Open Access Article
Open Access ArticleEnhancement of protein stability by an additional disulfide bond designed in human neuroglobin†
Hai-Xiao Liu a,
Lianzhi Li
a,
Lianzhi Li b,
Xin-Zhi Yangc,
Chuan-Wan Weia,
Hui-Min Chenga,
Shu-Qin Gaoc,
Ge-Bo Wenc and
Ying-Wu Lin
b,
Xin-Zhi Yangc,
Chuan-Wan Weia,
Hui-Min Chenga,
Shu-Qin Gaoc,
Ge-Bo Wenc and
Ying-Wu Lin *ac
*ac
aSchool of Chemistry and Chemical Engineering, University of South China, Hengyang 421001, China. E-mail: linlinying@hotmail.com; ywlin@usc.edu.cn
bSchool of Chemistry and Chemical Engineering, Liaocheng University, Liaocheng 252059, China
cLaboratory of Protein Structure and Function, University of South China, Hengyang 421001, China
First published on 31st January 2019
Abstract
Human neuroglobin (Ngb) forms an intramolecular disulfide bond between Cys46 and Cys55, with a third Cys120 near the protein surface, which is a promising protein model for heme protein design. In order to protect the free Cys120 and to enhance the protein stability, we herein developed a strategy by designing an additional disulfide bond between Cys120 and Cys15 via A15C mutation. The design was supported by molecular modeling, and the formation of Cys15–Cys120 disulfide bond was confirmed experimentally by ESI-MS analysis. Molecular modeling, UV-Vis and CD spectroscopy showed that the additional disulfide bond caused minimal structural alterations of Ngb. Meanwhile, the disulfide bond of Cys15–Cys120 was found to enhance both Gdn·HCl-induced unfolding stability (increased by ∼0.64 M) and pH-induced unfolding stability (decreased by ∼0.69 pH unit), as compared to those of WT Ngb with a single native disulfide bond of Cys46–Cys55. Moreover, the half denaturation temperature (Tm) of A15C Ngb was determined to be higher than 100 °C. In addition, the disulfide bond of Cys15–Cys120 has slight effects on protein function, such as an increase in the rate of O2 release by ∼1.4-fold. This study not only suggests a crucial role of the artificial disulfide in protein stabilization, but also lays the groundwork for further investigation of the structure and function of Ngb, as well as for the design of other functional heme proteins, based on the scaffold of A15C Ngb with an enhanced stability.
Introduction
Metalloproteins and metalloenzymes play crucial roles in biological systems, including electron transfer, gas transport, and catalysis.1–6 In order to expand their functionalities, researchers have made great efforts in the design of artificial metalloproteins and metalloenzymes.1,7–17 Since enzyme activity is affected by many factors, increasing the protein stability may ensure the artificial enzymes to be active under harsh reaction conditions, such as in presence of denaturing agents, with high temperature and in different pH solutions. For this purpose, various strategies have been developed in the last decade, such as rational design,18 directed evolution,19 and computational design,20 by improving the hydrophobic and hydrogen(H)-bonding interactions within the protein scaffold. Moreover, covalent interactions such as disulfide bonds play a crucial role in protein stabilization, and design of de novo disulfide bond has shown to be useful for enhancing the protein stability.21–27Heme proteins are a major class of metalloproteins and metalloenzymes.4 To increase the protein stability, especially for the heme-binding affinity of heme proteins, a common approach is to construct a covalent heme to protein cross-link, as occurred in natural cytochrome c (Cyt c) and some Cyt P450s.28–30 For example, a Cyt c-like covalent link (Cys-heme cross-link) has been constructed in Cyt b5,31–33 Cyt b562,34 ascorbate peroxidase,35 and myoglobin (Mb).36 Moreover, other heme-protein cross-links such as Met-Heme,37 His-Heme,38 and Tyr-heme,39,40 can also be designed in artificial heme proteins. In addition to heme-protein cross-links, design of de novo disulfide bond has shown to be powerful, not only to increase the protein stability but also fine-tune the reactivity of heme proteins.30 For example, Morishima and co-workers constructed a de novo disulfide bond in Mb that was found to affect ligand binding dynamics.41 In recent studies, we designed a de novo disulfide bond in Mb, which enhances both protein stability toward guanidine hydrochloride (Gdn·HCl)-inducing unfolding, and reactivity, such as peroxidase and nitrite reductase activities.42,43
Neuroglobin (Ngb) was discovered in 2000 as a new member of globins, highly expressed in nerve tissues.44 In addition to the feature of a bis-His (His64/His96) heme coordination, human Ngb forms an intramolecular disulfide bond (Cys46–Cys55), which regulates the coordination of His64 and the binding of exogenous ligands, such as O2 and CO, to the heme iron.45–48 Moreover, human Ngb has a third cysteine (Cys120), which is located near the protein surface, and is thus prone to be modified by reactive oxygen/nitrogen species (ROS/RNS) in oxidative stress conditions.49 The single free Cys120 near the protein surface may also lead to protein dimerization in purification of the recombinant Ngb protein. Therefore, Cys120 was usually replaced by a Ser to simplify protein purification and prevent protein dimerization, and functional studies were performed based on the C120S Ngb mutant.50–52 Meanwhile, Ser120 forms an H-bonding network with His23 and Glu22 that is missing in wild-type (WT) Ngb, which may affect ligand migration from the protein matrix.53 Although Cys120 has a biological role and contributes to the interface between helix G and the A–B loop, from a perspective of heme protein design and application, Ngb is a promising model protein. For example, H64Q Ngb can be a ligand-trap antidote for CO poisoning in practical medicine,54 and H64A Ngb is an artificial nitrite reductase with the fastest reported activity.55
With a motivation to design Ngb mutants with advanced properties and potential applications, we anticipate that if an intramolecular disulfide bond could be constructed between Cys120 and another Cys introduced nearby, Cys120 might be protected, and the protein stability could be enhanced as well. To this end, we inspected the X-ray structure of human Ngb (PDB code 4MPM48), and found that Ala15 was a potential target for introducing a Cys residue, which was further supported by molecular modeling studies. As shown in this study, we found that Cys15 could indeed form an intramolecular disulfide with Cys120, as confirmed by protein mass spectrometry analysis. The additional disulfide bond (C15–C120) in A15C Ngb mutant was found to enhance both chemical and pH stabilities compared to those of WT Ngb with a single disulfide bond (C46–C55). Moreover, the thermal stability of A15C Ngb was found to be more than 100 °C, suggesting a crucial role of C15–C120 in protein stabilization.
Results and discussion
Molecular modeling studies
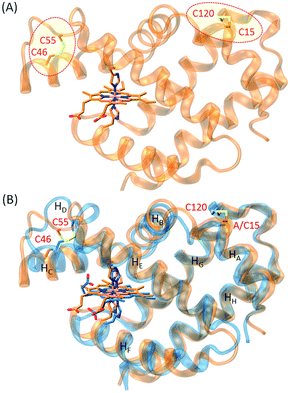
In order to construct an additional disulfide bond in human Ngb to protect Cys120 and stabilize the protein, we inspected the X-ray structure of human Ngb and found that Ala15 was a potential target for introducing a Cys residue. To reveal the possibility of formation of a disulfide bond between C15 and C120, we first performed a molecular modeling study for A15C Ngb with two intramolecular disulfide bonds. With minimization and equilibration, the simulated structure of A15C Ngb (Fig. 1A) showed that the two disulfide bonds could be well formed, with a disulfide bond distance of 2.11 Å (C15–C120) and 2.05 Å (C46–C55), respectively, in agreement with the X-ray structure of WT Ngb (2.02 Å for C46–C55).48 Moreover, as shown in Fig. 1B, the structure overlapped well with the X-ray structure of WT Ngb, which suggests that the formation an disulfide bond of C15–C120 causes minimal structural alterations.Spectroscopic studies
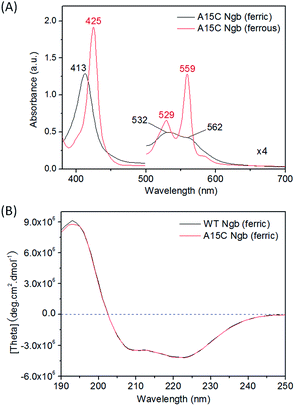
With support by molecular modeling, we introduced an additional Cys residue at position 15 in Ngb by A15C mutation. The A15C Ngb mutant was expressed and purified using the same procedure as that for WT Ngb. UV-Vis studies showed that A15C Ngb exhibits the spectra (ferric, 413, 532 and 562 nm; ferrous, 425, 529 and 559 nm, Fig. 2A) with the maximum absorbance identical to those of WT Ngb in both ferric and ferrous states (Fig. S1†).45,56 Moreover, A15C Ngb was found to exhibit the CD spectrum almost identical to that of WT Ngb (Fig. 2B). These observations suggest that introduction of Cys15 does not alter the heme coordination state and the protein secondary structure.To reveal whether Cys15 forms an intramolecular with Cys120 in A15C Ngb, we performed mass spectrometry studies using an electrospray ionization (ESI) technique, with WT Ngb as controls. As shown in Fig. 3A, the mass spectrum of WT Ngb exhibits a mass of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 931.5 Da, in agreement with previous observation for oxidized Ngb (16
931.5 Da, in agreement with previous observation for oxidized Ngb (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 930.9 Da),46 which corresponds to the apo-protein of WT Ngb with an intramolecular disulfide bond (Cys46–Cys55) (calculated, 16
930.9 Da),46 which corresponds to the apo-protein of WT Ngb with an intramolecular disulfide bond (Cys46–Cys55) (calculated, 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 931 Da). Note that the heme was dissociated under the detection conditions. Upon treatment by TCEP, a mass of 16
931 Da). Note that the heme was dissociated under the detection conditions. Upon treatment by TCEP, a mass of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 933.5 Da was observed, with an increase mass of 2 Da, which indicates the reduction of disulfide bond of Cys46–Cys55, as shown for reduced Ngb (16
933.5 Da was observed, with an increase mass of 2 Da, which indicates the reduction of disulfide bond of Cys46–Cys55, as shown for reduced Ngb (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 932.2 Da).46 In the mass spectrum of A15C Ngb (Fig. 3B), it was interesting to observe two mass peaks (16
932.2 Da).46 In the mass spectrum of A15C Ngb (Fig. 3B), it was interesting to observe two mass peaks (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 961.5 D and 17
961.5 D and 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 576.5 Da), which correspond to the calculated apo-protein of A15C Ngb with two intramolecular disulfide bonds (16
576.5 Da), which correspond to the calculated apo-protein of A15C Ngb with two intramolecular disulfide bonds (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 961 Da), and the holo-protein with heme not dissociated (17
961 Da), and the holo-protein with heme not dissociated (17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 576 Da), respectively. After treatment with TCEP, a single mass of 16
576 Da), respectively. After treatment with TCEP, a single mass of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 965.5 Da of the apo-protein was observed, with an increase mass of 4 Da, suggesting the reduction of two intramolecular disulfide bonds. The disappearance of mass peak for the holo-protein upon TCEP treatment also suggested that the heme dissociated from the protein matrix in mass determination, which, in turn, suggested that the formation of two intramolecular disulfide bonds, C46–C55 and C15–C120, may enhance the heme-binding stability of A15C Ngb under these conditions.
965.5 Da of the apo-protein was observed, with an increase mass of 4 Da, suggesting the reduction of two intramolecular disulfide bonds. The disappearance of mass peak for the holo-protein upon TCEP treatment also suggested that the heme dissociated from the protein matrix in mass determination, which, in turn, suggested that the formation of two intramolecular disulfide bonds, C46–C55 and C15–C120, may enhance the heme-binding stability of A15C Ngb under these conditions.
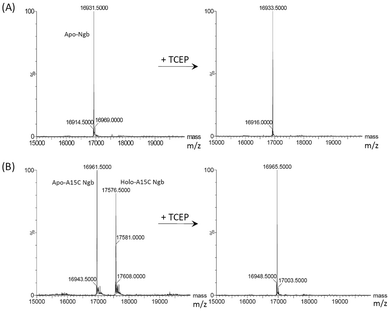 | ||
| Fig. 3 ESI-MS analysis of WT Ngb (A) and A15C Ngb (B) as purified (left) and treated with reducing agent TCEP (right). | ||
Chemical-induced unfolding studies
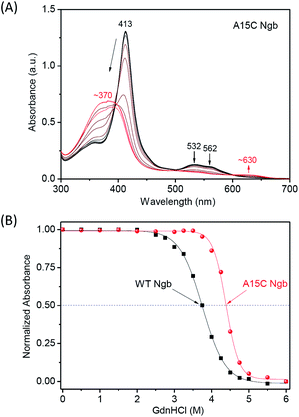
Since it is not possible to investigate A15C Ngb with one disulfide bond intact and the other cleaved, to probe the contribution of an additional disulfide bond of C15–C120 to the protein chemical stability, we first performed Gdn·HCl-induced unfolding studies for both A15C Ngb and WT Ngb. As shown in Fig. 4A and S2,† the intensity of Soret band (413 nm) of both proteins decreased with increasing the concentration of Gdn·HCl to 6 M, and resulted in a shoulder peak at ∼370 nm. Concurrently, the visible bands (532 nm and 562 nm) of bis-His coordinated heme decreased with an appearance of ∼630 nm. These observations suggest the release of heme from the heme pocket upon protein unfolding. Moreover, there was an isosbestic point observed in both UV-Vis spectroscopic changes, which indicates that the Gdn·HCl-induced unfolding was a two-state process for both proteins.To compare the half denaturation concentration (Cm), we plotted the spectral changes of the Soret band versus the concentration of Gdn·HCl and fitted to a normalized two-state model (Fig. 4B). The results showed that A15C Ngb exhibited a Cm value of 4.39 ± 0.01 M, which is ∼0.64 M higher than that of WT Ngb (3.75 ± 0.01 M). This observation suggest that the disulfide bond of C15–C120 in A15C Ngb enhances the protein chemical stability of Gdn·HCl-induced unfolding. Remarkably, the enhancement of ∼0.64 M is much larger than that of a disulfide bond constructed in other heme proteins. For example, we observed an increase of ∼0.32 M for V21C/V66C Mb, with a disulfide bond mimicking that in cytoglobin (Cgb), as compared to that of WT Mb upon Gdn·HCl-inducing unfolding.43 Fasan and co-workers showed that construction of an artificial thioether bond between a non-natural amino acid of O-2-bromoethyl Tyr and Cys in Mb may increase ∼0.24 M for Cm upon Gdn·HCl-induced unfolding.57 Moreover, Hollenberg and co-workers constructed a de novo disulfide bond close to the heme center in Cyt P450 2B1 by a double mutation of Y309C/S360C, which reduced the protein plasticity and fine-tuned the metabolic activity.58 These observations suggest that the location for engineering a covalent bond is crucial for the enhancement of stability, as well as functional regulation.
To further evaluate the chemical stability of A15C Ngb, we performed an acid titration experiment, and compared to that of WT Ngb under the same conditions. It was shown that by titration of concentrated HCl into the protein solution, the Soret band decreased and shifted to ∼370 nm, as well as for the visible bands (Fig. 5A and S3†). The spectral changes were similar to those occurred for Gdn·HCl-induced unfolding, with appearance of an isosbestic point, which indicates that acid-induced unfolding of both proteins was also a two-state process. This was further confirmed by the well-fitting of the plots of Soret band changes versus the pH values (Fig. 5B). The results showed that the half denaturation pH value was decreased by ∼0.69 pH unit for A15C Ngb (2.62 ± 0.02) compared to that of WT Ngb (3.31 ± 0.01), which indicates that the formation of an additional disulfide bond of C15–C120 increases the acid tolerance of A15C Ngb.
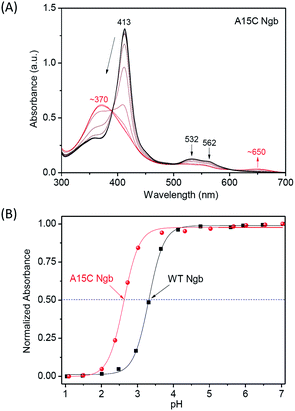 | ||
| Fig. 5 UV-Vis spectra of acid-induced unfolding of (A) A15C Ngb (10 μM). (B) Normalized absorbance of the Soret band versus pH values. | ||
Thermal-induced unfolding studies
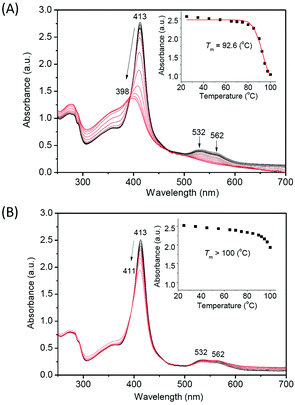
In addition to the protein chemical stability, we investigated the thermal stability of A15C Ngb, with WT Ngb as a control. We first performed the far-UV CD spectroscopy studies to monitor the changes in secondary structure upon increasing temperature. As shown in Fig. 6A and S4† with the temperature increasing from 25 °C to 100 °C, the negative Cotton effects at 208 nm and 222 nm gradually decreased, suggesting the unfolding of α-helix structure. When the temperature reached 100 °C, ∼50% α-helix was retained in A15C Ngb, while less than 25% was observed for WT Ngb. The plot of absorbance at 220 nm versus temperature showed that WT Ngb exhibits a half denaturation temperature (Tm) of 88.5 °C (Fig. 6B). Note that since A15C Ngb was only partially unfolded at 100 °C, Tm could not be determined and was estimated to be higher than 100 °C. These observations thus suggest that the second disulfide bond of C15–C120 stabilizes the secondary structure of A15C Ngb.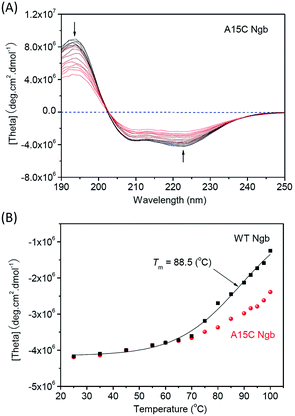 | ||
| Fig. 6 CD spectra of thermal-induced unfolding of (A) A15C Ngb (10 μM protein in 100 mM potassium phosphate buffer, pH 7.0). (B) Plots of absorbance at 222 nm versus temperature. | ||
Moreover, we monitored the UV-Vis spectral changes upon increasing temperature (Fig. 7), which provides valuable information for the heme binding and the overall protein structure. The results showed that with the temperature increasing from 25 °C to 100 °C, the Soret band of WT Ngb gradually decreased from 413 nm and blue shifted to ∼398 nm, with disappearance of visible bands at 532 nm and 562 nm (Fig. 7A). The isosbestic point also indicated that the thermal-induced unfolding of WT Ngb was a two-state process. The spectral changes were similar to those of Gdn·HCl or acid-induced unfolding, whereas the ultimate spectrum was different, with a Soret band of ∼398 nm instead of ∼370 nm of the free heme. These observations suggest that although the heme was released from the heme pocket at high temperature, it was still associated with the protein peptide. The plot of Soret band at 413 nm versus temperature showed that WT Ngb exhibits a half denaturation temperature (Tm) of 92.6 °C (Fig. 7A, inset). Note that this value is ∼4 °C higher than that determined by monitoring the changes in secondary structure, suggesting a more stability for the heme active center. In contrast to WT Ngb, A15C Ngb underwent only partial unfolding when the temperature was increased to 100 °C, with slight decreases in the intensity of Soret and visible bands (Fig. 7B). Therefore, the Tm was estimated to be higher than 100 °C, in agreement with the CD result. These results indicate that with an additional intramolecular disulfide bond of C15–C120, A15C Ngb exhibits extremely high thermal stability, and retains almost half α-helix structure at 100 °C, as well as most of the overall protein structure, where the heme was stabilized in the heme pocket.
Kinetic studies
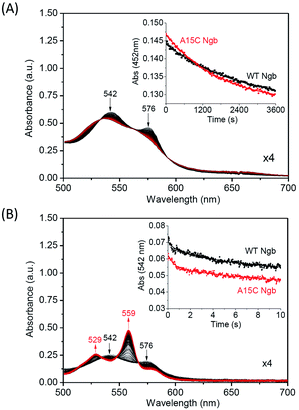
With an enhanced protein stability of A15C Ngb, we were further interested in testing the functional consequence of the formation of an additional disulfide bond. We first studied the autoxidation of A15C Ngb and compared to that of WT Ngb. As shown in Fig. 8A, the visible bands of the oxy-form (542 nm and 576 nm) decrease in intensity over time due to the autoxidation. The plot of the absorption change at 542 nm versus time obeys a single-exponential decay function (Fig. 8A, inset). The rate constant of autoxidation was determined to be 5.5 × 10−4 s−1 for A15C Ngb at pH 7.0, 25 °C, which is ∼1.38-fold higher than that for WT Ngb under the identical conditions (4.0 × 10−4 s−1). Note that these values (pH 7.0, 25 °C) are lower than that determined for WT Ngb in previous studies at higher pH value (2.8 × 10−3 s−1, pH 7.5, 25 °C)59 or higher temperature (3.8 × 10−3 s−1, pH 7.4, 37 °C).55Moreover, we determined the O2 release rate using stopped-flow spectroscopy upon reaction of oxy-Ngbs (5 μM) with sodium dithionite (1 mM) in 100 mM potassium phosphate buffer (pH 7.0) at 25 °C. As shown in Fig. 8B, the oxy-form (542 nm and 576 nm) of A15C Ngb was rapidly converted to the reduced form (529 nm and 559 nm). From the plot of the absorption change for the oxy-form at 542 nm versus time (Fig. 8B, inset), the rate constant of O2 release was calculated to be 0.32 ± 0.01 s−1, which is ∼1.39-fold higher than that for WT Ngb under the identical conditions (0.23 ± 0.01 s−1). These observations suggest that the formation of a second disulfide bond of C15–C120 in A15C Ngb slightly alters the protein function, with an increase in the rate of O2 release by ∼1.4-fold.
It was interesting to observe that A15C Ngb with a more stable structure releases O2 easier than that of WT Ngb. The explanation for this observation may include two aspects. (1) The de novo disulfide bond of C15–C120 is remote from the heme center, i.e., ∼23 Å according to the modeling structure (Fig. 1A). It may leads to long-range structural changes that impact on the heme iron coordination, as suggested by Astudillo et al. in study of C120S Ngb mutant;53 (2) the free C120 may interfere with the heme reduction process and thereby the O2 release. In a previous study, Hirota et al. introduced a cysteine to various sites on the surface of sperm whale Mb, and showed that the heme reduction rate constant decreased in general as the heme-cysteine distance was increased.60 Therefore, the formation of C15–C120 disulfide bond in A15C Ngb may eliminate such an effect. Note that although dithionite is commonly used to determine O2 release of heme proteins in vitro,39,61 for in vivo reduction of Ngb, many biological reductants could be involved, such as GSH and redox partners Cyt b5 and Cyt c.62,63
Conclusions
Ngb is a promising model protein for heme protein design, and in order to protect the free Cys120 in human Ngb and to enhance the protein stability, we rationally designed an additional disulfide of Cys15–Cys120 in Ngb by introduction of Cys15 in the vicinity of Cys120. As predicted by molecular modeling, Cys15 and Cys120 formed an intramolecular disulfide bond, which was confirmed experimentally by ESI-MS analysis, with WT Ngb as a control. The formation of Cys15–Cys120 disulfide bond was found to have minimal structural alterations of Ngb, meanwhile, it enhanced both chemical and pH stabilities with respect to those of WT Ngb with a single native disulfide bond of Cys46–Cys55. Thermal-induced unfolding studies revealed that A15C Ngb retained most of the overall protein structure as high as 100 °C, indicating an ultra-high thermal stability. In addition, the disulfide bond of Cys15–Cys120 remote from the heme center has only slight effects on protein function, such as an increase in the rate of O2 release by ∼1.4-fold. These results suggest that an additional disulfide can be rationally designed in Ngb, which plays a crucial role in protein stabilization. Moreover, this study lays the groundwork for further investigation of the structure and function of Ngb, as well as for the design of other functional heme proteins, based on the scaffold of A15C Ngb with an enhanced stability.Materials and methods
Protein preparation
The pET3a plasmid DNA containing the gene of human Ngb was a gift from Prof. T. Burmester, Gutenberg University of Mainz, Germany. Ngb was expressed in E. Coli. BL21(DE3) cells and purified as previously reported.45 The A15C Ngb gene was constructed using the QuikChange Site Directed Mutagenesis Kit (Stratagene) with two primers: forward,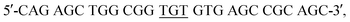 and reverse,
and reverse, 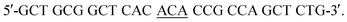 The mutation was confirmed by DNA sequencing assay, and the mutant was expressed and purified using the same procedure as that for WT Ngb.45
The mutation was confirmed by DNA sequencing assay, and the mutant was expressed and purified using the same procedure as that for WT Ngb.45
Molecular modeling studies
The initial structure of A15C Ngb was constructed based on the X-ray crystal structure of human Ngb (PDB code 4MPM, molecule A48) using program VMD 1.9. A patch of disulfide bond was applied to Cys46–Cys55 and Cys15–Cys120, respectively. The protein was solvated in a cubic box of TIP3 water, which extended 10 Å away from any given protein atom. Counter ions (Na+ and Cl−) were further added to obtain the physiological ionic strength of 0.15 M by using the autoionize plug-in of VMD 1.9.64 The resulting system was minimized with NAMD2.9 (Nanoscale Molecular Dynamics),65 using 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 minimization steps with conjugate gradient method at 0 K, and equilibrated for 15
000 minimization steps with conjugate gradient method at 0 K, and equilibrated for 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 molecular dynamics steps (1 fs per step, 15 ns in total) at 310 K, then further minimized for 50
000 molecular dynamics steps (1 fs per step, 15 ns in total) at 310 K, then further minimized for 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps at 0 K. Visualization and data analysis were done with VMD 1.9.
000 steps at 0 K. Visualization and data analysis were done with VMD 1.9.
UV-Vis and CD studies
UV-Vis spectra of A15C Ngb and WT Ngb were collected on an Agilent 8453 spectrometer in 100 mM potassium phosphate buffer (pH 7.0) at 25 °C. Circular dichroism (CD) spectra were collected on a Jasco 1500 spectropolarimeter equipped with a MCB-100 mini circulation bath (25 °C), using a cuvette with a path length of 0.1 cm and 1 cm for 190–250 nm and 250–700 nm, respectively. The protein concentrations of WT Ngb and A15C Ngb were determined with an extinction coefficient of ε413 nm = 129 mM−1 cm−1,49 and ε413 nm = 139 mM−1 cm−1, respectively, as calculated using the standard hemochromogen method.66Mass spectrometry
Protein mass spectrum was measured on G2-XS QToF mass spectrometer (Waters). The desalted protein solution was mixed with 50% acetonitrile solution (acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water, 1
water, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) containing 1% formic acid, and transferred into the mass spectrometer chamber for measurement by a direct injection method without performing the liquid chromatography under positive mode. The multiple m/z peaks were transformed to the protein molecular weight by using software MaxEnt1. For reduction studies, the reducing agent tris-(2-carboxyethyl)-phosphine (TCEP) was added to the protein solution at a final concentration of 10 mM, and the mixture was cultured at 37 °C for 30 min before mass measurements.
1) containing 1% formic acid, and transferred into the mass spectrometer chamber for measurement by a direct injection method without performing the liquid chromatography under positive mode. The multiple m/z peaks were transformed to the protein molecular weight by using software MaxEnt1. For reduction studies, the reducing agent tris-(2-carboxyethyl)-phosphine (TCEP) was added to the protein solution at a final concentration of 10 mM, and the mixture was cultured at 37 °C for 30 min before mass measurements.
Gdn·HCl-induced unfolding studies
Gdn·HCl-induced unfolding of A15C Ngb was studied by Agilent 8453 spectrometer. 10 μL of 2 mM protein sample was added into 2 mL of a guanidine hydrochloride (Gdn·HCl) solution (pH 7.0) with various concentrations (0–6.0 M). After incubation for 30 min, the UV-Vis spectrum was recorded. Control experiment was performed for WT Ngb under the same conditions. The unfolding transition was monitored at different Gdn·HCl concentrations through the changes of the absorbance of the Soret band. The denaturation midpoint (Cm) was calculated by fitting the absorbance of Soret band versus the concentration of Gdn·HCl to a two-state Boltzmann function (eqn (1)).| A = A2 + (A1 − A2)/(1 + e(C−Cm)/dC) | (1) |
Acid-induced unfolding studies
The acidic stability of A15C Ngb upon pH-induced unfolding was carried out on an Agilent 8453 spectrometer using a cell with a path length of 1 cm. The temperature was controlled at 25 °C with a circulating bath. The protein sample was titrated with increasing amounts of 10 M HCl to adjust the pH values. After incubation at different pH values for 10 min, the UV-Vis spectrum was recorded. Control experiment was performed for WT Ngb under the same conditions. The denaturation midpoint was calculated by fitting the absorbance of Soret band versus the pH values to a two-state Boltzmann function (eqn (2)).| A = A2 + (A1 − A2)/(1 + e(X−X0)/dX) | (2) |
Thermal-induced unfolding studies
Thermal-induced unfolding studies of WT Ngb and A15C Ngb were performed with a Jasco 1500 spectropolarimeter equipped with a MCB-100 minicirculation bath. The protein was dissolved in potassium phosphate buffer (100 mM, pH 7.0) using a cuvette with a path length of 0.1 cm and 1 cm with their corresponding covers. The temperature was increased stepwise over the range of 25–100 °C. At each temperature, the protein sample was allowed to equilibrate for 10 min before collecting the CD spectrum (190–250 nm) at a speed of 100 nm min−1 for five times, and the UV-Vis spectrum (250–700 nm) at a speed of 200 nm min−1 for six times. The denaturation midpoint (Tm) was calculated by fitting the ellipticity at 222 nm or the absorbance of Soret band versus the temperature to a two-state Boltzmann function (eqn (3)).| A = A2 + (A1 − A2)/(1 + e(T−Tm)/dT) | (3) |
Kinetic studies
The autoxidation rates of oxy-Ngb and oxy-A15C Ngb were determined using an Agilent 8453 spectrometer. The ferric protein was reduced by excess dithionite, which was removed from the solution by a PD-10 column (GE Healthcare). The reduced protein was then diluted into O2-saturated 100 mM potassium phosphate (pH 7.0) to generate the oxy-form protein. The changes in intensity of the oxy-form band at 542 nm were monitored for 60 min at 25 °C, and fitted to a single-exponential decay function. The O2 dissociation rates of oxy-Ngb and oxy-A15C Ngb were determined using a dual mixing stopped-flow spectrophotometer (SF-61DX2 Hi-Tech KinetAsyst™). The prepared oxy-Ngbs (10 μM protein in 100 mM potassium phosphate, pH 7.0) was mixed with an equal volume of 1 mM sodium dithionite. The changes in intensity of the oxy-form band at 542 nm were monitored for 10 s at 25 °C, and fitted to a single-exponential decay function to obtain the observed rate constant (kobs, s−1).Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully thank Prof. T. Burmester of the Gutenberg University of Mainz, Germany, for graciously providing the gene of human Ngb. This work was supported by the National Natural Science Foundation of China, NSFC (31370812; 21701081) and Hunan province college students research learning and innovative experiment project (136).References
- Y. Lu, N. Yeung, N. Sieracki and N. M. Marshall, Nature, 2009, 460, 855–862 CrossRef CAS PubMed.
- J. Du, M. Sono and J. H. Dawson, Coord. Chem. Rev., 2011, 255, 700–716 CrossRef CAS PubMed.
- U. Liebl, J.-C. Lambry and M. H. Vos, Biochim. Biophys. Acta, 2013, 1834, 1684–1692 CrossRef CAS PubMed.
- T. L. Poulos, Chem. Rev., 2014, 114, 3919–3962 CrossRef CAS PubMed.
- J. Liu, S. Chakraborty, P. Hosseinzadeh, Y. Yu, S. Tian, I. Petrik, A. Bhagi and Y. Lu, Chem. Rev., 2014, 114, 4366–4469 CrossRef CAS PubMed.
- P. C. E. Moody and E. L. Raven, Acc. Chem. Res., 2018, 51, 427–435 CrossRef CAS PubMed.
- Y.-W. Lin, E. B. Sawyer and J. Wang, Chem.–Asian J., 2013, 8, 2534–2544 CrossRef CAS PubMed.
- I. D. Petrik, J. Liu and Y. Lu, Curr. Opin. Chem. Biol., 2014, 19, 67–75 CrossRef CAS PubMed.
- K. Oohora and T. Hayashi, Curr. Opin. Chem. Biol., 2014, 19, 154–161 CrossRef CAS PubMed.
- O. Shoji and Y. Watanabe, J. Biol. Inorg Chem., 2014, 19, 529–539 CrossRef CAS PubMed.
- F. Nastri, M. Chino, O. Maglio, A. Bhagi-Damodaran, Y. Lu and A. Lombardi, Chem. Soc. Rev., 2016, 45, 5020–5054 RSC.
- Y.-W. Lin, Coord. Chem. Rev., 2017, 336, 1–27 CrossRef CAS.
- S. Hirota and Y. W. Lin, J. Biol. Inorg Chem., 2018, 23, 7–25 CrossRef CAS PubMed.
- Y. Yu, C. Hu, L. Xia and J. Wang, ACS Catal., 2018, 8, 1851–1863 CrossRef CAS.
- F. Schwizer, Y. Okamoto, T. Heinisch, Y. Gu, M. M. Pellizzoni, V. Lebrun, R. Reuter, V. Kohler, J. C. Lewis and T. R. Ward, Chem. Rev., 2018, 118, 142–231 CrossRef CAS PubMed.
- E. N. Mirts, I. D. Petrik, P. Hosseinzadeh, M. J. Nilges and Y. Lu, Science, 2018, 361, 1098–1101 CrossRef CAS PubMed.
- L.-L. Yin, H. Yuan, C. Liu, B. He, S.-Q. Gao, G.-B. Wen, X. Tan and Y.-W. Lin, ACS Catal., 2018, 8, 9619–9624 CrossRef CAS.
- M. Lehmann and M. Wyss, Curr. Opin. Biotechnol., 2001, 12, 371–375 CrossRef CAS PubMed.
- A. Gershenson and F. H. Arnold, Genet. Eng., 2000, 22, 55–76 CAS.
- T. J. Magliery, Curr. Opin. Struct. Biol., 2015, 33, 161–168 CrossRef CAS PubMed.
- L. J. Perry and R. Wetzel, Science, 1984, 226, 555 CrossRef CAS PubMed.
- R. Wetzel, L. J. Perry, W. A. Baase and W. J. Becktel, Proc. Natl. Acad. Sci. U. S. A., 1988, 85, 401–405 CrossRef CAS.
- M. Matsumura, G. Signor and B. W. Matthews, Nature, 1989, 342, 291–293 CrossRef CAS PubMed.
- J. M. Chalker, G. J. Bernardes, Y. A. Lin and B. G. Davis, Chem.–Asian J., 2009, 4, 630–640 CrossRef CAS PubMed.
- J. Lee and M. Blaber, J. Mol. Biol., 2009, 393, 128–139 CrossRef CAS PubMed.
- A. A. Dombkowski, K. Z. Sultana and D. B. Craig, FEBS Lett., 2014, 588, 206–212 CrossRef CAS PubMed.
- T. Liu, Y. Wang, X. Luo, J. Li, S. A. Reed, H. Xiao, T. S. Young and P. G. Schultz, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 5910–5915 CrossRef CAS PubMed.
- C. Colas and P. R. Ortiz de Montellano, Chem. Rev., 2003, 103, 2305–2332 CrossRef CAS PubMed.
- Y.-W. Lin, Biochim. Biophys. Acta, 2015, 1854, 844–859 CrossRef CAS PubMed.
- Y.-W. Lin, Arch. Biochem. Biophys., 2018, 641, 1–30 CrossRef CAS PubMed.
- P. D. Barker, J. C. Ferrer, M. Mylrajan, T. M. Loehr, R. Feng, Y. Konishi, W. D. Funk, R. T. MacGillivray and A. G. Mauk, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 6542–6546 CrossRef CAS.
- Y.-W. Lin, W.-H. Wang, Q. Zhang, H.-J. Lu, P.-Y. Yang, Y. Xie, Z.-X. Huang and H.-M. Wu, ChemBioChem, 2005, 6, 1356–1359 CrossRef CAS PubMed.
- S. Hu, B. He, K.-J. Du, X.-J. Wang, S.-Q. Gao and Y.-W. Lin, ChemistryOpen, 2017, 6, 325–330 CrossRef CAS PubMed.
- P. D. Barker, E. P. Nerou, S. M. V. Freund and I. M. Fearnley, Biochemistry, 1995, 34, 15191–15203 CrossRef CAS PubMed.
- C. L. Metcalfe, O. Daltrop, S. J. Ferguson and E. L. Raven, Biochem. J., 2007, 408, 355–361 CrossRef CAS PubMed.
- H.-M. Cheng, H. Yuan, X.-J. Wang, J.-K. Xu, S.-Q. Gao, G.-B. Wen, X. Tan and Y.-W. Lin, J. Inorg. Biochem., 2018, 182, 141–149 CrossRef CAS PubMed.
- C. L. Metcalfe, M. Ott, N. Patel, K. Singh, S. C. Mistry, H. M. Goff and E. L. Raven, J. Am. Chem. Soc., 2004, 126, 16242–16248 CrossRef CAS PubMed.
- S. L. Rice, M. R. Preimesberger, E. A. Johnson and J. T. Lecomte, J. Inorg. Biochem., 2014, 141, 198–207 CrossRef CAS PubMed.
- D.-J. Yan, W. Li, Y. Xiang, G.-B. Wen, Y.-W. Lin and X. Tan, ChemBioChem, 2015, 16, 47–50 CrossRef CAS PubMed.
- D.-J. Yan, H. Yuan, W. Li, Y. Xiang, B. He, C.-M. Nie, G.-B. Wen, Y.-W. Lin and X. Tan, Dalton Trans., 2015, 44, 18815–18822 RSC.
- T. Uchida, M. Unno, K. Ishimori and I. Morishima, Biochemistry, 1997, 36, 324–332 CrossRef CAS PubMed.
- L.-B. Wu, H. Yuan, H. Zhou, S.-Q. Gao, C.-M. Nie, X. Tan, G.-B. Wen and Y.-W. Lin, Arch. Biochem. Biophys., 2016, 600, 47–55 CrossRef CAS PubMed.
- L.-L. Yin, H. Yuan, K.-J. Du, B. He, S.-Q. Gao, G.-B. Wen, X. Tan and Y.-W. Lin, Chem. Commun., 2018, 54, 4356–4359 RSC.
- T. Burmester, B. Weich, S. Reinhardt and T. Hankeln, Nature, 2000, 407, 520–523 CrossRef CAS PubMed.
- S. Dewilde, L. Kiger, T. Burmester, T. Hankeln, V. Baudin-Creuza, T. Aerts, M. C. Marden, R. Caubergs and L. Moens, J. Biol. Chem., 2001, 276, 38949–38955 CrossRef CAS PubMed.
- D. Hamdane, L. Kiger, S. Dewilde, B. N. Green, A. Pesce, J. Uzan, T. Burmester, T. Hankeln, M. Bolognesi, L. Moens and M. C. Marden, J. Biol. Chem., 2003, 278, 51713–51721 CrossRef CAS PubMed.
- L. Astudillo, S. Bernad, V. Derrien, P. Sebban and J. Miksovska, Biophys. J., 2010, 99, L16–L18 CrossRef CAS PubMed.
- B. G. Guimaraes, D. Hamdane, C. Lechauve, M. C. Marden and B. Golinelli-Pimpaneau, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2014, 70, 1005–1014 CrossRef CAS PubMed.
- S. Nicolis, E. Monzani, C. Ciaccio, P. Ascenzi, L. Moens and L. Casella, Biochem. J., 2007, 407, 89–99 CrossRef CAS PubMed.
- M. Tiso, J. Tejero, S. Basu, I. Azarov, X. Wang, V. Simplaceanu, S. Frizzell, T. Jayaraman, L. Geary, C. Shapiro, C. Ho, S. Shiva, D. B. Kim-Shapiro and M. T. Gladwin, J. Biol. Chem., 2011, 286, 18277–18289 CrossRef CAS PubMed.
- A. Bocahut, V. Derrien, S. Bernad, P. Sebban, S. Sacquin-Mora, E. Guittet and E. Lescop, J. Biol. Inorg Chem., 2013, 18, 111–122 CrossRef CAS PubMed.
- S. Trashin, M. de Jong, E. Luyckx, S. Dewilde and K. De Wael, J. Biol. Chem., 2016, 291, 18959–18966 CrossRef CAS PubMed.
- L. Astudillo, S. Bernad, V. Derrien, P. Sebban and J. Miksovska, Biochemistry, 2012, 51, 9984–9994 CrossRef CAS PubMed.
- I. Azarov, L. Wang, J. J. Rose, Q. Xu, X. N. Huang, A. Belanger, Y. Wang, L. Guo, C. Liu, K. B. Ucer, C. F. McTiernan, C. P. O'Donnell, S. Shiva, J. Tejero, D. B. Kim-Shapiro and M. T. Gladwin, Sci. Transl. Med., 2016, 8, 368ra173 CrossRef PubMed.
- J. Tejero, C. E. Sparacino-Watkins, V. Ragireddy, S. Frizzell and M. T. Gladwin, Biochemistry, 2015, 54, 722–733 CrossRef CAS PubMed.
- H.-X. Liu, L. Li, B. He, S.-Q. Gao, G.-B. Wen and Y.-W. Lin, Dalton Trans., 2018, 47, 10847–10852 RSC.
- E. J. Moore, D. Zorine, W. A. Hansen, S. D. Khare and R. Fasan, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 12472–12477 CrossRef CAS PubMed.
- H. Zhang, C. Kenaan, D. Hamdane, G. H. Hoa and P. F. Hollenberg, J. Biol. Chem., 2009, 284, 25678–25686 CrossRef CAS PubMed.
- A. Fago, C. Hundahl, S. Dewilde, K. Gilany, L. Moens and R. E. Weber, J. Biol. Chem., 2004, 279, 44417–44426 CrossRef CAS PubMed.
- S. Hirota, K. Azuma, M. Fukuba, S. Kuroiwa and N. Funasaki, Biochemistry, 2005, 44, 10322–10327 CrossRef CAS PubMed.
- S. Nagao, H. Osuka, T. Yamada, T. Uni, Y. Shomura, K. Imai, Y. Higuchi and S. Hirota, Dalton Trans., 2012, 41, 11378–11385 RSC.
- P. Ascenzi, A. di Masi, L. Leboffe, M. Fiocchetti, M. T. Nuzzo, M. Brunori and M. Marino, Mol. Aspects Med., 2016, 52, 1–48 CrossRef CAS PubMed.
- M. Fiocchetti, M. Cipolletti, V. Brandi, F. Polticelli and P. Ascenzi, J. Mol. Recognit., 2017, e2654 CrossRef PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14(33–38), 27–38 Search PubMed.
- L. Kalé, R. Skeel, M. Bhandarkar, R. Brunner, A. Gursoy, N. Krawetz, J. Phillips, A. Shinozaki, K. Varadarajan and K. Schulten, J. Comput. Phys., 1999, 151, 283–312 CrossRef.
- M. Morrison and S. Horie, Anal. Biochem., 1965, 12, 77–82 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Additional UV-Vis, CD and stopped-flow spectra. See DOI: 10.1039/c8ra10390a |
| This journal is © The Royal Society of Chemistry 2019 |