Open Access Article
Open Access ArticleBifunctional nickel–iminodiacetic acid-core–shell silica nanoparticles for the exclusion of high molecular weight proteins and purification of His-tagged recombinant proteins
Sergio G. Hernandez-Leon a,
Jose A. Sarabia-Sainz
a,
Jose A. Sarabia-Sainz b,
Gabriela Ramos-Clamont Montfort
b,
Gabriela Ramos-Clamont Montfort a,
José Ángel Huerta-Ocampo
a,
José Ángel Huerta-Ocampo c,
Ana M. Guzman-Partida
c,
Ana M. Guzman-Partida a,
Maria del Refugio Robles-Burgueñoa,
Alexel J. Burgara-Estrella
a,
Maria del Refugio Robles-Burgueñoa,
Alexel J. Burgara-Estrella b and
Luz Vazquez-Moreno*a
b and
Luz Vazquez-Moreno*a
aCentro de Investigación en Alimentación y Desarrollo A.C. Carretera Gustavo Enrique Astiazarán Rosas, No. 46, col. La Victoria, C.P. 83304, Hermosillo, Sonora, Mexico. E-mail: lvazquez@ciad.mx
bUniversidad de Sonora. Blvd. Luis Encinas y Rosales S/N, Col. Centro, Hermosillo, Sonora, Mexico
cCONACYT-Centro de Investigación en Alimentación y Desarrollo A.C. Carretera Gustavo Enrique Astiazarán Rosas, No. 46, col. La Victoria, C.P. 83304, Hermosillo, Sonora, Mexico
First published on 9th April 2019
Abstract
Herein, silica nanoparticles were synthesized and chemically modified with iminodiacetic acid (IDA) and Ni2+ ions surrounded by a bis-acrylamide polymeric shell to obtain a new core–shell immobilized metal affinity chromatography (IMAC) based material. These Ni2+–IDA-core–shell silica nanoparticles (Ni2+–IDA-CSS-NP) represent a new alternative for purification of His-tagged proteins and exclusion of high molecular weight (HMW) proteins at the same time. Nanoparticles presented a final size of 479.6 ± 6.9 nm determined by dynamic light scattering (DLS) and a surface charge of −37.2 ± 0.5 mV. Successful incorporation of the different compounds at every phase of synthesis was evidenced by ATR-FTIR analysis. Ni2+–IDA-CSS-NP were used for isolation of His-tagged spo0F (6His-spo0F) from E. coli lysate. Ni2+–IDA-CSS-NP presented a capacity of 4.16 ± 0.45 μg mg−1. Purification of 6His-spo0F with high selectivity and the effective exclusion of HMW proteins were evidenced by SDS-PAGE and validated through mass spectrometry analysis.
1. Introduction
Currently, in most studies regarding protein structure and function, recombinant protein technology is widely used.1 To produce these heterologous proteins different hosts such as Escherichia coli, Pichia pastoris, Saccharomyces cerevisiae, and insect or mammalian cells have been employed.2 Biotechnology enables proteins to be expressed with a certain tag and histidine tags, mainly hexa-histidine (6His-tag), are the most used affinity tags for recombinant protein purification.1,3Immobilized metal ion affinity chromatography (IMAC) was first developed by Porath in 1975 (ref. 4) and is one of the most effective procedures for purification of His-tagged proteins5 and is based on the metal coordination interaction between the imidazole ring of histidine and divalent transition metal ions such as Ni2+, Cu2+, Zn2+ or Co2+.3,5,6 The most widespread IMAC supports use either iminodiacetic acid (IDA) or nitrilotriacetic acid (NTA) as metal chelating ligands.2
IDA is a tridentate ligand that coordinates a divalent ion with three coordination sites leaving two coordination sites for the imidazole ring (from histidines) interaction while it is unclear if the third is sterically able to interact.7 In a typical IMAC process, a complex sample such as cell lysate is passed over the IMAC matrix, washed and recombinant protein or native proteins of interest are eluted by variation of pH or preferably with high concentration of imidazole.7–9
However, most of conventional IMAC based affinity methods present high leaching of metal ions from ligands in the equilibration and washing steps which result in reduced purity since leaching metal generates charged groups which could act as a cation exchanger and bind charged groups on the surface of proteins. Also, since the purification is based on the interaction of metal ions with the imidazole group on the histidine, proteins with a histidine-rich region may also interact and contaminate His-tagged recombinant proteins.10 Also, copurification of proteins with affinity to agarose which is the most widely used IMAC support may occur and therefore additional steps could be needed, decreasing the recovery of proteins of interest.10
Alternatively, other studies use titanium dioxide11,12 or silica nanoparticles13–15 with anchored nickel nanoparticles at the surface for histidine-rich proteins adsorption.
Therefore, materials to improve purity of His-tagged proteins in a single-step purification process is crucial. In this context, silica nanoparticles have appealing physical and chemical properties as a sorbent, due to its large surface area, low toxicity, high stability and particularly because it can be chemically modified with several compounds.1,16–19 The aim of this work was to synthetize functionalized core–shell silica nanoparticles with IDA and a polyacrylamide shell, thus only low molecular weight (LMW) His-tagged proteins that pass through the polymeric shell to interact with the immobilized metal ions, while other HMW host proteins with inherent histidine-rich region, are excluded.
2. Materials and methods
2.1 Synthesis of IDA-core–shell silica nanoparticles
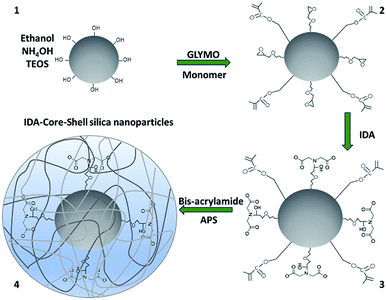
The synthesis of IDA-core–shell silica nanoparticles was performed in four phases that consisted in formation of the support material followed by three chemical modifications. Phase 1—silica nanoparticles were obtained according to the modified Stöber method reported previously.16 Briefly, 40 mL of absolute ethanol, ammonium hydroxide (1.81 M) and tetraethyl orthosilicate (77 mM) were placed into a round-bottom and kept under stirring at room temperature for 24 h.Phase 2—silica nanoparticles solution formed in phase 1 were modified by adding the monomer 3-(trimethoxysilyl)propyl methacrylate (4.8 mM) and (3-glycidyloxypropyl)trimethoxysilane, GLYMO, (2.58 mM). The solution was then incubated at room temperature under magnetic stirring for 24 h. Consecutive washes with ethanol 80% (v/v) and carbonate buffer (50 mM, pH 9.5) were performed using centrifugation at 2422 × g for 12 min (Sorvall Lynx 4000, Thermo Scientific, San Jose, CA, USA) supernatants were discarded and the particle pellet was suspended in 30 mL of carbonate buffer.
Phase 3—iminodiacetic acid (25.04 mM) was added to the previous solution and kept under magnetic stirring at 55 °C for 24 h. The particles were then washed as aforementioned. The solution was purged with nitrogen for 1.5 h.
Phase 4—N-N′-methylene-bis-acrylamide (8 mM) and ammonium persulfate (8 mM) were suspended in 20 mL of ultrapure water (Milli-Q, Merck KGaA, Darmstadt, Germany) and added to the previous solution. Reaction was maintained under nitrogen environment for additional 6 h. IDA-core–shell nanoparticles were washed once with ultrapure water, centrifuged and supernatant was discarded. Finally, the resultant pellet was suspended in 20% ethanol. Aliquots of 1 mL were transferred to new microtubes and then completely dried by centrifugation in vacuum (Centrivap, Labconco, Kansas City, MO, USA) and stored at 4 °C. As control, two simultaneous synthesis of nanoparticles were included, one without the bis-acrylamide shell (Fig. 3-A) and core–shell silica nanoparticles without the IDA molecules (data not shown).
2.2 Characterization
Dynamic light scattering (DLS) and zeta potential (ζ-potential) analysis using a nano-zetasizer (Nano-ZS 90, Malvern instrument, Malvern, UK) was performed to estimate nanoparticles size and surface charge. Measurements were performed at every phase of synthesis (1.5 mL) following manufacturer instructions and recorded as the average of three test run. Attenuated total reflection Fourier transformed infrared (ATR-FTIR) spectra of samples at every phase of synthesis, previously dried under vacuum centrifugation, were collected on a Cary 630 spectrometer (Agilent, Cary 630 FTIR Spectrometer, Santa Clara, CA, USA) with 4 cm−1 resolution over a spectral range of 650–4000 cm−1.Morphological characterization at every phase of synthesis was performed by Atomic Force Microscopy (AFM, XE-Bio, Park Systems Corp, Suwon, Korea). The Atomic force microscope was operated in the non-contact cantilever mode and the morphology analysis of the nanoparticles (one drop air-dried) was performed using scans of 20 × 20 μm followed by 5 × 5 μm scan images. 3D mode AFM images were obtained and analysed by the XEI software (version 1.8) by Park systems.
2.3 Ni2+ attachment onto functionalized IDA-core–shell silica nanoparticles
To obtain Ni2+–IDA-core–shell silica nanoparticles (Ni2+–IDA-CSS-NP), five milligram of IDA-core–shell silica nanoparticles were incubated with 1 mL of 0.25 M NiSO4·6H2O solution under gentle stirring at room temperature for 2 h. Ni2+ ions coordinates IDA with three coordination sites via metal carboxyl chemistry. Obtained Ni2+–IDA-CSS-NP were washed three times with ultrapure water to remove free Ni2+ ions using centrifugation at 5585 × g for 5 min (Costar mini centrifuge, Cambridge scientific, Watertown, MA, USA). Supernatant was discarded, and the particle pellet was then equilibrated with binding buffer (20 mM phosphate buffer, 100 mM NaCl, pH 7.3) three times, using centrifugation at 5585 × g for 5 min.2.4 Production of recombinant protein and cell lysate
In this work, sporulation initiation phosphotransferase F (spo0F) from Bacillus thuringiensis20 that presents a theoretical pI/Mw of 4.61/13801.2 Da, and the sequence MEGKILIVDDQYGIRVLLHEVFQKEGYQTFQAANGFQALDIVKKDNPDLVVLDMKIPGMDGIEILKHVKEIDESIKVILMTAYGELDMIQEAKDLGALMHFAKPFDIDEIRQAVRNELAVEA was selected as the model 6His-tagged recombinant protein. For 6His-spo0F overexpression, the procedure reported by Cabrera in 2014 was applied.20 Briefly, E. coli strain BL21 Gold (DE3) carrying Pet11aΩspo0F was used for overexpression. Cultures were incubated overnight at 30 °C. The inoculum was prepared in ampicillin containing Terrific Broth (TB). When OD600nm = 0.4 was reached, spo0F was induced with 0.5 mM IPTG for 4 h. After incubation, cells were centrifugated and cell pellet was suspended in lysis buffer (100 mM NH2PO4, 100 mM NaCl, 5 mM imidazole, 10% glycerol, 1 mg mL−1 lysozyme and 1 mM PMSF) and sonicated. Cell debris was removed by centrifugation and resultant cell lysate (supernatant) was stored at −40 °C.2.5 His-tagged recombinant protein purification by the Ni2+–IDA-core–shell silica nanoparticles
Five milligrams of Ni2+–IDA-core silica nanoparticles (without the polymeric shell) or Ni2+–IDA-CSS-NP were equilibrated as described in Section 2.3. Nanoparticles were suspended in 0.3 mL of binding buffer and incubated with 0.3 mL of E. coli lysate (∼486 μg of protein) for 1 h under gentle stirring. After incubation, particles were centrifuged at 5585×g for 5 min (Costar mini centrifuge, Cambridge scientific, Watertown, MA, USA) and supernatant (non-binding fraction) was transferred into a new vial. To remove non-specifically adsorbed proteins, particles were consecutively washed once with 0.4 mL of phosphate buffer (PB) 20 mM, 100 mM NaCl, 100 mM imidazole (wash solution 1) and 0.4 mL of 20% (v/v) isopropanol, 100 mM NaCl (wash solution 2) performing centrifugation at each wash as above-mentioned. Subsequently, His-tagged protein (6His-spo0F) was eluted with 0.2 mL of elution solution (300 mM imidazole, 100 mM NaCl). Washes and elutions were dried by centrifugation under vacuum, suspended in ultrapure water and stored at 4 °C for further analysis. Protein concentration was estimated by Bradford assay,21 using bovine serum albumin (BSA) as standard. Protein electrophoretic profile was analysed using 15% SDS-PAGE according to Laemmli procedure.22Five micrograms of protein from each purification fraction were loaded in the polyacrylamide gel. To visualize bands, silver staining protocol was performed according to Shevchenko.23
2.6 Protein digestion and liquid chromatography-tandem mass spectrometry analysis (LC-MS/MS)
Washed proteins with wash solution 1 (PB 20 mM, 100 mM NaCl, 100 mM imidazole) and eluted with 300 mM imidazole, 100 mM NaCl solution were selected for LC-MS/MS analysis. Samples were filtered, separately, and then washed two times with ultrapure water using a MWCO 3 kDa cut-off Centricon (Merck Millipore, Darmstadt, Germany) following manufacturer's instructions. Then, 6 μg of total protein from the retained fractions (proteins > 3 kDa) of each sample were reconstituted in 1 M urea, reduced with 10 mM dithiothreitol, alkylated with 50 mM iodoacetamide and digested overnight at 37 °C with sequencing grade trypsin (Promega, Madison, WI, USA). Tryptic peptides were dried by centrifugation in vacuum, suspended in 0.1% trifluoroacetic acid and purified with ZipTip (Merck Millipore, Darmstadt, Germany).Tryptic peptides were subjected to reverse phase ultra-performance liquid chromatography using the 1290 Infinity LC System (Agilent Technologies, Santa Clara, CA, USA) equipped with an analytical column ZORBAX 300SB-C8 (5 μm × 2.1 mm × 150 mm, Agilent Technologies, Santa Clara, CA, USA) coupled to a Dual AJS ESI ionization source (Agilent Technologies, Santa Clara, CA, USA) and analysed by tandem mass spectrometry through a data-dependent analysis in the 6530 Accurate-Mass Quadrupole Time-of-Flight (Q-TOF) LC/MS system (Agilent Technologies, Santa Clara, CA, USA) with the chromatographic and MS/MS conditions reported by Morales-Amparano24 with slight modifications. Briefly, MS and MS/MS scans were obtained at a rate of 3 spectra per s and a maximum of five precursors per cycle were selected in MS mode for further peptide fragmentation by collision-induced dissociation.
2.7 Protein identification
Mass spectrometry data was search against an in-house database created with the E. coli BL21-Gold (DE3) subset of the RefSeq protein database (4359 sequences, January 2019) plus the sequence of spo0F from Bacillus thuringiensis Bt407 reported at NCBI (accession number AFV21188) and Chicken Lysozyme C from Swiss-Prot database (accession number P00698). Protein identification was performed using the Spectrum Mill MS Proteomics Workbench server (Agilent technologies, Santa Clara, CA, USA). Trypsin was selected as specific protease allowing one missed cleavage. Mass error tolerance for parent and fragment ions was set at 20 ppm and 0.1 Da, respectively. Carbamidomethyl cysteine was selected as fixed modification, while methionine oxidation was set as variable modification. Individual ion scores ≥ 12 and scored peak intensity (SPI) ≥ 60 were considered as good matches, whereas protein score ≥ 25 and at least two peptides were necessary for a confident protein identification.3. Results and discussion
3.1 Synthesis and characterization of nanoparticles
Silica nanoparticles with a uniform distribution were synthesized by the hydrolysis of TEOS with aqueous ammonia as reported previously.16 The matrix silica nanoparticles were then functionalized with randomly distributed chemical groups for the addition of IDA by conjugation of imine groups of IDA to the epoxy groups of GLYMO1 and formation of the polymeric shell by free radical polymerization of the monomer and bis-acrylamide in presence of APS.16 Synthesis of IDA-core–shell silica nanoparticles is shown in Scheme 1.Size and surface charge characterization at each modification step was performed by DLS and zeta potential, respectively, to confirm the successful modifications (Table 1).
| Phase of synthesis and compounds added | ||||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| Silica nanoparticles | GLYMO and monomer | IDA | Bis-acrylamide | |
| a Values are expressed as mean ± standard deviation (SD). GLYMO = (3-glycidyloxypropyl)trimethoxysilane; monomer = 3-(trimethoxysilyl)propyl methacrylate; IDA = iminodiacetic acid; bis-acrylamide = N-N′-methylene-bis-acrylamide. | ||||
| Size ± SD (nm) | 205.8 ± 1.9 | 239.9 ± 0.5 | 398.3 ± 14.4 | 479.6 ± 6.9 |
| Zeta potential ± SD (mV) | −42 ± 2 | −33.2 ± 2.6 | −43.1 ± 0.6 | −37.2 ± 0.5 |
Average size of silica nanoparticles (first stage) resulted in 205.8 ± 1.9 nm, that was within the expected size range from 50 to 900 nm, as reported by Stöber in 1968.25 Nanoparticles size increased after the addition of GLYMO and the monomer 3-(trimethoxysilyl)propyl methacrylate (239.9 ± 0.5 nm), which provides epoxy and ester functional groups, respectively, on the silica nanoparticles surface (core), and are necessary for the following chemical modifications.
A major increase in size was observed when IDA was added (398.3 ± 14.4 nm). This behaviour may be attributed to the hydrophilic nature of the organic compound attached to the core of nanoparticles, which form a dense layer that increases electrostatic repulsion at the surface and, therefore, increases particle size.16,26
Finally, due to growth of the polymer chains27,28 in shell formation, nanoparticles reached 479.6 ± 6.9 nm in size.
Changes at the surface charge (zeta potential) after every functionalization step can be attributed to the functional groups at the surface and confirms the successful chemical modification at every phase29 as well as the stability of the nanoparticles since nanoparticles with a zeta potential greater than +30 mV or less than −30 mV are considered as strongly anionic or cationic and therefore stable to remain in solution.30
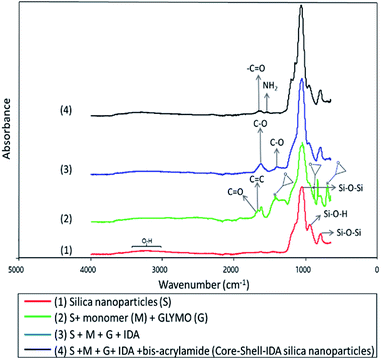
Characterization of nanoparticles by ATR-FTIR was also performed. ATR-FTIR spectra of silica nanoparticles and its consecutive modifications are shown in Fig. 1.
Typical spectrum of silica nanoparticles is shown in Fig. 1(1).16,31 In Fig. 1(2), new absorption peaks appeared at 1628 cm−1, which correspond to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching vibrations32 and another peak at around 1701 cm−1 associated to C
C stretching vibrations32 and another peak at around 1701 cm−1 associated to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups of the monomer.16,33 Also, adsorption bands at 693, 890 and 1410 cm−1 were assigned to vibrations of epoxide rings of the GLYMO molecule.34 Fig. 1(3) shows the infrared spectrum after incorporation of IDA. Peaks at 1398 and 1615 cm−1 are characteristic adsorption bands of C–O symmetric stretching vibration of IDA.35 Also, epoxide ring adsorption bands, which are involved in the interaction with imine groups of IDA, disappeared indicating that IDA was successfully attached to nanoparticles. Finally, in Fig. 1(4), formation of the polymeric shell after addition of bis-acrylamide was evidenced by appearance of two new absorption bands at 1649 and 1535 cm−1 which were assigned to the –C
O groups of the monomer.16,33 Also, adsorption bands at 693, 890 and 1410 cm−1 were assigned to vibrations of epoxide rings of the GLYMO molecule.34 Fig. 1(3) shows the infrared spectrum after incorporation of IDA. Peaks at 1398 and 1615 cm−1 are characteristic adsorption bands of C–O symmetric stretching vibration of IDA.35 Also, epoxide ring adsorption bands, which are involved in the interaction with imine groups of IDA, disappeared indicating that IDA was successfully attached to nanoparticles. Finally, in Fig. 1(4), formation of the polymeric shell after addition of bis-acrylamide was evidenced by appearance of two new absorption bands at 1649 and 1535 cm−1 which were assigned to the –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration of amide group and –NH2 bending vibrations of amine group from amide I and amide II, respectively.16,36,37
O stretching vibration of amide group and –NH2 bending vibrations of amine group from amide I and amide II, respectively.16,36,37
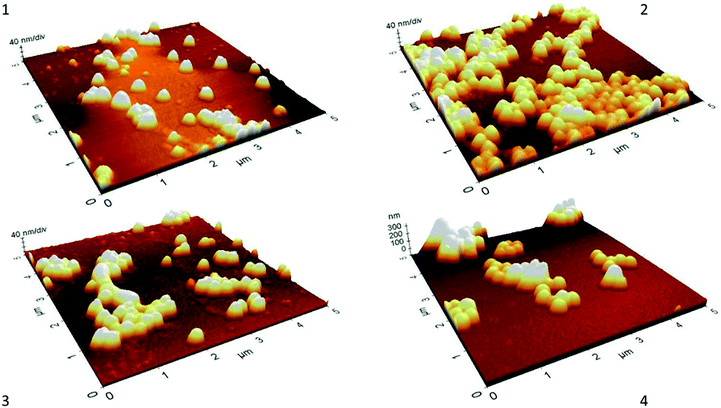
Morphological characterization at every phase of synthesis performed by AFM (Fig. 2) shows the spherical shape of nanoparticles with a homogeneous distribution. An increased in size after every chemical modification was observed, however, all scale-estimated values in AFM were smaller than the sizes obtained by DLS. This could be attributed to sample treatments, in DLS the hydrodynamic diameter of the particles is measured, and this includes hydration layers or polymeric shells which leads to a larger particle size.38 On the other hand, AFM requires observation of nanoparticles in a dry state,39 which is a more compact state where no solvent molecules are associated with nanoparticles and therefore smaller sizes in comparison to DLS might be expected.
3.2 6His-spo0F recombinant protein purification by the Ni2+–IDA-core–shell silica nanoparticles
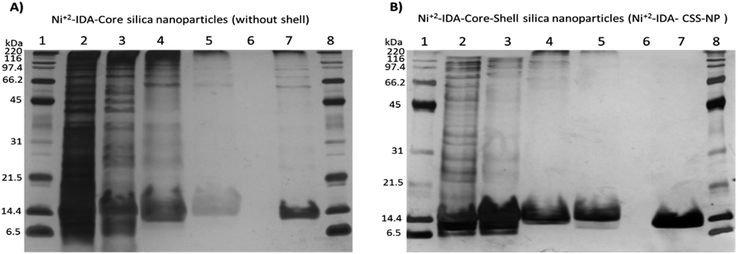
To evaluate the performance of both Ni2+–IDA-core silica nanoparticles and Ni2+–IDA-CSS-NP on the purification of His-tagged target protein, expression of 6His-spo0F was induced in E. coli strain BL21 Gold (DE3) carrying Pet11aΩspo0F and cell lysate was then obtained and incubated with the nanoparticles.Purification process for the 6His-spo0F was analysed by SDS-PAGE (Fig. 3-A and B). As seen in lane 2 (E. coli cell lysate) a prominent wide band of around 13–14 kDa was observed. These bands might correspond to lysozyme of ∼14.7 kDa, which was used in the cell lysis and/or the target 6His-spo0F recombinant protein with a molecular weight of ∼13.8 kDa. Excess of lysozyme and other proteins with high molecular weight were removed consecutively in the non-binding fraction (Fig. 3-B, lane 3) and two additional washes. Wash 1 (lane 4) containing PB 20 mM, 100 mM NaCl and 100 mM imidazole was used to remove host proteins with inherent histidine-rich regions,10 while wash 2 (Fig. 3-B, lane 5) containing 20% (v/v) isopropanol and 100 mM NaCl was used to disrupt possible hydrophobic interaction between proteins and some rarely free epoxide rings from the GLYMO molecule.
On the other hand, the 6His-spo0F band (∼13.8 kDa) was distinguishable in elution with 300 mM of imidazole (lane 7) as it migrated slightly lower than those of lysozyme (∼14.7 kDa, lanes 4 and 5). It is important to note that, incorporation of IDA in the core of nanoparticles is crucial for the capture of 6His-spo0F, since nanoparticles synthesized without IDA molecule did not interact with His-tagged recombinant protein (data not shown). Likewise, nanoparticles without the polymeric shell (Ni2+–IDA-core silica nanoparticles) presented HMW proteins in the elution (Fig. 3-A, lane 7). On the other hand, using the Ni2+–IDA-CSS-NP (Fig. 3-B, lane 7) HMW proteins were not retained, thus indicating the effectiveness of the polymeric shell as a molecular sieve, as it hinders the entrance of these proteins, but allows the entrance of the small ones to interact with the pseudo-affinity molecule attached in the core of the nanoparticles.
Exclusion of HMW proteins with the Ni2+–IDA-CSS-NP is more evident compared to other reports of IMAC-based core–shell sorbents.10,40
In our work, HMW proteins were not observed, even with a more sensitive staining protocol (silver staining), whereas in other studies10,40 HMW proteins were abundant, as they were observed using a less sensitive staining protocol such as Coomassie staining.41
Adsorption capacity of Ni2+–IDA-CSS-NP was determined by estimating the amount of protein in the elutions by Bradford assay21 from three different purifications, and resulted in 4.16 ± 0.45 μg mg−1 of sorbent and this capacity was higher than that reported by Li and colleagues in 2016,42 where they used modified Ni2+–IDA magnetic silica nanoparticles and presented a capacity of 3.6 μg mg−1 of sorbent.
Capture of His-tagged recombinant proteins by the Ni2+–IDA-CSS-NP via interaction of Ni2+ ions attached to the core of nanoparticles with the imidazole ring from the histidine residues (6His-tag) of the recombinant protein is illustrated in Fig. 4.
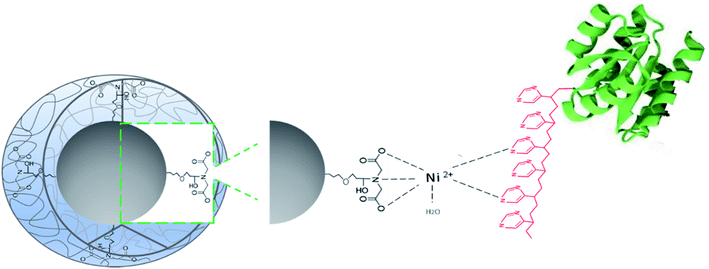 | ||
| Fig. 4 Proposed interaction of target 6His-tagged recombinant protein with the Ni2+–IDA-core–shell-silica nanoparticles. | ||
3.3 6His-spo0F recombinant protein purification by the Ni2+–IDA-core–shell silica nanoparticles
To confirm that the protein eluted was the 6His-spo0F recombinant protein a LC-MS/MS analysis was conducted and further identification of the proteins in washes and elutions was performed. Ni2+–IDA-CSS-NP were incubated with cell lysate as described in the previous section.Proteins from wash 1 and elution samples were digested with trypsin and analysed by LC-MS/MS (Table 2). MS data of the samples were further analysed using the Spectrum Mill MS Proteomics Workbench server and protein identification was performed by searching against an in-house data base created for E. coli BL21-Gold (DE3), spo0F and lysozyme proteins sequences.
| Peptide sequence | m/z measured (Da) | MH + matched (Da) | MH + error (Da) | Protein MW (Da) | Score | SPIa | NCBI accession number | NPM/SCb (%) |
|---|---|---|---|---|---|---|---|---|
| a Scored peak intensity.b Number of peptides matched/sequence coverage. | ||||||||
| Wash 1 (PB 20 mM, 100 mM NaCl, 100 mM imidazole) | ||||||||
| Lysozyme C (Gallus gallus) | ||||||||
| FESNFNTQATNR | 714.8220 | 1428.6500 | −0.0135 | 14769.3 | 16.24 | 63.3 | P00698 | 3/28.2 |
| NTDGSTDYGILQINSR | 877.4127 | 1753.8350 | −0.0170 | 14769.3 | 13.26 | 60.6 | P00698 | |
| GTDVQAWIR | 523.2705 | 1045.5430 | −0.0088 | 14769.3 | 12.9 | 70.2 | P00698 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||||
| Elution (300 mM imidazole, 100 mM NaCl) | ||||||||
| Sporulation initiation phosphotransferase F (Bacillus thuringiensis Bt407) | ||||||||
| ILIVDDQYGIR | 652.8556 | 1304.7210 | −0.0170 | 13800.8 | 17.78 | 93.2 | AFV21188 | 4/45.9 |
| EGYQTFQAANGFQALDIVK | 1050.5082 | 2100.0400 | −0.0305 | 13800.8 | 17.38 | 62.2 | AFV21188 | |
| VLLHEVFQK | 556.8198 | 1112.6460 | −0.0139 | 13800.8 | 15.45 | 64.7 | AFV21188 | |
| VILMTAYGELDMIQEAK | 962.9778 | 1924.9760 | −0.0276 | 13800.8 | 14.61 | 70.1 | AFV21188 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||||
| Trigger factor (E. coli BL21 Gold, DE3) | ||||||||
| NVALEEQAVEAVLAK | 792.4319 | 1583.8640 | −0.0074 | 48191.8 | 12.83 | 62.5 | 447121130 | 2/6.2 |
| VVVGLLLGEVIR | 633.9071 | 1266.8140 | −0.0075 | 48191.8 | 14.69 | 75.1 | 447121130 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||||
| DNA-directed RNA polymerase subunit alpha (E. coli BL21 Gold, DE3) | ||||||||
| EGVQEDILEILLNLK | 863.4725 | 1725.9630 | −0.0255 | 36739.3 | 13.22 | 64.5 | 447084838 | 2/8.2 |
| LLVDACYSPVER | 711.3501 | 1421.7090 | −0.0165 | 36739.3 | 14.89 | 66.8 | 447084838 | |
As hypothesized, mainly the 6His-spo0F recombinant protein was obtained and identified in the elution. Thus, confirming the strong interaction with the imidazole rings from the recombinant protein with the Ni2+ ions immobilized in the IDA molecule in the core of nanoparticles,1,10,40 that required a high concentration of imidazole to be disrupted (300 mM imidazole, 100 mM NaCl). Identification of four distinct peptides in the elution led to the confirmation of spo0F (accession number AFV21188) with a sequence coverage of 45.9% (Table 2).
On the other hand, lysozyme was only present in the wash; indicating that it did not interact with Ni2+ ions attached to IDA or that the interaction was not strong enough since it was removed with a lower concentration of imidazole (100 mM imidazole). Lysozyme (accession number P00698) was identified in the wash with three distinct peptides and a sequence coverage of 28.6%.
Two distinct proteins, from the, E. coli BL21 (DE3), were also identified in the elution content (Table 2). Two peptides led to identification of the protein trigger factor (accession number 447121130), whereas another two distinct peptides led to identification of DNA-directed RNA polymerase subunit alpha (accession number 447084838). These results and Fig. 3-B data, indicate that the latter two proteins were in a very low abundance, not observed with silver staining, but recognized using a more sensitive technique, mass spectrometry.
4. Conclusions
A new material based on functionalized core–shell silica nanoparticles was synthesized for His-tagged protein purification. In one step, Ni2+–IDA-core–shell silica nanoparticles allowed capture of 6His-spo0F with high selectivity from an E. coli lysate and a more effective exclusion of high molecular weight proteins with respect to other core–shell based nanoparticles. Based on these results, Ni2+–IDA-core–shell silica nanoparticles could also be applied for purification of other low molecular weight His-tagged proteins from different biological sources.Conflicts of interest
Authors declare no conflict of interestAcknowledgements
We are grateful to the Institutional Analytical Platform of CIAD, for the support for this research, under Project PAI-10363. Hernandez-Leon thanks the Mexican National Council for Science and Technology (CONACYT) for the scholarship for PhD studies award. Special thanks to PhD Rosina Cabrera for providing E. coli cells for the overexpression of spo0F.References
- K. Salimi, D. D. Usta, İ. Koçer, E. Çelik and A. Tuncel, RSC Adv., 2017, 7, 8718–8726 RSC.
- J. Arnau, C. Lauritzen, G. E. Petersen and J. Pedersen, Protein Expression Purif., 2006, 48, 1–13 CrossRef CAS PubMed.
- P. Li, L. Li, Y. Zhao, L. Sun and Y. Zhang, J. Inorg. Biochem., 2016, 156, 49–54 CrossRef CAS PubMed.
- J. Porath, J. Carlsson, I. Olsson and G. Belfrage, Nature, 1975, 258, 598–599 CrossRef CAS PubMed.
- S. V. Wegner and J. P. Spatz, Angew. Chem., Int. Ed., 2013, 52, 7593–7596 CrossRef CAS PubMed.
- A. Kumar, M. Kamihira, I. Y. Galaev, S. Iijima and B. Mattiasson, Langmuir, 2003, 19, 865–871 CrossRef CAS.
- H. Block, B. Maertens, A. Spriestersbach, N. Brinker, J. Kubicek, R. Fabis, J. Labahn and F. Schäfer, Methods Enzymol., 2009, 463, 439–473 CAS.
- J. A. Bornhorst and J. J. Falke, Methods Enzymol., 2000, 326, 245–254 CAS.
- J. Porath, Protein Expression Purif., 1992, 3, 263–281 CrossRef CAS PubMed.
- S. Li, K. Yang, L. Liu, B. Zhao, Y. Chen, X. Li, L. Zhang and Y. Zhang, Anal. Chim. Acta, 2018, 997, 9–15 CrossRef CAS PubMed.
- Y. Ling, M. Zhang, X. Li, J. Zheng and J. Xu, Dalton Trans., 2018, 47, 10093–10101 RSC.
- M. Zhang, M. Wakeel, N. S. Alharbi, J. Zheng, T. Miao, J. Xu and J. Yang, Mater. Chem. Front., 2018, 3, 224–232 RSC.
- Y. Zhang, M. Zhang, J. Yang, L. Ding, J. Zheng, J. Xu and S. Xiong, Nanoscale, 2016, 8, 15978–15988 RSC.
- M. Zhang, J. Yang, L. Ding, J. Xu, J. Zheng and Y. Zhang, Green Chem., 2016, 18, 6282–6290 RSC.
- X. Yang, M. Zhang, J. Zheng, W. Li, W. Gan, J. Xu, T. Hayat, N. S. Alharbi and F. Yang, Appl. Surf. Sci., 2018, 439, 128–138 CrossRef CAS.
- S. G. Hernandez-Leon, J. A. I. Sarabia-Sainz, G. R. C. Montfort, A. M. Guzman-Partida, M. Del Refugio Robles-Burgueño and L. Vazquez-Moreno, Molecules, 2017, 22, 1712 CrossRef PubMed.
- T. Pham, J. B. Jackson, N. J. Halas and T. R. Lee, Langmuir, 2002, 18, 4915–4920 CrossRef CAS.
- A. Watermann and J. Brieger, Nanomaterials, 2017, 7, 189 CrossRef PubMed.
- A. Liberman, N. Mendez, W. C. Trogler and a. C. Kummel, Surf. Sci. Rep., 2014, 69, 132–158 CrossRef CAS PubMed.
- R. Cabrera, J. Rocha, V. Flores, L. Vázquez-Moreno, G. Guarneros, G. Olmedo, A. Rodríguez-Romero and M. de la Torre, Appl. Microbiol. Biotechnol., 2014, 98, 9399–9412 CrossRef CAS PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- U. K. Laemmli, Nature, 1970, 227, 680–685 CrossRef CAS PubMed.
- A. Shevchenko, M. Wilm, O. Vorm and M. Mann, Anal. Chem., 1996, 68, 850–858 CrossRef CAS PubMed.
- M. B. Morales-Amparano, G. Ramos-Clamont Montfort, I. Baqueiro-Peña, M. del R. Robles-Burgueño, L. Vázquez-Moreno and J. Á. Huerta-Ocampo, Food Sci. Biotechnol., 2018, 1–10 Search PubMed.
- W. Stöber, A. Fink and E. Bohn, J. Colloid Interface Sci., 1968, 26, 62–69 CrossRef.
- G. Ferreira, A. R. Hernandez-Martinez, H. Pool, G. Molina, M. Cruz-Soto, G. Luna-Barcenas and M. Estevez, Mater. Sci. Eng., C, 2015, 57, 49–57 CrossRef CAS PubMed.
- C. Pinto Reis, R. J. Neufeld, A. J. Ribeiro and F. Veiga, Nanomedicine, 2006, 2, 8–21 CrossRef PubMed.
- D. S. Achilleos and M. Vamvakaki, Materials, 2010, 3, 1981–2026 CrossRef CAS.
- M. A. Morini, M. B. Sierra, V. I. Pedroni, L. M. Alarcon, G. A. Appignanesi and E. A. Disalvo, Colloids Surf., B, 2015, 131, 54–58 CrossRef CAS PubMed.
- J. D. Clogston and A. K. Patri, Zeta Potential Measurement, in Characterization of Nanoparticles Intended for Drug Delivery, ed. S. E. McNeil, Humana Press, Switzerland, 2011, ch. 6, vol. 697, pp. 63–70, DOI:10.1007/978-1-60327-198-1_6.
- M. A. Mousavi, S. Hassanajili and M. R. Rahimpour, Appl. Surf. Sci., 2013, 273, 205–214 CrossRef CAS.
- N. A. Rangel-Vázquez and T. Leal-García, J. Mex. Chem. Soc., 2010, 54, 192–197 Search PubMed.
- D. H. Zhang, N. Chen, M. N. Yang, Y. F. Dou, J. Sun, Y. D. Liu and G. Y. Zhi, J. Mol. Catal. B: Enzym., 2016, 133, 136–143 CrossRef CAS.
- S. F. Reza Eslami-Farsani and H. Khosravi, International Journal of Chemical, Molecular, Nuclear, Materials and Metallurgical Engineering, 2015, 9, 1455–1458 Search PubMed.
- W. Wei, B. Zhao, M. He, B. Chen and B. Hu, RSC Adv., 2017, 7, 8504–8511 RSC.
- L. T. Chiem, L. Huynh, J. Ralston and D. A. Beattie, J. Colloid Interface Sci., 2006, 297, 54–61 CrossRef CAS PubMed.
- Y. Zhao, M. Li, Q. Lu and Z. Shi, Langmuir, 2008, 24, 12651–12657 CrossRef CAS PubMed.
- H. Fissan, S. Ristig, H. Kaminski, C. Asbach and M. Epple, Anal. Methods, 2014, 6, 7324–7334 RSC.
- C. M. Hoo, N. Starostin, P. West and M. L. Mecartney, J. Nanopart. Res., 2008, 10, 89–96 CrossRef CAS.
- Y. Zhou, D. Yan, S. Yuan, Y. Chen, E. E. Fletcher, H. Shi and B. Han, Protein Expression Purif., 2018, 144, 5–11 CrossRef CAS PubMed.
- G. Candiano, M. Bruschi, L. Musante, L. Santucci, G. M. Ghiggeri, B. Carnemolla, P. Orecchia, L. Zardi and P. G. Righetti, Electrophoresis, 2004, 25, 1327–1333 CrossRef CAS PubMed.
- S. Li, K. Yang, B. Zhao, X. Li, L. Liu, Y. Chen, L. Zhang and Y. Zhang, J. Mater. Chem. B, 2016, 4, 1960–1967 RSC.
| This journal is © The Royal Society of Chemistry 2019 |