosensors.25,26 Due to their adsorption of DNA with appropriate affinity and the ability to quench different types of fluorophores, graphene-based nanomaterials can act as unique platforms for fluorescence sensing.27 Luo et al. demonstrated that different DNA conformations have distinguishable affinities for GO, and this can be used for pH detection.28 Biosensors with novel DNA structures have been developed, along with multi-stranded DNA structures. Triplex DNA is easily formed from a duplex and a third strand via Hoogsteen or reverse Hoogsteen hydrogen base pairing. TA.T and CG.C+ are two typical Hoogsteen bonds and are commonly utilized for sensor design. Two labelled DNA probes contain a certain amount of TAT under acidic conditions, and the probes fold into triplex DNA structures, which are not absorbed onto the GO surface and so retain strong fluorescence. In contrast, under alkaline conditions the probes contain a single stranded tail, which can be absorbed onto the GO, and so substantially quenching the fluorescence (Fig. 2).
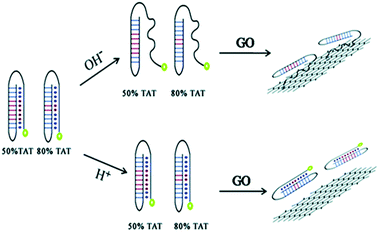 | ||
| Fig. 2 pH detection by applying the different conformations of a DNA duplex and triplex and their varying affinities to the GO surface. This figure has been adapted from ref. 28, with permission from the Royal Society of Chemistry. | ||
A sensitive signal readout strategy has made great contribution to the application of DNA biosensors for various targets, including heavy metal ions, toxins, pathogens, as well as in cancer biomarkers. Metal ion detection is a typical application in DNA based biosensors. Many heavy metal ions are harmful when a certain threshold is exceeded, leading to a great risk to human health and to the environment.29–32 However, traditional detection methods severely limit the on-site and real-time detections.33,34 The interaction between DNA and the metal ions is vital for sensitive sensor design. Based on the conformation transition of the DNA, highly sensitive DNA biosensors have been designed.35,36 Furthermore, nanomaterials such as polymers, quantum dots and noble metal nanoclusters, can be modified onto the electrode to improve DNA biosensor performances. Recently, CdSe@CdS quantum dot core–shell nanoparticles were labelled on hemin/G-quadruplex DNA to detect Pb2+. CDs and GO modified DNA fluorescent biosensors were designed for Hg2+ detection by forming a T–Hg2+–T duplex,37,38 and Ag+ was detected based on a similar mechanism involving C–Ag+–C formation.39 There are also artificial bases reported to form stabilized pairs with Cu2+.40–42 In order to solve the ionic strength dependent problem of DNA-based sensors, Liu43 et al. used 2-aminopurine with high robust fluorescent signalling to label DNA homopolymers for Hg2+ and Ag+ detections. To date, K+,44–47 Mg2+,48–50 Zn2+,51–53 Co2+,54 Mn2+,48,55 and Cd2+ metal-ion dependent DNA biosensors have been obtained through combinatorial selection and rational design of DNA structures, and these biosensors have been applied further in real-time environmental monitoring and for on-site medical diagnostics.
Furthermore, DNA-based biosensors have also been developed for carcinogen and antibiotic detection in food safety. A series of sensitive aptamer-based biosensors were reported to detect aflatoxin B1 (AFB1) using quadruplex DNA on a gold electrode or by cy5-modified DNA probes with electrochemical and fluorescent readouts.56,57 Liu et al.58 detected microcystin-LR via a multiple amplified enzyme-free electrochemical immunosensor, in which SiO2@MSN-functionalized DNAzyme concatemers load hemin and MB as a mimic enzyme. In addition, the quantitative detection of streptomycin residues in blood serum, milk and animal derived foods has been reported based on aptamer-complimentary strand conjugates and fluorescence quenching by gold nanoparticles (AuNPs).59,60 Another important antibiotic in food safety and clinical diagnosis, kanamycin, has also been detected by a fluorescent aptasensor based on the catalytic recycling activity of exonuclease and AuNPs.61
Most importantly, DNA biosensors have made a great contribution to the early diagnosis and efficient therapy of various diseases. By combining different nanomaterials, such as silver nanoclusters, and gold nanorods, sensitive fluorescence sensors were used for human immunodeficiency virus (HIV) and hepatitis B virus (HBV) detections through the hybridization chain reaction (HCR).62,63 Different electrochemical/fluorescent DNA sensors have been designed for cancer biomarker detection, involving typical miRNAs, prostate specific antigen (PSA),64 and carcinoembryonic antigen (CEA).65 For example, ECL biosensors were constructed for cancer relevant DNA/miRNA detection via programmable DNA cyclic amplification.66–68 A dual-mode AuNPs@MoS2 electrochemical sensor was also constructed for miRNA-21 detection.69 In addition, the fluorescent DNA-AgNCs biosensor with a cell-specific internalization aptamer could be internalized into tumor cells specifically, realizing multiple tumor-related miRNA detections and imaging.70 The activity of human intracellular telomerase is an important biomarker in primary human tumors for early clinical diagnosis. With high intracellular stability, biocompatibility and good specificity, a silver pyramid probe-based biosensor detecting telomerase activity in situ in living cells has been reported.71 A porphyrin-functionalized graphene ECL technique has also been constructed for label-free ultrasensitive telomerase activity detection, according to the amount of DNA loading on the electrode surface.72 Our group has also demonstrated an electrochemically triggered click reaction strategy for an aptamer-based biosensor that effectively detected VEGF165, which is another important biomarker in cancer and neuron diseases.73
In addition, the non-invasive or minimally-invasive nuclear imaging detections were usually employed to specifically identify diseased tissues at the molecular level, and they play critical roles in biomedical research.74–76 For example, fluorescence labelled DNA nanostructures with high biocompatibility and flexibility were designed to perform sentinel lymph node imaging77,78 and dual modality targeted imaging detections in cancer cells.79 In order to enhance the imaging detection quality, AuNRs were also assembled on the surface of the DNA origami structure, along with an in vivo photothermal therapy function at the same time.80
3 Biomedical therapies using DNA nanotechnology
Cancer is a leading cause of death worldwide. At present, chemotherapy,81,82 radiotherapy83 and immunotherapy are three important therapeutic strategies, but they are also subject to many challenges. Numerous efforts have been made for more effective therapies. In recent years, the emergence of molecular medicine based on DNA nanotechnology has attracted tremendous interest. In this section, the recent research progresses in chemotherapy and immunotherapy involving DNA nanostructures will mainly be summarized.Chemotherapy as a primary therapy cancer strategy has made great contributions to cancer treatment over the last several decades. However, typical chemotherapy agents exhibit poor drug selectivity and side effects,84 therefore it is necessary to develop new chemotherapeutic drug-loading platforms to solve these problems. DNA nanostructures with unique features85–87 have been used to design heterogeneous anticancer agents (including molecular drugs, RNA88 and proteins89) for targeted cancer treatments.90–93 A wide range of DNA nanostructures have been used to create chemotherapeutic drug-loading platforms, as well as for aptamer-based delivery for the specific uptake of drugs in vivo.94 Doxorubicin (DOX) as a typical chemotherapeutic anticancer drug is widely used in cancer treatment, as it interacts with DNA and restrains macromolecular biosynthesis.95 A DNA icosahedra nanostructure has been used as the nanocarrier of DOX by assembling motifs, demonstrating it as an efficient and specific strategy for epithelial cancer cells.96 Furthermore, the release of DOX can be rationally controlled through adjusting the twisting degree of the DNA tubular carrier. With the development of DNA nanotechnology, DNA origami technology has also been created with abundant and complicated nanostructures as drug-loading platforms for DOX (Fig. 3A), showing promising cancer therapeutic efficiency in vivo.97,98 For maximum loading of DOX, multiple GC-pair sequences were embedded in a self-degradable cocoon-like DNA nanoclew (Fig. 3B).99 An open-caged pyramidal DNA@DOX nanostructure was developed for drug delivery, which exhibited a significant enhancement in cytotoxicity to breast and liver cancer cells compared to the free DOX.100 By combining an aptamer and GC-rich dsDNA, Ma et al.101 developed a multifunctional drug delivery platform with high stability, an efficient payload capacity and reduced side effects in order to solve multi-drug resistance through targeted drug delivery. Meanwhile, a novel self-assembled DNA nanostructure was able to specifically recognize cancer cells and further release the loaded anticancer drug at pH 5.0 (Fig. 3C).102
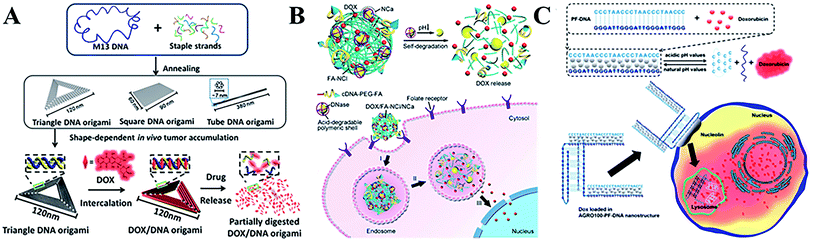 | ||
| Fig. 3 (A) The triangular, square, and tube DNA origami used as DOX vehicles for cancer therapy. This figure has been adapted from ref. 98, with permission from the American Chemical Society. (B) Cocoon-like self-degradable DNA nanoclew for anticancer drug delivery. This figure has been adapted from ref. 99, with permission from the American Chemical Society. (C) A self-assembled DNA nanostructure for targeted and pH-triggered DOX delivery. This figure has been adapted from ref. 102, with permission from the Royal Society of Chemistry. | ||
With the advantages of a well-defined 3D shape, a uniform size, good biocompatibility and precise spatial addressability, 3D polyhedral structures based on DNA self-assembling promise novel application to in vivo molecular cargo delivery processes.103,104 A tetrahedral DNA nanostructure (TDN) is a novel type of 3D DNA nanostructure that has also been rapidly explored in biomedical fields. It was reported that TDN was able to promote cell proliferation. The expression of cyclin-dependent kinase like-1 gene was up-regulated in the presence of TDN by activating the Wnt/β-catenin pathway.105 Walsh106 et al. investigated the access of the DNA tetrahedron cage to cultured mammalian cells without transfection reagent for at least 48 h. Furthermore, it was investigated, utilizing the DNA nanostructure, for effective drug delivery. The endocytic and transport of tetrahedral DNA nanostructures through a caveolin dependent pathway in mammalian cells was demonstrated. Moreover, Krishnan and co-workers107 employed a novel DNA icosahedron-based intelligent nanovehicle with chemically responsive behaviour for FITC dextran encapsulation and delivery (Fig. 4A). In addition, Gianneschi et al.108 presented a reversible nanoscale anticoagulant by combining thrombin and DNA-aptamer polymer amphiphilic spherical NPs (DAPA-NPs), which effectively inhibited human plasma clotting. However, this effect can rapidly be reversed by adding the complementary oligonucleotides, in order to control blood circulation times in vivo. In addition, the nano-size DNA tetrahedron carrying MB provided an effective photodynamic therapy (Fig. 4B).109
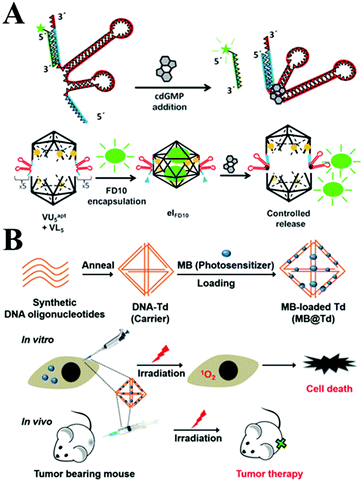 | ||
| Fig. 4 (A) Controlled release of encapsulated cargo from a DNA icosahedron using a chemical trigger. This figure has been adapted from ref. 107, with permission from John Wiley and Sons. (B) Loading of MB onto DNA Td constructed with four DNA strands for in vivo photodynamic therapy. This figure has been adapted from ref. 109, with permission from the Royal Society of Chemistry. | ||
Small interfering RNA is known to interfere with protein synthesis by binding to complementary messenger RNA molecules, which then silences the specific genes being expressed.110–112 DNA nanostructures have been enhanced for use as powerful delivery carriers of the therapeutic nucleic agents for cancer targeted therapy. Anderson et al.103 assembled DNA/siRNA tetrahedral nanoparticles using six ssDNA fragments and six double-stranded siRNAs to deliver siRNA into the target genes. The introduction of nanoparticles improves the stability of tethered RNA molecules, and this greatly enhances their interference efficacy in biomedicine. Recently, it has been reported that DNA nanoparticles (DNPs) with DNA bricks could be easily constructed as delivery vehicles of siRNA, which would target anti-apoptotic protein Bcl2 and significantly inhibit cell growth in vitro and in vivo.113
Moreover, immunotherapy has provided another strategy due to its specificity and minimal side effects compared with chemotherapy. Cytosino-phosphate-guanine (CpG) DNA as a ligand for toll-like receptor 9 (TLR9) has been used for the immunotherapy of cancers, infectious diseases, asthma etc., by triggering the innate immune responses.114 However, ssDNA containing CpG sequences is unstable and can be digested in physiological conditions. Much effort has been made to develop CpG DNA nanostructures with high delivery efficiency and low toxicity for clinical applications.92 Various complicated DNA nanostructures (TDN, DNA origami and DNA polypod structures etc.) have been fabricated and have significantly enhanced the CpG loading efficiency of the immunostimulatory effect in vivo (Fig. 5A).115,116 CpG-tripodna and CpG-hexapodna were also designed to induce a higher tumor necrosis factor alpha (TNF-a) release from RAW 264.7 cells, thus increasing the potency of CpG-SS in vivo.117 Fan's group118 has assembled a functional and multivalent DNA tetrahedral nanostructure with unmethylated CpG motifs, and this has enhanced the immunostimulatory effect when entering macrophage-like RAW264.7 cells even without transfection agents. The group has also used a DNA rolling circle amplification (RCA) strategy to assemble DNA-origami, such as nanoribbons, for carrying CpG ODNs. It was demonstrated that this strategy was readily internalized by mammalian cells, and thus enhanced immunostimulatory activity.119 Taking advantage of the strong and long-lasting specific antibody response in vivo, the antigen-adjuvant DNA complex has also been demonstrated as a potential candidate to recognize various vaccines.120 Recently, a DNA vaccine containing a CpG motif was developed, and the cytotoxicity and survival life time were greatly increased. Liu et al. constructed a bicistronic plasmid to extract the fusion genes of survivin/MUC1 (MS) and IL-2, which showed good effects on colorectal cancer and successfully simplified the whole industrial production process. Furthermore, the anti-tumour effects of this fusion gene could be enhanced by combining it with oxaliplatin.121 Meanwhile, the dendrimer-like DNA and nano-centipedes structures were considered as efficient vehicles to deliver immunostimulatory CpG sequences, which could be internalized by cancer cells and trigger the corresponding stronger immune response (Fig. 5B).122,123
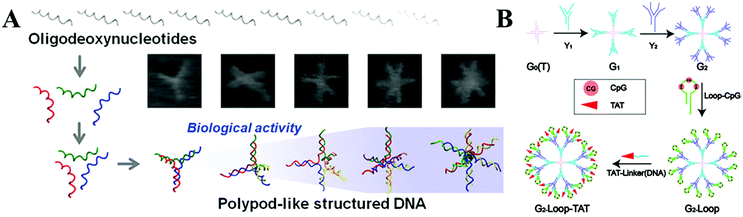 | ||
| Fig. 5 (A) Nanosized DNA assemblies in polypod-like structures as efficient vehicles for immunostimulatory CpG motifs. (B) A self-assembled DNA dendrimer nanoparticle for efficient delivery of the immunostimulatory CpG motifs. These figures have been adapted from ref. 116 and 122, with permission from the American Chemical Society. | ||
4 DNA molecular logic gates and computing
With the development of technology, the conventional electronic computer based on Boolean logic encounters great technical bottlenecks for complex problems and manufacturing.124 However, with the development of DNA nanotechnology, DNA as a basic and promising alternative material for the construction of molecular logic gates has attracted remarkable attention. DNA can perform various logical operations by transforming inputs into defined outputs in logic circuit systems due to its outstanding data-storage capacity, highly specific hybridizations and flexibility in design.125,126 Numerous DNA computer applications will be briefly introduced.A great deal of effort has been devoted to designing and developing typical DNA logic gates, including YES, NOT, AND, NAND, OR, NOR, XOR, XNOR, IMPLY and N-IMPLY etc. The inputs of DNA/RNA, metal ions or light will activate the logic gates and then produce output signals based on Watson–Crick base-pairing, DNA strand displacement, DNAzyme cleavage,127 nucleic acid ligand binding,128 i-motif transformation, DNA hairpins129 and PCR.130 Ghadiri et al.131 successfully created three photonic logic gates, AND, NAND and INHIBIT, using the unique base-pairing properties of DNA and FRET between fluorescent molecules. Subsequently, ssDNA was exploited to invade and displace local sections of complementary dsDNA. A series of entirely AND, OR and XOR logic gates and a half-adder were assembled by AND and XOR gates.132
Recently, numerous novel DNA logic gates have been reported. By taking advantage of biocatalysis, colorimetric logic gates and multi-level circuits have been developed based on a typical triplex-helix molecular switch machine and DNAzyme (Fig. 6A), and this paved a simple path for designing complex DNA-based logic devices.133 Based on the previous work involving DNA interactions with carbon nanotubes, our group has reported novel photoluminescence spermine-functionalized C-dots for the induction of the DNA B–Z structure transition. Several DNA logic gates under physiological low salt conditions were constructed using a FRET design.134 In addition, a multi-level logic gate was built from sequential INHIBIT and AND gates for the analysis of a net XOR gate with an electrochemical signal as the output. In addition, Fan's team135 constructed a series of on-chip DNA logic gates via DNA strand displacement reactions. By regulating the ssDNA inputs into mammalian cells, the behavior of the cells can be accurately controlled. This has provided an alternative route to programming cell movements and functions in biomedical applications. With the help of hemin–G-quadruplex complexes, a set of optical DNA logic gates and a Boolean logic circuitry were constructed based on strand displacement and magnetic separation. The coloured ABTS product can be formed in response to three microRNA input signals from cancer cells (Fig. 6B).136 Moreover, a sequential logic DNA gate was established utilizing a two-ring DNA structure (Fig. 6C), and this recognizes two DNA signals with different triggering sequences via the “loop-open” mechanism.137,138
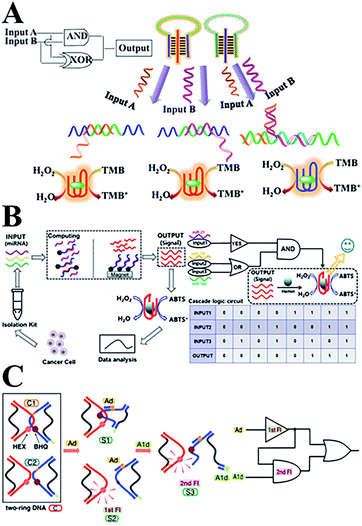 | ||
| Fig. 6 (A) DNA colorimetric logic gates based on a triplex-helix molecular switch. This figure has been adapted from ref. 133, with permission from the American Chemical Society. (B) An optical DNA logic gate based on strand displacement and magnetic separation, which responds to multiple microRNAs. This figure has been adapted from ref. 136, with permission from Springer Nature. (C) A DNA sequential logic gate using two-ring DNA. This figure has been adapted from ref. 138, with permission from the American Chemical Society. | ||
With more sophisticated precision in their uses, DNA molecular logic gates can hold great potential for diagnosis and therapy at the molecular level. In comparison to conventional single input and output states (0 or 1), DNA-based molecular logic gates usually depend on the change in fluorescent or colorimetric signals to realize multiple detections of small molecules or macromolecules in bioanalysis and biological diagnosis. A series of AND, OR and SET-RESET logic gates have been constructed by employing ion-driven conformational changes of a DNA G-quadruplex.139 In addition, an INHIBIT logic gate based on K+ or Pb2+-switched DNA structures, a chemical XOR gate and a three-input molecular logic gate was also designed.140,141 With relentless electronic signals and reusable sensing elements, this device can be applied for continuous monitoring, particularly in heavy metal ion, pathogen, and genetic biomedicine detections.142 Furthermore, the ECL signals have been used as new outputs using energy or an electron transfer-quenching path.143 Recently, Macdonald144 arranged user-friendly deoxyribozyme-based molecular logic gate networks into visual displays, and then further developed a prototype molecular automation. This was able to comprehensively differentiate seven different genotypes of the Lyssavirus genus, and an AND-NOT gate was designed to prevent non-specific activation. The DNAzyme-DNA complexes have been applied in the design of powerful colorimetric biosensors with logic gate operations to identify DNA,145 and the target can be viewed directly by colour changes during the OR logic gate operation (Fig. 7A).
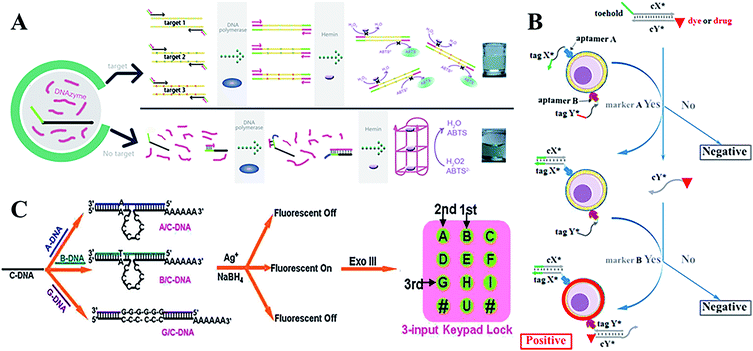 | ||
| Fig. 7 (A) A colorimetric biosensor based on DNAzyme logic gate operations for DNA screening. This figure has been adapted from ref. 145, with permission from Elsevier. (B) A programmable and multi-parameter DNA-based logic platform for cancer recognition and targeted therapy. This figure has been adapted from ref. 153, with permission from the American Chemical Society. (C) DNA-templated Ag nanoclusters as signal transducers for a resettable keypad lock. This figure has been adapted from ref. 159, with permission from the Royal Society of Chemistry. | ||
Numerous research on the properties and advantages of 3D DNA tetrahedral nanostructures has been reported.118,146–148 Using a series of reconfigurable DNA tetrahedrons that respond to a target, various scaffold logic gates were constructed.149 DNA origami nanostructures also provided programmable platforms for logic gates, which could be operated to capture or release small molecules, proteins, nanoparticles or external light signals.150 Kjems and Birkedal demonstrated that the logic gate complexes can control the lid of a 3D DNA origami box with high reliability and high fidelity.151 The unique DNA origami structures provide masses of potential applications in nanomedicine for diagnostics and therapeutics. For multiple cancer cell-surface markers, a “Nano-Claw” has been reported to perform autonomous logic analysis.152 Meanwhile, by combining the special structure-transition of DNA aptamers with toehold-mediated strand displacement reactions, a diagnostic signal can be produced and targeted photodynamic therapy can be realized. High-order multiple cell-surface markers have been successfully identified via integrating AND, OR and NOT logic gates (Fig. 7B).153
Furthermore, researchers have paid much attention to the molecular-scale keypad lock, which is a novel data security device. Its output signals rely on the proper combination of inputs or the inputting sequence. Until now, the fluorescein-linker-pyrene assembly system and photonic keypad locks using ions as inputs had only been reported.154–156 Qu's group first constructed a nucleic acid-based molecular keypad lock with sequence specific recognition of DNA and solid-phase substrates.157 Recently, the novel nanomaterial AgNCs as signal transducers that convert inputs into fluorescence output, can be constructed for various DNA-based logic gates and a sequential logic gate.158 These AgNC-based logic systems have been used to fabricate a biomolecular resettable keypad for crossword puzzles, which provides a universal design and biocompatible operation. These versatile logic devices with new signal transducer materials are simple in practical applications (Fig. 7C).159 Wang et al. further developed a resettable DNA-based keypad lock160 and a visible multi-digit DNA keypad lock based on cleavable G-quadruplex DNAzyme and silver microspheres.161 Zhou et al. first constructed concatenated logic circuits as a visible keypad-lock security system based on toehold-mediated strand displacement and a three-way DNA junction. This system could reset and cycle for a greater number of times automatically, without an external stimulus, compared to previous reports.162
Since 1994, Adleman has used DNA oligonucleotides to perform logical operations for solving complex mathematical problems.163 Qian et al. experimentally demonstrated several scaled up digital logic circuits with a four-bit square-root circuit consisting of 130 DNA strands for threshold and catalysis, achieving digital signal restoration within logical operation.164 Furthermore, researchers have demonstrated the autonomous smart behaviors of molecular systems using a simple DNA gate architecture for multilayer digital circuits based on the vastness of DNA computing and strand displacement cascades.165
5 DNA molecular machines
In the design of artificial molecular machines, DNA represents a smart building material with specific base-pairing, predictable assembly, abundant polymorphisms and unique physical/chemical properties.166 By the rational design of a DNA sequence and stimulus conditions, the transformation of DNA structures can be manipulated and can endow DNA with controllable motion or special functions. It is important to study DNA molecular machines during the conversion of energy and the interaction of biomolecules at the nanometer level, as this is important for the development of biosensors, transporters and controlled drug delivery systems.167 Over the past few years, the construction of artificial DNA machines has attracted much attention.168,169 A great deal of machine-like functioning DNA nanostructures have been reported,146 such as DNA tweezers,170,171 gears,129 walkers,172–175 cranes,176,177 catenanes,178 rotaxanes,179 metronome operations180 and so on.181–187 For instance, a contractile DNA machine has been developed based on the crosslinking of circular DNA into nanowires, and this can be used in target medicine delivery or non-viral gene delivery and will transduce DNA hybridization energy into controlled movement energy on the biomolecular level.188 Molecular walking machines have transported cargoes on the nanometer scale in the presence of DNA fuels.189 Yuan and Xiang have designed a restriction enzyme-powered, autonomous DNA walking machine to detect sequence-specific target DNA with high sensitivity and selectivity.190 Tao et al.191 engineered a phage T4 DNA packaging machine by incorporating vaccine/reporter genes, functional enzymes and target ligands to deliver genes and proteins into mammalian cells with a near 100% efficiency. This has promoted the development of efficacious vaccines against complex HIV-1, malaria and TB infectious agents.With high sensitivity and selectivity, DNA machines also have astonishing applications in biosensors and screening.192 The duplicator-like DNA molecular machines offer a number of DNA templates to fabricate AgNCs for Hg2+ ion detection,193 and this autonomous bio-barcode DNA machine has carried out exponential DNA circular amplification for Hg2+, DNA or small biomolecule detection.194 Another RCA-based DNA machine has been fabricated by coupling miRNA and a catalytic hairpin for the formation of DNAzyme.195 By integrating strand displacement and nicking enzyme signal amplification into a one-step system, polymerase-nicking enzyme synergetic quadratic DNA machines have been designed to sense Pb2+, miRNA and DNA methyltransferase.196,197 In addition, powered by the hydrolysis of NAD+, a repair ligation-mediated light-producing DNA machine has been universally developed for DNA, thrombin, adenosine, K+ and endonuclease detection in human serum.198 Yuan199 has constructed an Fc-switched ECL “off–on” sensor for the sensitive detection of cardiac troponin with a DNA walking machine. Willner's group has spent much effort on constructing a set of DNA machines based on the activation of replication and scission reactions or the aggregation of different nanomaterials.200–203 In addition, Hg2+/cysteine as well as H+/OH− triggers can activate bipedal DNA devices by walking through and stepping on the template track.204,205 Depending on the addition of different fuels and anti-fuels, reconfigurable stimuli-responsive DNA machines can be triggered.206,207
Very recently, a variety of novel and dynamic DNA–hydrogel machines have been constructed. Linked bridge hybridizing was induced to develop split G-quadruplex DNA machines for uracil-DNA glycosylase detection.208 The energetic and mechanical performances of compact (fold) and extended (unfold) DNA motifs have been explored, by tethering small DNA hairpins onto the central seam of a DNA origami frame. Yang and Zhan209 have constructed a central axis and an external sliding tube on a DNA origami mechanical device with controllable regulation of the microcosmic structural rigidity of liposome. The used fuel and anti-fuel strands are important for cell proliferation. A novel proton-driven DNA nanopump has also been developed by incorporating a pH-sensitive i-motif structure into a surface-confined 3D DNA tetrahedron (Fig. 8A).210 A plasmonic nanorod decorated with DNA strands has been designed to carry out directional, progressive and reverse walking on a DNA origami, as an efficient walker robot.211 Friedrich's group has developed a DNA-based molecular platform with a robotic arm (with a length range from 25 to 400 nanometers) that will be actuated under external electric fields. The robotic arm can rotate in arbitrary positions on the platform within milliseconds, in a precise and computer-controlled way (Fig. 8B). Combined with lithographic patterning and self-assembly techniques, multiple DNA robot arms or extended lattices have been created. The robot arm arrays with defined platform orientation have been applied for electrically driven delivery of molecules or nanoparticles within tens of nanometers.212
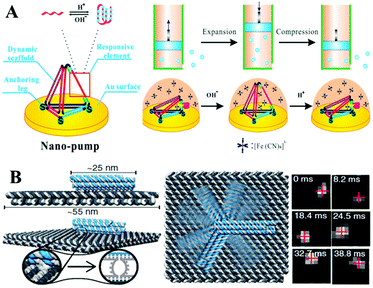 | ||
| Fig. 8 (A) A surface-confined proton-driven DNA pump using a rigid DNA tetrahedron nanostructure as a dynamic scaffold. This figure has been adapted from ref. 210, with permission from John Wiley and Sons. (B) A molecular platform with an integrated rotatable positioning arm controlled by electric fields. This figure has been adapted from ref. 212, with permission from Science Publishing Group. | ||
On the basic research of multi-component DNA origami nanostructures directing folding pathways,213 Su et al. designed a complex 3D DNA origami mechanism to analyse and manufacture DNA machines and robots by assembling basic building blocks together, such as six links or five kinematic joins.214 Stimuli-responsive hydrogels as functional matrices can be integrated with DNA hairpin structures, exhibiting switchable mechanical, structural and chemical properties. The DNA–hydrogel complex could be used for drug delivery, in valves or actuators, and in biosensors and information process.215 Cangialosi et al. demonstrated that a 100-fold volumetric expansion was realized by DNA in a hydrogel system upon external stimuli. Then the controllable bend, twist, or actuated shape changes were realized with the successive extension of cross-links with different DNA hairpins. Moreover, the author has further fabricated dynamic DNA “flower” and “crab” hydrogel devices with the petals, antennae, claws, and legs responding to different sequences (Fig. 9). This strategy provided the possibility of building soft DNA robots, programmable devices and smart medicine via diverse biochemical inputs.216
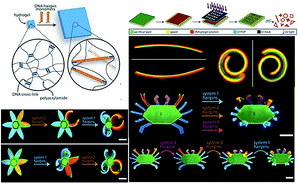 | ||
| Fig. 9 DNA sequence-directed shape changes of photopatterned hydrogels of six-petal flower and crab structures. This figure has been adapted from ref. 216, with permission from Science Publishing Group. | ||
6 Conclusions and perspectives
Over the past decades, much effort has been made towards DNA biomedical applications with the rapid development of DNA nanotechnology. In this mini-review, we have mainly summarized the recent progresses in the design and applications of DNA nanomaterials, including their use in novel biosensors, bioimaging and therapies, DNA logic gates and molecular nanomachines, covering a wide range of aspects of potential future uses of DNA. Notably, despite substantial applications quickly blooming, several issues remain that need to be addressed. One major challenge is that, until now, the developed applications are still highly limited at the bench research stage. Some restrictions are urgently needed to be overcome in order to make a breakthrough, including the difficult construction, unstable and high-cost chemical synthesis, poor reproducibility and being susceptible to interference, especially for the promising clinical applications. In the near future, DNA-based biosensors and biocomputing applications have the potential to be improved by overcoming false interference at the molecular level with a more rigid and stable nanostructure design, or by introducing variable DNA analogues to inhibit degradation in a complex medium. We positively believe that DNA nanomaterials will provide various novel and powerful methodologies in biomedical applications, as well as for exploring new paths to address more complex mathematical logic problems and mechanical behaviours.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (NSFC 21705106), the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning (No. TP2016023), the Shanghai Sailing Program (17YF1406400) and the Shanghai Natural Science Foundation (No. 18ZR1415400). We also thank Wenxia Liu for the useful discussion.Notes and references
- J. D. Watson and F. H. Crick, Nature, 1953, 171, 964–967 CrossRef CAS PubMed.
- K. Gehring, J. L. Leroy and M. Gueron, Nature, 1993, 363, 561–565 CrossRef CAS PubMed.
- Z. G. Suo, J. Q. Chen, Z. H. Hu, Y. H. Liu, F. F. Xing and L. Y. Feng, Nanofabrication, 2018, 4, 32–52 Search PubMed.
- W. Xu, X. J. Xue, T. H. Li, H. Q. Zeng and X. G. Liu, Angew. Chem., 2010, 48, 6742 CrossRef.
- L. Y. Feng, C. Q. Zhao, Y. Xiao, L. Wu, J. S. Ren and X. G. Qu, Chem. Commun., 2012, 48, 6900–6902 RSC.
- Y. G. Ke, S. Lindsy, Y. Chang, Y. Liu and H. Yan, Science, 2008, 319, 180–183 CrossRef CAS PubMed.
- Q. Liu, Z. L. Ge, X. H. Mao, G. B. Zhao, X. L. Zuo, J. W. Shen, J. Y. Shi, J. Li, L. H. Wang, X. Q. Chen and C. H. Fan, Angew. Chem., 2018, 57, 7131–7135 CrossRef CAS PubMed.
- L. P. Qiu, H. Zhou, Z. S. Wu, G. L. Shen and R. Q. Yu, New J. Chem., 2014, 38, 4711–4715 RSC.
- T. Kang, S. M. Yoo, I. Yoon, S. Y. Lee and B. Kim, Nano Lett., 2010, 10, 1189–1193 CrossRef CAS PubMed.
- P. Mehrotra, J. Oral. Biol. Craniofac. Res., 2016, 6, 153–159 CrossRef PubMed.
- C. H. Fan, K. W. Plaxco and A. J. Heeger, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 9134–9137 CrossRef CAS PubMed.
- F. Li, L. M. Yang, M. Q. Chen, Y. Qian and B. Tang, Biosens. Bioelectron., 2013, 41, 903–906 CrossRef CAS PubMed.
- D. Li, S. P. Song and C. H. Fan, Acc. Chem. Res., 2010, 43, 631–641 CrossRef CAS PubMed.
- K. J. Huang, H. L. Shuai and Y. X. Chen, Sens. Actuators, B, 2016, 225, 391–397 CrossRef CAS.
- Y. P. Wei, X. P. Wei, C. J. Mao, H. L. Niu, J. M. Song and B. K. Jin, Biosens. Bioelectron., 2017, 103, 99–103 CrossRef PubMed.
- N. Arroyo-Curras, K. Scida, K. L. Ploense, T. E. Kippin and K. W. Plaxco, Anal. Chem., 2017, 89, 12185–12186 CrossRef CAS PubMed.
- H. Li, P. Dauphin-Ducharme, N. Arroyo-Curras, C. H. Tran, P. A. Vieira, S. G. Li, C. Shin, J. Somerson, T. E. Kippin and K. W. Plaxco, Angew. Chem., 2017, 56, 7492–7495 CrossRef CAS PubMed.
- H. Li, N. Arroyo-Curras, D. Kang, F. Ricci and K. W. Plaxco, J. Am. Chem. Soc., 2016, 138, 15809–15812 CrossRef CAS PubMed.
- Z. L. Ge, M. H. Lin, P. Wng, H. Pei, J. Yan, J. Y. Sho, Q. Huang, D. N. He, C. H. Fan and X. L. Zuo, Anal. Chem., 2014, 86, 2124–2130 CrossRef CAS PubMed.
- E. A. Hemmig, C. Fitzgerald, C. Maffeo, L. Hecker, S. E. Ochmann, A. Aksimentiev, P. Tinnefeld and U. F. Keyser, Nano Lett., 2018, 8, 1962–1971 CrossRef PubMed.
- J. W. Liu, Z. H. Cao and Y. Lu, Chem. Rev., 2009, 109, 1948–1998 CrossRef CAS PubMed.
- C. Lu, F. Chen, M. Li, X. D. Guo, S. L. Li, N. Zheng and J. Q. Wang, Food Chem., 2017, 215, 377–382 CrossRef PubMed.
- T. X. Chen, F. Ning, H. S. Liu, K. F. Wu, W. Li and C. B. Ma, Chin. Chem. Lett., 2017, 28, 1380–1384 CrossRef CAS.
- C. Y. Sun, R. F. Su, J. X. Bie, H. J. Sun, S. N. Qiao, X. Y. Ma, R. Sun and T. H. Zhang, Dyes Pigm., 2017, 149, 867–875 CrossRef.
- P. Zheng and N. Q. Wu, Chem. - Asian J., 2017, 12, 2343–2353 CrossRef CAS PubMed.
- Z. B. Liu, B. W. Liu, J. S. Ding and J. W. Liu, Anal. Bioanal. Chem., 2014, 406, 6885–6902 CrossRef CAS PubMed.
- H. Zhang, H. L. Zhang, A. Aldalbahi, X. L. Zuo, C. H. Fan and X. Q. Mi, Biosens. Bioelectron., 2017, 89, 96–106 CrossRef CAS PubMed.
- F. Y. Luo, Q. Xi, J. H. Jiang and R. Q. Yu, Anal. Methods, 2016, 8, 6982–6985 RSC.
- G. L. Turdean, Int. J. Electrochem., 2011, 139, 343125 Search PubMed.
- C. Gao, X. Y. Yu, S. Q. Xiong, J. H. Liu and X. J. Huang, Anal. Chem., 2013, 85, 2673–2680 CrossRef CAS PubMed.
- K. Tag, K. Riedel, H. J. Bauer, G. Hanke, K. H. R. Baronian and G. Kunze, Sens. Actuators, B, 2007, 122, 403–409 CrossRef CAS.
- A. Singh, R. K. Sharma, M. Agrawal and F. M. Marshall, Food Chem. Toxicol., 2010, 48, 611–619 CrossRef CAS PubMed.
- Y. Xiang and Y. Lu, Inorg. Chem., 2014, 53, 1925 CrossRef CAS PubMed.
- W. H. Zhou, R. Saran and J. W. Liu, Chem. Rev., 2017, 117, 8272–8325 CrossRef CAS PubMed.
- T. Li, S. J. Dong and E. K. Wang, J. Am. Chem. Soc., 2010, 132, 13156–13157 CrossRef CAS PubMed.
- F. Sun, Z. Y. Wang, Y. Q. Feng, Y. X. Cheng, H. X. Ju and Y. W. Quan, Biosens. Bioelectron., 2017, 100, 28–34 CrossRef PubMed.
- X. Cui, L. Zhu, J. Wu, Y. Hou, P. Y. Wang, Z. N. Wang and M. Yang, Biosens. Bioelectron., 2015, 63, 506–512 CrossRef CAS PubMed.
- A. Ono and H. Togashi, Angew. Chem., 2010, 43, 4300–4302 CrossRef PubMed.
- A. Ono, S. Cao, H. Togashi, M. Tashiro, T. Fujimoto, T. Machinami, S. Oda, Y. Miyake, I. Okamato and Y. Tanaka, Chem. Commun., 2008, 39, 4825–4827 RSC.
- P. Scharf and J. Muller, Chempluschem, 2013, 78, 20–34 CrossRef CAS.
- N. Megger, J. Johannsen, J. Muller and R. K. O. Sigel, Chem. Biodiversity, 2012, 9, 2050–2063 CrossRef CAS PubMed.
- C. Kaul, M. Müller, M. Wagner, S. Schneider and T. Carell, Nat. Chem., 2011, 3, 794–800 CrossRef CAS PubMed.
- W. H. Zhou, J. S. Ding and J. W. Liu, Biosens. Bioelectron., 2017, 87, 171–177 CrossRef CAS PubMed.
- I. Smirnov and R. H. Shafer, J. Mol. Biol., 2000, 296, 1–5 CrossRef CAS PubMed.
- F. He, Y. L. Tang, S. Wang, Y. L. Li and D. B. Zhu, J. Am. Chem. Soc., 2005, 127, 12343–12346 CrossRef CAS PubMed.
- B. Kim, I. H. Jung, M. Kang, H. K. Shim and H. Y. Woo, J. Am. Chem. Soc., 2012, 134, 3133–3138 CrossRef CAS PubMed.
- H. J. Sun, X. H. Li, Y. C. Li, L. Z. Fan and H. B. Kraatz, Analyst, 2013, 138, 856–862 RSC.
- E. Freisinger and R. K. O. Sigel, Coord. Chem. Rev., 2007, 251, 1834–1851 CrossRef CAS.
- X. Y. Gao, H. M. Huang, S. Y. Niu, H. Z. Ye, Z. Y. Lin, B. Qiu and G. N. Chen, Anal. Methods, 2012, 4, 947–952 RSC.
- D. Mazumdar, N. Nagraj, H. K. Kim, X. L. Meng, A. K. Brown, Q. Sun, W. Li and Y. Lu, J. Am. Chem. Soc., 2009, 131, 5506–5515 CrossRef CAS PubMed.
- B. Cuenoud and J. W. Szostak, Nature, 1995, 375, 611–614 CrossRef CAS PubMed.
- S. W. Santoro, G. F. Joyce, K. Sakthivel, S. Gramatikova and C. F. Barbas, J. Am. Chem. Soc., 2000, 122, 2433–2439 CrossRef CAS PubMed.
- J. Li, W. C. Zheng, A. H. Kwon and Y. Lu, Nucleic Acids Res., 2000, 28, 481–488 CrossRef CAS PubMed.
- K. E. Nelson, H. E. Ihms, D. Mazumdar, P. J. Bruesehoff and Y. Lu, ChemBioChem, 2012, 13, 381–391 CrossRef CAS PubMed.
- M. Chandra, A. Sachdeva and S. K. Silverman, Nat. Chem. Biol., 2009, 5, 718–720 CrossRef CAS PubMed.
- G. Castillo, K. Spinella, A. Poturnayová, M. Šnejdárková, L. Mosiello and T. Hianik, Food Control, 2015, 52, 9–18 CrossRef CAS.
- W. B. Shim, M. J. Kin, H. Mun and M. G. Kim, Biosens. Bioelectron., 2014, 62, 288–294 CrossRef CAS PubMed.
- C. F. Gan, B. F. Wang, J. Y. Huang, A. Qileng, Z. Y. He, H. T. Lei, W. P. Liu and Y. J. Liu, Biosens. Bioelectron., 2017, 98, 126–133 CrossRef CAS PubMed.
- N. M. Danesh, M. Ramezani, A. S. Emrani, K. Abnous and S. M. Taghdisi, Biosens. Bioelectron., 2015, 75, 123–128 CrossRef PubMed.
- A. S. Emrani, N. M. Danesh, P. Lavaee, M. Ramezani, K. Abnous and S. M. Taghdisi, Food Chem., 2016, 190, 115–121 CrossRef CAS PubMed.
- M. Ramezani, N. M. Danesh, P. Lavaee, K. Abnous and S. M. Taghdisi, Sens. Actuators, B, 2016, 222, 1–7 CrossRef CAS.
- S. Q. Zhang, K. Wang, K. B. Li, W. Shi, W. P. Jia, X. Y. Chen, T. Sun and D. M. Han, Biosens. Bioelectron., 2017, 91, 374–379 CrossRef CAS PubMed.
- X. C. Lu, X. Dong, K. Y. Zhang, X. W. Han, X. Fang and Y. Z. Zhang, Analyst, 2013, 138, 642–650 RSC.
- M. Souada, B. Piro, S. Reisberg, G. Anquetin, V. Noel and M. C. Pham, Biosens. Bioelectron., 2015, 68, 49–54 CrossRef CAS PubMed.
- N. Shahbazi, S. Hosseinkhani and B. Ranjbar, Biosens. Bioelectron., 2017, 253, 794–803 CAS.
- Q. M. Feng, Y. H. Guo, J. J. Xu and H. Y. Chen, ACS Appl. Mater. Interfaces, 2017, 9, 17638–17645 Search PubMed.
- X. N. Yao, Z. H. Guo and X. W. Zheng, Anal. Methods, 2016, 9, 312–321 RSC.
- P. Zhang, Z. Y. Li, H. J. Wang, Y. Zhuo, R. Yuan and Y. Q. Chai, Nanoscale, 2017, 9, 2310–2316 RSC.
- S. Su, W. Cao, W. F. Liu, Z. W. Lu, D. Zhu, J. Chao, L. X. Weng, L. H. Wang, C. H. Fan and L. H. Wang, Biosens. Bioelectron., 2017, 94, 552–559 CrossRef CAS PubMed.
- J. J. Li, J. You, Y. P. Zhuang, C. P. Han, J. F. Hu, A. Wang, K. Xu and J. J. Zhu, Chem. Commun., 2014, 54, 7107–7110 RSC.
- L. G. Xu, S. Zhao, W. Ma, X. L. Wu, S. Li, H. Kuang, L. B. Wang and C. L. Xu, Adv. Funct. Mater., 2016, 26, 1602–1608 CrossRef CAS.
- L. Wu, J. S. Wang, L. Y. Feng, J. S. Ren, W. L. Wei and X. G. Qu, Adv. Mater., 2012, 24, 2447–2452 CrossRef CAS PubMed.
- L. Y. Feng, Z. Z. Lyu, A. Offenhäusser and D. Mayer, Eng. Life Sci., 2016, 16, 550–559 CrossRef.
- M. Rudin and R. Weissleder, Nat. Rev. Drug Discovery, 2003, 2, 123–131 CrossRef CAS PubMed.
- J. K. Willmann, N. V. Bruggen, L. M. Dinkelborg and S. S. Gambhir, Nat. Rev. Drug Discovery, 2003, 3, 123–131 Search PubMed.
- T. Hussain and Q. T. Nguyen, Nat. Rev. Drug Discovery, 2008, 7, 591–607 CrossRef PubMed.
- K. R. Kim, Y. D. Lee, T. Lee, B. S. Kim, S. Kim and D. R. Ahn, Biomaterials, 2013, 34, 5226–5235 CrossRef CAS PubMed.
- M. Torabi, S. L. Aquino and M. G. Harisinghani, J. Nucl. Med., 2004, 45, 1509–1518 Search PubMed.
- D. W. Jiang, Y. H. Sun, J. Li, Q. Li, M. Lv, B. Zhu, T. Tian, D. F. Cheng, J. Y. Xia, L. Zhang, L. H. Wang, Q. Huang, J. Y. Shi and C. H. Fan, ACS Appl. Mater. Interfaces, 2016, 8, 4378–4384 CrossRef CAS PubMed.
- Y. Du, Q. Jiang, N. Beziere, L. L. Song, Q. Zhang, D. Peng, C. W. Chi, X. Yang, H. B. Guo, G. Diot, V. Ntziachristos, B. Q. Ding and J. Tian, Adv. Mater., 2016, 28, 10000–10007 CrossRef CAS PubMed.
- U. Emmenegger and R. S. Kerbel, Nature, 2010, 468, 637–638 CrossRef CAS PubMed.
- D. Weycker, H. S. Wu, M. Hagiwara, X. Y. Li and R. L. Barron, Blood, 2014, 124, 2167–2175 Search PubMed.
- D. C. Lombe and B. Jeremic, Clin. Lung Cancer, 2015, 16, 406–412 CrossRef PubMed.
- H. G. Keizer, H. M. Pinedo, G. J. Schuurhuis and H. Joenje, Pharmacol. Ther., 1990, 47, 219–231 CrossRef CAS PubMed.
- P. W. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- X. B. Shen, Q. Jiang, J. Y. Wang, L. R. Dai, G. Z. Zuo, Z. G. Wang, W. Q. Chen, W. Jiang and B. Q. Ding, Chem. Commun., 2012, 48, 11301–11303 RSC.
- G. Z. Zhu, J. Zheng, E. Q. Song, M. Donoxan, K. J. Zhang, C. Liu and W. H. Tan, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 7998–8003 CrossRef CAS PubMed.
- D. R. Han, S. Pal, J. Nangreave, Z. T. Deng, Y. Liu and H. Yan, Science, 2011, 332, 342–346 CrossRef CAS PubMed.
- R. Nunez-Lozano, M. Cano, B. Pimentel and G. da la Cueva-Mendz, Curr. Opin. Biotechnol., 2015, 35, 135–140 CrossRef CAS PubMed.
- S. Pal, A. T. Deng, H. N. Wang, S. L. Zou, Y. Liu and H. Yan, J. Am. Chem. Soc., 2011, 133, 17606–17609 CrossRef CAS PubMed.
- B. Sacca and C. M. Niemeyer, Chem. Soc. Rev., 2011, 40, 5910–5921 RSC.
- R. Jia, T. Wang, Q. Jiang, Z. G. Wang, C. Song and B. Q. Ding, Chin. J. Chem., 2016, 34, 265–272 CrossRef CAS.
- Y. L. Wen, L. Y. Li, L. L. Li, L. Xu, W. Liang, S. Z. Ren and G. Liu, Chem. Inform., 2016, 34, 283–290 CAS.
- S. K. Soininen, J. K. Repo, V. Karttunen, S. Auriola, K. H. Vahakangas and M. Ruponen, Toxicol. Lett., 2015, 239, 108–114 CrossRef CAS PubMed.
- M. Chang, C. S. Yang and D. M. Huang, ACS Nano, 2011, 5, 6156–6163 CrossRef CAS PubMed.
- L. H. Tan, K. G. Neoh, E. T. Kang, W. S. Choe and X. D. Su, Macromol. Biosci., 2011, 11, 1331–1335 CrossRef CAS PubMed.
- Q. Jiang, C. Song, J. Nangreave, X. W. Liu, L. Lin, D. L. Qiu, Z. G. Wang, G. Z. Zou, X. J. Liang, H. Yan and B. Q. Ding, J. Am. Chem. Soc., 2012, 134, 13396–13403 CrossRef CAS PubMed.
- Q. Zhang, Q. Jiang, N. Li, L. R. Dai, Q. Liu, L. L. Song, J. Y. Wang, Y. Q. Li, J. Tian, B. Q. Bao and Y. Du, ACS Nano, 2014, 8, 6633–6643 CrossRef CAS PubMed.
- W. J. Sun, T. Y. Jiang, Y. Lu, M. Reiff, R. Mo and Z. Gu, J. Am. Chem. Soc., 2014, 136, 14722–14725 CrossRef CAS PubMed.
- V. Kumar, S. Bayda, M. Hadla, I. Caligiuri, C. Russo Spena, S. Palazzolo, S. Kempter, G. Corona, G. Toffoli and F. Rizzolio, J. Cell. Physiol., 2016, 231, 106–110 CrossRef CAS PubMed.
- J. Liu, T. Wei, J. Zhao, Y. Y. Huang, H. Deng, A. Kumar, C. X. Wang, Z. C. Liang, X. W. Ma and X. J. Liang, Biomaterials, 2016, 91, 44–56 CrossRef CAS PubMed.
- J. Liu, X. W. Ma, C. N. Lei, X. D. Xue, T. Wei, J. Zhao, S. Y. Li and X. J. Liang, J. Mater. Chem., 2016, 4, 3854–3858 RSC.
- H. Lee, A. K. R. Lytton, Y. Chen, K. T. Love, A. I. Park, E. D. Karagiannis, A. Sehgal, Q. Querbes, C. S. Zurenko, M. Jayaraman, C. G. Peng, K. Charisse, A. Borodovsky, M. Manoharan, J. S. Donhoe, J. Truelove, M. Nahrendorf, R. Langer and D. G. Anderson, Nat. Nanotechnol., 2012, 7, 389–393 CrossRef CAS PubMed.
- D. Bhatia, S. Chakraborty and Y. Krishnan, Nat. Nanotechnol., 2012, 7, 344–346 CrossRef CAS PubMed.
- Q. Peng, X. R. Shao, J. Xie, S. R. Shi, X. Q. Wei, T. Zhang, X. X. Cai and Y. F. Lin, ACS Appl. Mater. Interfaces, 2016, 8, 12733–12739 CrossRef CAS PubMed.
- A. S. Walsh, H. F. Yin, C. M. Erben and M. J. A. Wood, ACS Nano, 2011, 5, 5427–5432 CrossRef CAS PubMed.
- A. Banerjee, D. Bhatia, A. Saminathan, S. Chakraborty, S. Kar and Y. Krishnan, Angew. Chem., 2013, 125, 6992–6995 CrossRef.
- A. Roloff, A. S. Carlini, C. E. Callmann and N. C. Gianneschi, J. Am. Chem. Soc., 2017, 139, 16442–16445 CrossRef CAS PubMed.
- K. R. Kim, D. Bang and D. R. Ahn, Biomater. Sci., 2016, 4, 605–609 RSC.
- E. J. Lee, S. J. Lee, Y. S. Kang, J. H. Ryu, K. C. Kwon, E. Jo, J. Y. Yhee, I. C. Kwon, K. Kim and J. Lee, Adv. Funct. Mater., 2015, 25, 1279–1286 CrossRef CAS.
- Y. D. Kim, T. E. Park, B. Singh, S. Maharjan, Y. J. Choi, P. H. Choung, R. B. Arote and C. S. Cho, Nanomedicine, 2015, 10, 1165–1188 CrossRef CAS PubMed.
- M. Saraswathy and S. Q. Gong, Mater. Today, 2014, 17, 298–306 CrossRef CAS.
- M. A. Rahman, P. F. Wang, Z. X. Zhao, D. S. Wang, S. Nannapaneni, C. Zhang, Z. J. Chen, C. C. Griffith, S. J. Hurwitz, Z. G. Chen, Y. G. Ke and D. M. Shin, Angew. Chem., Int. Ed., 2017, 56, 16023–16027 CrossRef CAS PubMed.
- K. Mohri, M. Nishikawa, Y. Takahashi and Y. Takakura, Eur. J. Pharm. Sci., 2014, 58, 26–33 CrossRef CAS PubMed.
- V. J. Schuller, S. Heidegger, N. Sandholzer, P. C. Nickels, N. A. Suhartha, S. Endres, C. Bourquin and T. Liedl, ACS Nano, 2011, 5, 9696–9702 CrossRef PubMed.
- K. Mohri, M. Nishikawa, N. Takahashi, T. Shiomi, N. Matsuoka, K. Ogawa, M. Endo, K. Hidaka, H. Sugiyama, Y. Takahashi and Y. Takakura, ACS Nano, 2012, 6, 5931–5940 CrossRef CAS PubMed.
- Y. Takahashi, T. Maezawa, Y. Araie, Y. Takahashi, Y. Takakura and M. Nishikawa, J. Pharm. Sci., 2017, 106, 2457–2462 CrossRef CAS PubMed.
- J. Li, H. Pei, B. Zhu, L. Liang, M. Wei, Y. He, N. Chen, D. Li, Q. Huang and C. H. Fan, ACS Nano, 2011, 5, 8783–8789 CrossRef CAS PubMed.
- X. Y. Ouyang, J. Li, H. J. Liu, B. Zhao, J. Yan, Y. Z. Ma, S. J. Xiao, S. P. Song, Q. Huang, J. Chao and C. H. Fan, Small, 2013, 9, 3082–3087 CrossRef CAS PubMed.
- X. W. Liu, Y. Xu, T. Yu, C. Clifford, Y. Liu, H. Yan and Y. Chang, Nano Lett., 2012, 12, 4254–4259 CrossRef CAS PubMed.
- C. Liu, Y. Xie, B. Sun, F. Geng, F. Zhang, Q. Guo, H. Wu, B. Yu, J. Wu, X. Yu, W. Kong and H. Zhang, Scand. J. Immunol., 2017, 87, 63–72 CrossRef PubMed.
- Y. J. Qu, J. J. Yang, P. F. Zhang, S. L. Liu, K. Zhang, Q. Jiang, C. Li and B. Q. Ding, ACS Appl. Mater. Interfaces, 2017, 9, 20324–20329 CrossRef CAS PubMed.
- W. S. Li, L. Luo, J. Huang, Q. Wang, J. B. Liu, X. Xiao, H. M. Fang, X. H. Yang and K. M. Wang, Chem. Commun., 2017, 53, 5565–5568 RSC.
- V. Balzani, A. Credi and M. Venturi, ChemPhysChem, 2003, 4, 49–59 CrossRef CAS PubMed.
- Y. Jing, L. J. Shen, J. J. Ma, H. I. Schlaberg, S. Liu, J. Xu and C. Zhang, ACS Appl. Mater. Interfaces, 2013, 5, 5392–5396 CrossRef PubMed.
- T. Carell, Nature, 2011, 469, 45–46 CrossRef CAS PubMed.
- M. N. Stojanovic, T. E. Mitchell and D. Stefanovic, J. Am. Chem. Soc., 2002, 124, 3555–3561 CrossRef CAS PubMed.
- L. Y. Feng, Z. Z. Lyu, A. Offenhausser and D. Mayer, Angew. Chem., 2015, 54, 7693–7697 CrossRef CAS PubMed.
- J. Elbaz, Z. G. Wang, R. Orbach and I. Willner, Nano Lett., 2009, 9, 4510–4514 CrossRef CAS PubMed.
- T. Nojima, T. Yamamoto, H. Kimura and T. Fujii, Chem. Commun., 2008, 32, 3771–3773 RSC.
- A. Saghatelian, N. H. Volcker, K. M. Guckian, V. S. Y. Lin and M. R. Ghadiri, J. Am. Chem. Soc., 2003, 125, 346–347 CrossRef CAS PubMed.
- N. H. Voelcker, K. M. Guckian, A. Saghatelian and M. R. Ghadiri, Small, 2008, 4, 427–431 CrossRef CAS PubMed.
- W. Gao, L. Zhang, Y. M. Zhang, R. P. Liang and J. D. Qiu, J. Phys. Chem. C, 2014, 118, 14410–14417 CrossRef CAS.
- L. Y. Feng, A. D. Zhao, J. S. Ren and X. G. Qu, Nucleic Acids Res., 2013, 41, 7987–7996 CrossRef CAS PubMed.
- X. M. Qu, S. P. Wang, Z. L. Ge, J. B. Wang, G. B. Yao, J. Li, X. L. Zuo, J. Y. Shi, S. P. Song, L. H. Wang, L. Li, H. Pei and C. H. Fan, J. Am. Chem. Soc., 2017, 139, 10176–10179 CrossRef CAS PubMed.
- X. T. Ji, H. Y. Lv, M. H. Ma, B. L. Lv and C. F. Ding, Microchim. Acta, 2017, 184, 2505–2513 CrossRef CAS.
- Y. Jing, Z. C. Song, S. Liu, Q. Zhang and C. Zhang, ACS Appl. Mater. Interfaces, 2016, 8, 22451–22456 CrossRef PubMed.
- C. Zhang, L. J. Shen, C. Liang, Y. F. Dong, J. Yang and J. Xu, ACS Appl. Mater. Interfaces, 2016, 8, 9370–9376 CrossRef CAS PubMed.
- M. Moshe, J. Elbaz and I. Willner, Nano Lett., 2009, 9, 1196–1200 CrossRef CAS PubMed.
- X. L. Feng, X. R. Duan, L. B. Liu, F. D. Feng, S. Wang and D. B. Zhu, Angew. Chem., 2009, 48, 5316–5321 CrossRef CAS PubMed.
- T. Li, E. K. Wang and S. J. Dong, J. Am. Chem. Soc., 2009, 131, 15082–15083 CrossRef CAS PubMed.
- F. Xia, X. L. Zuo, R. Q. Yang, R. J. White, Y. Xiao, D. Kang, X. O. Gong and A. A. Lubin, J. Am. Chem. Soc., 2010, 132, 8557–8559 CrossRef CAS PubMed.
- X. M. Li, L. Sun and T. R. Ding, Biosens. Bioelectron., 2011, 26, 3570–3576 CrossRef CAS PubMed.
- P. Vijayakumar and J. Macdonald, ChemPhysChem, 2017, 18, 1735–1741 CrossRef CAS PubMed.
- C. G. Wang, N. Cheng, L. J. Zhu, Y. C. Xu, K. L. Huang, P. Y. Zhu, S. F. Zhu, W. Fu and W. T. Xu, Anal. Chim. Acta, 2017, 987, 111–117 CrossRef CAS PubMed.
- R. P. Goodman, I. A. T. Schaap, C. F. Tardin, C. M. Erben, R. M. Berry, C. F. Schmidt and A. J. Turberfield, Science, 2005, 310, 1661–1665 CrossRef CAS PubMed.
- R. P. Goodman, R. M. Berry and A. J. Turberfield, Chem. Commun., 2004, 12, 1372–1373 RSC.
- A. S. Walsh, H. F. Yin, C. M. Erben, M. J. A. Wood and A. J. Turberfield, ACS Nano, 2011, 5, 5427–5432 CrossRef CAS PubMed.
- H. Pei, L. Liang, G. B. Yao, J. Li, Q. Huang and C. H. Fan, Angew. Chem., 2012, 51, 9020–9024 CrossRef CAS PubMed.
- F. R. Li, H. R. Chen, J. Pan, T. G. Cha, I. L. Medintz and J. H. Choi, Chem. Commun., 2016, 52, 8369–8372 RSC.
- R. M. Zadegan, M. D. E. Jepsen, L. L. Hildebrandt, V. Birkedal and J. Kjems, Small, 2015, 11, 1811–1817 CrossRef CAS PubMed.
- M. X. You, L. Peng, N. Shao, L. Q. Zhang, L. P. Qiu, C. Cui and W. H. Tan, J. Am. Chem. Soc., 2014, 136, 1256–1259 CrossRef CAS PubMed.
- M. X. You, G. Z. Zhu, T. Chen, M. J. Donovan and W. H. Tan, J. Am. Chem. Soc., 2015, 137, 667–674 CrossRef CAS PubMed.
- D. Margulies, C. E. Felder, G. Melman and A. Shanzer, J. Am. Chem. Soc., 2007, 129, 347–354 CrossRef CAS PubMed.
- M. Suresh, A. Ghosh and A. Das, Chem. Commun., 2008, 3906–3908 RSC.
- J. Andreasson, S. D. Straight, T. A. Moore, A. L. Moore and D. Gust, Chemistry, 2009, 15, 3936–3939 CrossRef CAS PubMed.
- F. Pu, Z. Liu, X. J. Yang, J. S. Ren and X. G. Qu, Chem. Commun., 2011, 47, 6024–6026 RSC.
- Z. Z. Huang, Y. Tao, F. Pu, J. S. Ren and X. G. Qu, Chem. - Eur. J., 2012, 18, 6663–6669 CrossRef CAS PubMed.
- Z. X. Zhou, Y. Q. Liu and S. J. Dong, Chem. Commun., 2013, 49, 3107–3109 RSC.
- W. Hong, Y. Du, T. S. Wang, J. Y. Liu, Y. Q. Liu, J. Wang and E. K. Wang, Chemistry, 2012, 18, 14939–14942 CrossRef CAS PubMed.
- J. B. Zhu, X. Yang, L. B. Zhang, L. L. Zhang, B. H. Lou, S. J. Dong and E. K. Wang, Chem. Commun., 2013, 49, 5459–5461 RSC.
- J. H. Chen, S. G. Zhou and J. L. Wen, Angew. Chem., 2015, 54, 446–450 CAS.
- L. M. Adleman, Science, 1994, 266, 1021–1024 CrossRef CAS PubMed.
- L. Qian and E. Winfree, Science, 2011, 332, 1196–1201 CrossRef CAS PubMed.
- L. Qian, E. Winfree and J. Bruck, Nature, 2011, 475, 368–372 CrossRef CAS PubMed.
- C. Teller and I. Willner, Curr. Opin. Biotechnol., 2010, 21, 376–391 CrossRef CAS.
- C. H. Lu, B. Willner and I. Willner, ACS Nano, 2013, 7, 8320–8332 CrossRef CAS PubMed.
- M. K. Beissenhirtz and I. Willner, Org. Biomol. Chem., 2006, 4, 3392–3401 RSC.
- J. M. Abendroth, O. S. Bushuyev, P. S. Weiss and C. J. Barrett, ACS Nano, 2015, 9, 7746–7768 CrossRef CAS PubMed.
- I. Willner and V. Allen, Org. Biomol. Chem., 2006, 4, 3381–3382 RSC.
- R. Chhabra, J. Sharma, Y. Liu and H. Yan, Nano Lett., 2006, 6, 978–983 CrossRef CAS PubMed.
- Y. Tian and C. Mao, J. Am. Chem. Soc., 2004, 126, 11410–11411 CrossRef CAS PubMed.
- W. B. Sherman and N. C. Seeman, Nano Lett., 2004, 4, 1203–1207 CrossRef CAS.
- T. Omabegho, R. Sha and N. C. Seeman, Science, 2009, 324, 67–71 CrossRef CAS PubMed.
- J. S. Shin and N. A. Pierce, J. Am. Chem. Soc., 2004, 126, 10834–10835 CrossRef CAS.
- Y. Tian, Y. He, Y. Chen, P. Yin and C. D. Mao, Angew. Chem., Int. Ed., 2005, 44, 4355–4358 CrossRef CAS.
- S. K. Kufer, E. M. Puchner and H. E. Gaub, Science, 2008, 319, 594–596 CrossRef CAS PubMed.
- Z. G. Wang, J. Elbaz and I. Willner, Angew. Chem., Int. Ed., 2012, 51, 4322–4326 CrossRef CAS PubMed.
- J. Elbaz, Z. G. Wang, F. A. Wang and I. Willner, Angew. Chem., Int. Ed., 2012, 51, 2349–2353 CrossRef CAS PubMed.
- D. Ackermann, T. L. Schmidt, J. S. Hannam, C. S. Purohit, A. Heckel and M. Famulok, Nat. Nanotechnol., 2010, 5, 436–442 CrossRef CAS PubMed.
- C. Buranachai, S. A. Mckinney and T. Ha, Nano Lett., 2006, 6, 496–500 CrossRef CAS PubMed.
- D. Lubrich, J. Lin and J. Yan, Angew. Chem., 2008, 47, 7026–7028 CrossRef CAS PubMed.
- J. Bath, S. J. Green and A. J. Turberfield, Angew. Chem., 2005, 117, 4432–4435 CrossRef.
- S. Sahu, T. H. Labean and J. H. Reif, Nano Lett., 2008, 8, 3870–3878 CrossRef CAS PubMed.
- H. Zhang and N. C. Seeman, Nano Lett., 2006, 6, 2899–2903 CrossRef PubMed.
- Y. Chen, M. S. Wang and C. D. Mao, Angew. Chem., 2004, 43, 3554–3557 CrossRef CAS PubMed.
- S. Modi, M. G. Swetha, D. Goswami, G. D. Gupta, S. Mayor and Y. Krishnan, Nat. Nanotechnol., 2009, 4, 325–330 CrossRef CAS PubMed.
- K. Lund, A. J. Manzo, N. Dabby, N. Michelotti, A. Johnson-Buck, J. Nangreave, S. Taylor, R. J. Pei, M. N. Stojanovic, N. G. Walter, E. Winfree and H. Yan, Nature, 2010, 465, 206–210 CrossRef CAS PubMed.
- N. F. Wang, P. Y. Fu, J. Song and H. F. Li, Asian J. Chem., 2013, 25, 6666–6668 CrossRef CAS.
- Y. Chen, Y. Xiang, R. Yuan and Y. Q. Chai, Nanoscale, 2015, 7, 981–986 RSC.
- T. Pan, M. Mahalingam, B. S. Marasa, Z. H. Zhang, A. K. Chopra and V. B. Rao, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 5846–5851 CrossRef.
- H. L. Li, J. T. Ren, Y. Q. Liu and E. K. Wang, Chem. Commun., 2014, 50, 704–706 RSC.
- J. J. Yin, X. X. He, X. K. Jia, K. M. Wang and F. Z. Xu, Analyst, 2013, 138, 2350–2356 RSC.
- J. Wan, X. L. Ma and L. Xing, Sens. Actuators, B, 2013, 178, 615–620 CrossRef CAS.
- J. Y. Zhuang, W. Q. Lai, G. N. Chen and D. P. Tang, Chem. Commun., 2014, 50, 2935–2938 RSC.
- Y. Zhao, F. Chen, Q. Zhang, Y. Zhao, X. L. Zuo and C. H. Fan, NPG Asia Mater., 2014, 6, e131 CrossRef CAS.
- Q. Zhang, F. Chen, F. Xu, Y. X. Zhao and C. H. Fan, Anal. Chem., 2014, 86, 8098–8105 CrossRef CAS PubMed.
- Q. Xu, Y. Zhang and C. Y. Zhang, Chem. Commun., 2015, 51, 5652–5655 RSC.
- Z. Q. Xu, Y. W. Dong, J. Y. Li and R. Yuan, Chem. Commun., 2015, 51, 14369–14372 RSC.
- D. Li, A. Wieckowska and I. Willner, Angew. Chem., Int. Ed., 2008, 47, 3927–3931 CrossRef CAS PubMed.
- M. K. Beissenhirtz, R. Elnathan, Y. Weizmann and I. Willner, Small, 2007, 3, 375–379 CrossRef CAS PubMed.
- B. Shlyahovsky, D. Li, Y. Weizmann, R. Nowarski, M. Kotler and I. Willner, J. Am. Chem. Soc., 2007, 129, 3814–3815 CrossRef CAS PubMed.
- Y. Weizmann, Z. Cheglakov and I. Willner, J. Am. Chem. Soc., 2008, 130, 17224–17225 CrossRef CAS.
- Z. Wang, J. Elbaz and I. Willner, Nano Lett., 2011, 11, 304–309 CrossRef CAS PubMed.
- X. Q. Liu, A. Niazov-Elkan, F. A. Wang and I. Willner, Nano Lett., 2013, 13, 219–225 CrossRef CAS PubMed.
- X. Q. Liu, C. H. Lu and I. Willner, Acc. Chem. Res., 2014, 47, 1673–1680 CrossRef CAS PubMed.
- X. J. Qi, C. H. Lu, X. Q. Liu, S. Shimron, H. H. Yang and I. Willner, Nano Lett., 2013, 13, 4920–4924 CrossRef CAS.
- E. C. Schoeneweiss and B. Sacca, Nanoscale, 2017, 9, 4486–4496 RSC.
- N. Wan, Z. P. Hong, H. D. Wang, X. Fu, Z. Y. Zhang, C. Li, H. Xia, Y. Fang, M. T. Li, Y. Zhan and X. L. Yang, Small, 2017, 13, 1700866–1700869 CrossRef.
- D. Zhu, H. Pei, G. B. Yao, L. H. Wang, S. Su, J. Chao, L. H. Wang, A. Aldalbahi, S. P. Song, J. Y. Shi, J. Hu, C. H. Fan and X. L. Zuo, Adv. Mater., 2016, 28, 6860–6865 CrossRef CAS.
- C. Zhou, X. Duan and N. Liu, Nat. Commun., 2015, 6, 8102–8106 CrossRef CAS PubMed.
- E. Kopperger, J. List, S. Madhira, F. Rothfischer, D. C. Lamb and F. C. Simmel, Science, 2018, 359, 296–301 CrossRef CAS PubMed.
- A. E. Marras, L. Zhou, V. Kolliopoulos, H. J. Su and C. E. Castro, New J. Phys., 2016, 18, 055005–055009 CrossRef.
- H. J. Su, C. E. Castro, A. E. Marras and L. F. Zhou, J. Mech. Des., 2017, 139, 1590–1599 CrossRef.
- J. S. Kahn, Y. Hu and I. Willner, Acc. Chem. Res., 2017, 50, 680–690 CrossRef CAS.
- A. Cangialosi, C. Yoon, J. Liu, Q. Huang, J. K. Guo, T. D. Nguyen, D. H. Gracias and R. Schulman, Science, 2017, 357, 1126–1129 CrossRef CAS.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2019 |
