Open Access Article
Open Access ArticlemiR-425 suppresses EMT and the development of TNBC (triple-negative breast cancer) by targeting the TGF-β 1/SMAD 3 signaling pathway
Yingping Liua and
Jinglong Chen *b
*b
aDepartment of Obstetrics and Gynecology, Beijing Obstetrics and Gynecology Hospital, Capital Medical University, 251# Yaojia Yuan Road, Chaoyang District, Beijing 100026, P. R. China
bDepartment of Oncology, Beijing Ditan Hospital, Capital Medical University, Beijing 100015, PR China. E-mail: dtzlzx@sina.com
First published on 21st December 2018
Abstract
Background: EMT has a crucial effect on the progression and metastasis of tumors. This work will elucidate the role of miR-425 in EMT and the development of TNBC. Methods: The differential miRNA expression among non-tumor, para-tumor (adjacent tissue of tumor) and tumor tissues was analyzed. The luciferase activities of TGF-β1 3′UTR treated with miR-425 were determined. Then human breast cancer cell lines were treated with mimics or inhibitors of miR-425, and then the cell proliferation and migration, and invasion ability were assessed. The expression of TGF-β1 and markers of epithelial cells and mesenchymal cells were analyzed. The influences of miR-425 on the development of TNBC through inducing EMT by targeting the TGF-β1/SMAD3 signaling pathway in TNBC cell lines were investigated. Furthermore, xenograft mice were used to explore the potential roles of miR-425 on EMT and the development of TNBC in vivo. Results: Compared with non-tumor tissues, 9 miRNAs were upregulated and 3 miRNAs were down-regulated in tumor tissues. The relative expression of miR-425 in tumor tissues was obviously much lower than that in para-tumor and non-tumor tissues. MiR-425 suppressed TGF-β1 expression, and further inhibited expression of mesenchymal cell markers, while it exerted effects on cell proliferation and migration of TNBC cell lines. Moreover, the agomir of miR-425 could protect against the development process in a murine TNBC xenograft model. Conclusions: Our results demonstrated that miR-425 targets TGF-β1, and was a crucial suppressor on EMT and the development of TNBC through inhibiting the TGF-β1/SMAD3 signaling pathway. This suggests that aiming at the TGF-β1/SMAD3 signaling pathway by enhancing relative miR-425 expression, is a feasible therapy strategy for TNBC.
Introduction
The most usual cancer of women is BC (Breast Cancer) worldwide.1 Moreover, the morbidity of BC is the seventh among all cancers, and its mortality rate is third in women's world.2 According to different prognoses and options of treatment, BC is divided into several subtypes of disease associated with heterogeneity.3,4 Based on the expression of ERBB2/HER2 (epidermal growth factor receptor 2), PR (progesterone receptor), and ER (estrogen receptor), patients with BC are classified in three subtypes of triple-negative, HER2+, and ER+/PR+ in the clinic. The overall prognosis of patients with Her2+ or ER+/PR+ subtypes has been dramatically improved by treatment with targeted therapy at HER2 with hormone. However, the aggressiveness of TNBC (triple-negative breast cancer) and the lack of effective targeted therapy cause the worst overall survival rate of patients with TNBC.4 About 20% patients with BC have TNBC, and they have high risk of visceral organ metastasis, such as brain, liver, and lung, etc.5 Therefore, there is an urgent need to develop new therapeutic interventions for preventing and treating metastasis of TNBC.EMT (epithelial to mesenchymal transition) plays an important role in metastasis and progression of cancer. During this process, the invasion and migration abilities increase, epithelial polarity vanishes away together with epithelial markers down-expression and mesenchymal markers over-expression.6–13 Some transcription factors regulate the process of EMT, including Zeb1, Slug, Snail, TGFβ, and Twist.9,13 The biological process of EMT is complex, including loss of epithelial characteristics and acquire mesenchymal phenotypes. Moreover, the expressions of mesenchymal cell markers, such as Smad 3, α-SMA (alpha smooth muscle actin), etc., enhanced.14,15 The growth of tumor relies not only on tumor malignancy but also the relationship between stromal and tumor cells.14,15 The system of tumor vessel includes pericytes, smooth muscle cells and, epithelial cells (ECs), which plays an important role in tumors progression and angiogenesis activation, such as BC.16,17 The EMT process of ECs is conduced to the progression of cancer, that has been identified by recent researches.18–20 In this process, the properties of migration or invasion and markers of mesenchyma (e.g., α-SMA, FSP-1) are obtained, the epithelial markers (e.g., CD31, VE-cadherin) of ECs and junctions of cell–cell were losed.21 These investigations confirm that all kinds of pathological processes occurs with, for example, cancer.18,19,22 Therefore, inhibition of EMT process may be a great treatment strategy for tumor.
Lately, some scholars have revealed that TGF-β1 is a cytokine with multifunction, and plays a crucial role in pathophysiology of BC.23 The canonical and noncanonical TGF-β1 signaling pathways gene mutations encoding TGF-β1 or the change of TGF-β1 receptors associates with BC oncogenic activity. The progression of epithelial cell cycle of BC is inhibited by TGF-β1 at early phase while apoptosis is promoted, which indicates its suppressive effects on tumor.24 However, in the late phase, TGF-β1 promotes tumor progression, is associated with metastasis or invasiveness of tumor and higher motility of cancer cells. In addition, TGF-β1 induces EMT, and is related to modification of microenvironment in cancer.25 The well known “TGF-β1 paradox” is called to describe this phenomenon, which suggests TGF-β1 can be used for the novel therapy approaches on BC with better understanding its functions.26 By protecting against TGF-β1, several drugs for nonclinical and clinical investigation in early stages are developed based on these knowledge.
The current realization demonstrated that mesenchymal characteristics of TNBC cells with paclitaxel-resistance, and these cells has higher mesenchymal markers expression, including N-cadherin and vimentin. Thus, through inducing inhibition of TGF-β signaling pathway can promote chemotherapeutic effect on anti-tumor and reduce metastasis after paclitaxel therapy with kinase inhibitor for TGF-β1 receptor.27 Therefore, it indicates that the novel potential strategy for TNBC therapy through combination with kinase inhibitors of TGF-β and its receptor.
Many data confirmed that microRNAs have crucial effect on activity of epithelial or EMT.28,29 So, in our investigation, the relationship between down-expression of miR-425 and TNBC was identified by comparing miRNA expression profiles from breast cancer tissues of patients with TNBC. Over-expression of miR-425 played an important regulator in negative feedback for TGF-β signaling and canonical pathway through suppressing expression of TGF-β1 and its changes associated with EMT in development of TNBC.
Materials and methods
Specimens of patients
The tissues of patients with TNBC were obtained from January 2014 to December 2017. All these patients were performed with mastectomy for BC, and did not receive any treatment before hospital. At the operation day, samples of tumor tissues, para-tumor tissues and non-tumor tissues were collected and handled immediately, respectively. The para-tumor is adjacent tissue of tumor.This investigation and the utilization of human specimens in this work had been approved by the Ethics Committee of our hospitals (Beijing Obstetrics and Gynecology Hospital, Capital Medical University, Beijing, China) on the basis of the Declaration of Helsinki. We clearly confirmed that the informed consents were obtained from all patients. Moreover, the recorded and documented of participant consents were kept in our hospital.
Extraction of miRNAs and next-gener. sequencing
The miRNA-sequencing test was conducted after extraction of RNAs with TRIzol method and miRNeasy Mini Kit (Cat. no. 74104; Invitrogen Thermo Fisher Scientific, Inc., Waltham, MA, USA). Furthermore, total RNA was harvested from tissues for RT-qPCR assay with mirVana™ PARIS™ Kit (Cat. no. AM1556; Invitrogen Thermo Fisher Scientific, Inc, Waltham, MA, USA). Hybridization and ligation were conducted using Adaptor Mix. Moreover, microRNA sequencing RNAs using Illumina HiSeq 2000 platform was conducted after reversely transcribe of these RNAs.Data extraction and analysis
All the mature miRNAs sequences were identified through library preparation of reference sequences with miRBase (version 19.0) (http://www.mirbase.org/).30 Given that the length of minimum read was 50 nucleotides, specific adapters were used to anneal to their 3′ ends and extended the entire small RNAs during the library preparation. Thus, we removed the adapters and obtained the potential miRNAs with the length of 15–30 nucleotides for further analysis using cut adapt software in silico.31 Subsequently, the sequences were perfectly matched with the prepared reference library with Bowtie (version 0.12.7) (https://sourceforge.net/projects/bowtie-bio/files/bowtie/0.12.7/).32 Then the numbers of mapped reads were counted for every miRNA. Acquired data of all samples were normalized with RPM (reads per million).33RNA extraction
TRIzol method was used to extract total RNA from tissues based on the operation manual. Then, smaller RNA (<200 nt) was removed with mirVana RNA isolation kit (Cat. no. AM27828; Invitrogen Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the instructions for manufacturer.Culture of cell
The cell lines of human TNBC, including MDA-MB-231 (ATCC® HTB-26™) and Hs 578T (ATCC® HTB-126™) were purchased from ATCC (American Type Culture Collection), and then cultured in DMEM with 10% FBS accompanied by Streptomycin and Penicillin (bi-antibiotics) with atmosphere (5% CO2) at 37 °C.siRNA for TGF-β1
The well known siRNA of TGF-β1 supported by ref. 26 had been used in our study, which had been used as a positive in our study. The siRNA duplexes for TGF-β1 sequences are 5′-GAAGCGGACUACUAUGCUAUU-3′ (upstream) and 5′-UUCUUCGCCUGAUGAUACGAU-3′ (downstream).qRT-PCR
The miR-425 expression level was assessed with TaqMan miRNA assays in tissues. The data analysis was performed with 2−ΔΔCq method (Livak 2011). The qRT-PCR assay was conducted using SYBR Green kit (Cat. no. QR0100-1KT; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). The thermocycling conditions for qPcR were as follows: pre-enaturation at 95 °C for 30 s, followed by 40 cycles of denaturation at 95 °C for 5 s and extension at 60 °C for 30 s.The primer sequences of miR-425 were 5′-ACACTCCAGCTGGGAATGACACGATCACTCC-3′ (upstream) and 5′-TGGTGTCGTGGAGTCG-3′ (downstream), respectively.34 Moreover, the primer sequences of U6 were 5′-TGACCTGAAACATACACGGGA-3′ (upstream) and 5′-TATCGTTGTACTCCACTCCTTGAC-3′ (downstream), respectively.
The primer sequences of TGF-β1 were 5′-CAACACGATGCTTGAAGGTAACG-3′ (upstream) and 5′-TCCAGAGAGATGATTGCCGAGG-3′ (downstream), respectively. The primer sequences of VE-cadherin were 5′-GGATGCAGAGGCTCACAGAG-3′ (upstream) and 5′-CTGGCGGTTCACGTTGGACT-3′ (downstream), respectively. The primer sequences of E-cadherin were 5′-TACACTGCCCAGGAGCCAGA-3′ (upstream) and 5′-TGGCACCAGTGTCCGGATTA-3′ (downstream), respectively. The primer sequences of N-cadherin were 5′-GGACAGTTCCTGAGGGATCA-3′ (upstream) and 5′-GGATTGCCTTCCATGTCTGT-3′ (downstream), respectively. The primer sequences of vimentin were 5′-GGCTCAGATTCAGGAACAGC-3′ (upstream) and 5′-GCTTCAACGGCAAAGTTCTC-3′ (downstream), respectively. The primer sequences of α-SMA were 5′-TGTTCCAGCCGTCCTTCAT-3′ (upstream) and 5′-GGCGTAGTCTTTCCTGATG-3′ (downstream), respectively. The primer sequences of SMAD3 were 5′-AGCACACAATAACTTGGACC-3′ (upstream) and 5′-TAAGACACACTGGAACAGCGGATG-3′ (downstream), respectively. Moreover, the primer sequences of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were 5′-CTCATGACCACAGTCCATGCC-3′ (upstream) and 5′-GGCATGGACTGTGGTCATGAG-3′ (downstream), respectively.
Reporter gene assays
The online software Targetscan (version 7.2) (http://www.targetscan.org/vert_72/) was firstly used to predict the microRNA which could regulate the expression of TGF-β1. Therefore, miR-425 was acquired from the predict result. Furthermore, the 3′UTR mRNAs sequence of TGF-β1 was cloned into pmirGLO vector with luciferase reporter firefly and renilla. The control was used with reverse orientation of TGF-β1 3′UTRs mRNA. The complementary region sequence in TGF-β1 3′UTR was GGACUGCGGAUCUCUGUGUCAUU (Fig. 1), which located at position 151–157, and corresponding miR-425 seed sequence AGUUGCCCUCACUAGCACAGUAA. Site-Mutation kit (Cat. no. E0554S; New England Biolabs Ltd, UK) was used to constructed TGF-β1 3′UTR mutated vector. Finally, Dual-Glo® Luciferase Assay System (Cat. no. E2940; Promega Corporation, Madison, WI, USA) was utilized to analyze the activity of luciferase with analyzer VICTOR.The mimics of miR-425 and its inhibitors
The mimics of miR-425 and its inhibitors were transfected with Lipofectamine 3000. Moreover, the mimics sequences of miR-425 were 5′-AAUGACACGAUCACUCCCGUUGA-3′.34 Moreover, the control sequence of miR-425 mimics were 5′-UUCUCCGAACGUGUCACGUTT-3’. The sequence of miR-425 agomir was 5′-aSaSugacacgaucacucccguSuSgSaS-3′. The sequence of NC (negative control) was 5′-uSuScuccgaacgugucacgSuStStS-3′, while it was not consistent with sequences of human genome.Western blot
A lysis buffer (1% NP-40, 0.1% SDS, 150 mM NaCl, 0.02% NaN3, and 50 mM Tris pH 8.0) containing cocktail of inhibitor for protease and phenylmethylsulfonyl fluoride (1 mM) was used to lysate cells. Western blotting assays were performed with BioRad Protean II minigel system. Then, 50 μg protein sample was injected in every gel (12%) well, and then shifted to PVDF membrane after electrophoresis. 1× TBST (5% dry skimmed milk) was utilized to dispute the non-specific binding. Then these membranes were sequentially dealt with primary and secondary antibody, respectively. The membranes were incubated with primary antibodies at 4 °C overnight. The membranes were incubated with secondary antibodies with secondary antibodies goat anti-rabbit immunoglobulin G (IgG)-horseradish peroxidase (HRP) (sc-2004; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000 dilution; Santa Cruz Biotechnology, Inc.) or goat anti-mouse IgG-HRP (sc-2005; 1
5000 dilution; Santa Cruz Biotechnology, Inc.) or goat anti-mouse IgG-HRP (sc-2005; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000 dilution; Santa Cruz Biotechnology, Inc.) for 1 h at room temperature. Furthermore, these membranes were determined with enhanced ECL (chemiluminescence) reagent of Hyperfilm ECL kit (Cat. no. 28906835; Amersham GE Healthcare, Chicago, IL, USA), and exposed using X-ray film.
5000 dilution; Santa Cruz Biotechnology, Inc.) for 1 h at room temperature. Furthermore, these membranes were determined with enhanced ECL (chemiluminescence) reagent of Hyperfilm ECL kit (Cat. no. 28906835; Amersham GE Healthcare, Chicago, IL, USA), and exposed using X-ray film.
The primary antibodies included TGF-β1 (Santa Cruz, CA, USA; sc-130348, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1200), α-SMA (CST 19245, dilution 1
1200), α-SMA (CST 19245, dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1200) and SMAD 3 (C67H9) (CST 9523, dilution 1
1200) and SMAD 3 (C67H9) (CST 9523, dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1500), p-SMAD 3 (CST-9520, dilution 1
1500), p-SMAD 3 (CST-9520, dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), VE-cadherin (CST2158, dilution 1
1000), VE-cadherin (CST2158, dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), E-cadherin (24 × 1019) (CST 3195, dilution 1
1000), E-cadherin (24 × 1019) (CST 3195, dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800), N-cadherin (D4R1H) (CST 13116, dilution 1
800), N-cadherin (D4R1H) (CST 13116, dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1200), vimentin (D21H3) (CST 5741, dilution 1
1200), vimentin (D21H3) (CST 5741, dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), β-actin (13 × 105) (CST 4970S, dilution 1
1000), β-actin (13 × 105) (CST 4970S, dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4000) (Cell Signaling, Danvers, MA, USA).
4000) (Cell Signaling, Danvers, MA, USA).
Analysis of cell proliferation and migration
The proliferation ability of cells was assessed using MTS kit (Cat. no. G3580; Promega Corporation, Madison, WI, USA). Subculture the stock cultures of cells to 2 × 104 cells per ml in medium containing 50 μM 2-mercaptoethanol (2-ME), when a density of 2 × 105 cells per ml is reached, mimics or inhibitors of miR-425 and its control is added, respectively. Wash the cells twice with the above medium centrifugation at 300×g for 5 minutes. The number and viability of cells based on trypan blue stain, 50 μl resuspend cells (1 × 105 cells per ml), was added into all wells of 96-well plate prepared, then incubate at 37 °C for 48 h. Furthermore, 20 μl CellTiter 96® AQueous One Solution Reagent was added into per well, and then incubate at 37 °C for 2 h. Record the absorbance at 490 nm using a 96-well plate reader.The ability of cells migration was detected using wound scratch assay using CytoSelect™ 24-Well Wound Healing Assay kit (Cat. no. CBA-120-5; Cell Biolabs, Inc., San Diego, CA, USA). At first, 24-well plate with CytoSelect™ Wound Healing Inserts was warm up for 10 minutes at room temperature, then sterile forceps was used to orient the inserts direction. Secondly, 500 μl cell suspension (1 × 106 cells per ml) was added into each well. Then cultured in incubator overnight. The next day, the inserts were removed from wells, then slowly discarded the media. After washing, dead cells and debris in wells were further removed. Finally, hydrated cells were visualized with light microscope. The migration rate of the cells into the wound field were measured. Ten fields per experimental condition were randomly selected and micrographed with IX71 microscope (Olympus, Japan) to quantify cells of migration. Images represent at least three independent experiments.
Matrigel invasion assay
Matrigel was dissolved overnight at 4 °C and then diluted with serum-free DMEM (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3). The diluted Matrigel was added to the upper chamber of the Transwell to form the membrane in three additions (15, 7.5 and 7.5 μl), with 10 min between each addition. The Matrigel was evenly spread and covered all micropores at the bottom of the upper chamber. The two cell lines were collected at 48 h after transfection to prepare cell suspensions. Cell suspensions were seeded into the upper well, and 0.5 ml of DMEM containing 10% FBS was added to the lower chambers (24-well plates). The cells were cultured routinely for 48 h at 37 °C with 5% CO2. After using a cotton swab to gently wipe off the remaining free cells in the upper well, the well membrane was collected and fixed for 15–20 min in 95% ethanol. After rinsing with clean water, the membrane was stained for 10 min, and washed again with clean water. The membrane was then observed under a high-powered inverted microscope to count the number of cells on the membrane. The invasion ability of the cells was determined by counting the number of cells that passed through the Matrigel, and the average number from five high-magnification fields of view was used for each sample.
3). The diluted Matrigel was added to the upper chamber of the Transwell to form the membrane in three additions (15, 7.5 and 7.5 μl), with 10 min between each addition. The Matrigel was evenly spread and covered all micropores at the bottom of the upper chamber. The two cell lines were collected at 48 h after transfection to prepare cell suspensions. Cell suspensions were seeded into the upper well, and 0.5 ml of DMEM containing 10% FBS was added to the lower chambers (24-well plates). The cells were cultured routinely for 48 h at 37 °C with 5% CO2. After using a cotton swab to gently wipe off the remaining free cells in the upper well, the well membrane was collected and fixed for 15–20 min in 95% ethanol. After rinsing with clean water, the membrane was stained for 10 min, and washed again with clean water. The membrane was then observed under a high-powered inverted microscope to count the number of cells on the membrane. The invasion ability of the cells was determined by counting the number of cells that passed through the Matrigel, and the average number from five high-magnification fields of view was used for each sample.
Animals
Nude mice (purchased from Laboratory Animal Sciences, Capital Medical University, Beijing, China) were used to conduct our investigation at this work. A total of 12 nude mice, female, were randomly divided into 2 groups. The experiment was initiated with 6 week-old mice weighing 20–25 g. They were induced to generate xenograft model with injection of MDA-MB-231 cells, and then housed of Laboratory Animal Center, Capital Medical University, Beijing, China. Mice were maintained at 20–24 °C (temperature controlled) with 12 h light/dark cycle and standard laboratory diet in a pathogen-free environment. All experiments related to animals were performed based on Laboratory Animal Center, Capital Medical University, Beijing, China, Ethical Committee Acts. The experimental mice were dealt with according to the standards supported by the Animal Protection Committee of Capital Medical University. Moreover, the ethical approval was obtained for the animal experiments conducted in your study from Animal Protection Committee of Capital Medical University.Experiment in vivo
The experiment was initiated with 6 week-old of mice, which weight was 20–25 g. Then, 1 ml suspension of MDA-MB-231 cells (1 × 106 cells per ml) was injected into subcutaneous tissue of nude mice at right flanks (n = 6/group). While tumor size of mice reached 3–8 mm, the experiment initiated as the 1st day. The agomir of miR-425 were formulated (1.0 mg ml−1 in PBS). The nude mice were categorized into experiment and control groups at random, then injected with agomir of miR-425 (5 μl) or it control for 6 times once every three days and terminated on the 18th day. Then, these mice were sacrificed at the 21st day, and tumor mass was dissected from each mouse. The methods of anesthesia and euthanasia conducted in our study was that 20% urethane anesthetized (at 1 g kg−1) was used to execute mice at three weeks after injection of agomir of miR-425 or its control. These tumors were cut into two sections, then the volume of tumors were determined every 3 days base on the formulate width2 × length × 0.5. Moreover, the expression of TGF-β1 were determined with western blotting and immunohistochemical staining assay, respectively.The dose of urethane used for euthanasia of mice is (at 1 g kg−1), the following methods were used to evaluate consciousness or confirm death: lack of a heartbeat, lack of respiration, and lack of corneal reflex. The following conclusions can be drawn from this definition. A humane endpoint: (a) not necessarily means the humane killing of the animal, but could also result in interventions to alleviate the stressful/painful experimental procedure (e.g. performing surgery) or providing analgesics. (b) It is not necessarily based on clinical signs but could also start from pre-clinical signs or from physiological or molecular biomarkers predictive of pain/distress later on the disease process. (c) Should be balanced against the scientific endpoints to be met. Thus, pain and distress might be intrinsic to a certain experimental model (e.g. arthritis). However, in this case the humane endpoint should never be beyond the scientific endpoint. (d) Should never be beyond the level of moral justification.
Immunohistochemistry assay
Tissue samples were prepared using 10% neutral formalin, then embedded using paraffin after dehydration. Furthermore, it was cut into thick sections with 5 μM (Marcela et al. 2018), dewaxed and hydrated. After antigen retrieval, incubation with primary antibody for these sections was performed. The sections were incubated with primary antibodies TGF-β1 (Santa Cruz, CA, USA; sc-130348, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200), or SMAD 3 (C67H9) (CST 9523, dilution 1
200), or SMAD 3 (C67H9) (CST 9523, dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200) at 4 °C overnight, respectively. The sections were incubated with a DAB substrate kit (Cat. no. 34002; Thermo Scientific™ Pierce™, San Jose, CA, USA) after incubation with the secondary antibody. The membranes were incubated with secondary antibodies goat anti-rabbit immunoglobulin G (IgG)-horseradish peroxidase (HRP) (sc-2004; 1
200) at 4 °C overnight, respectively. The sections were incubated with a DAB substrate kit (Cat. no. 34002; Thermo Scientific™ Pierce™, San Jose, CA, USA) after incubation with the secondary antibody. The membranes were incubated with secondary antibodies goat anti-rabbit immunoglobulin G (IgG)-horseradish peroxidase (HRP) (sc-2004; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution; Santa Cruz Biotechnology, Inc.) or goat anti-mouse IgG-HRP, (sc-2005; 1
1000 dilution; Santa Cruz Biotechnology, Inc.) or goat anti-mouse IgG-HRP, (sc-2005; 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution; Santa Cruz Biotechnology, Inc.) for 1 h at room temperature. These sections were assessed, and then photographed with microscope at 200× (IX71; Olympus, Japan). Per sample was token for 3–5 shots. The software Image-Proplus (version 6.0) was used measure proteins expression through detecting optical density in each photograph.
1000 dilution; Santa Cruz Biotechnology, Inc.) for 1 h at room temperature. These sections were assessed, and then photographed with microscope at 200× (IX71; Olympus, Japan). Per sample was token for 3–5 shots. The software Image-Proplus (version 6.0) was used measure proteins expression through detecting optical density in each photograph.
Statistics
Welch t-test-paired was performed to compare isomiRs deregulation and selection of miRNAs between samples of patients and control. The errors were evaluated with false discovery rate (FDR) due to multiple comparisons. On the basis of the expression profiles, hierarchical clustering of selected miRNAs was conducted with Ward's agglomeration. Target Rank software (http://genes.mit.edu/targetrank/) was performed to identify the significantly deregulated each seed sequence of miRNA in target genes between samples of patients and control.35All data with normally distributed was represented as mean ± SD (standard deviation). The differences between two groups were analyzed with Student's t-test. Kruskal–Wallis ANOVA method was used to analyze abnormally distributed data among groups. When the variances were equal, the least significant difference post hoc test was used, whereas when the variances were not uniform, Dunnett's post hoc test was used. SPSS (version 18.0; SPSS, Inc., Chicago, IL, USA) soft was performed for entire statistical analyses. While P < 0.05 indicated statistically significant.
Results
Differential miRNA expression profiling
To assess the differential expression of miRNA profiles between tumorous and para-tumorous tissues, miRNA-sequencing experiments were conducted on the total RNA obtained from samples. Among the results of miRNAs-sequencing, 12 were differentially expressed between tumorous and para-tumorous tissues. Compared with para-tumorous tissues, 9 miRNAs were upregulated and 3 miRNAs were down-regulated in patients with TNBC (Table 1).| miRNA | Fold-change (tumorous/para-tumorous tissues) | t value | P value |
|---|---|---|---|
| miR-148b | 5.73 | 8.381 | 7.72 x 10−4 |
| miR-34a | 4.08 | 6.935 | 9.33 x 10−3 |
| miR-196a | 4.01 | 7.220 | 7.00 x 10−3 |
| miR-205 | 3.91 | 9.272 | 5.46 x 10−3 |
| miR-328 | 3.18 | 2.702 | 6.96 x 10−3 |
| miR-122 | 2.64 | 7.254 | 1.77 x 10−3 |
| miR-155 | 2.45 | 3.823 | 5.44 x 10−3 |
| miR-202 | 2.36 | 6.256 | 7.68 x 10−3 |
| miR-let-7a | 2.34 | 3.293 | 4.42 x 10−3 |
| miR-7 | 0.35 | −0.391 | 5.49 x 10−3 |
| miR-425 | 0.18 | −0.468 | 7.53 x 10−3 |
| miR-31 | 0.14 | −0.326 | 2.07 x 10−3 |
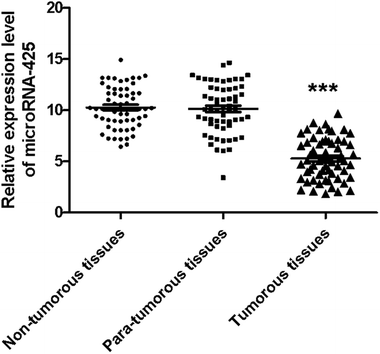
The expression level of miR-425 in non-tumorous, para-tumorous, and tumorous tissues
Furthermore, the relative miR-425 expression levels in non-tumorous, para-tumorous, and tumor tissues (n = 60) were determined using qRT-PCR method. Our work identified the relative miR-425 expression level was obviously less than that of non-tumorous or para-tumorous tissues. However, there was no obvious difference between para-tumorous and non-tumorous tissues (Fig. 2).Expression levels of miR-425 were determined in cell lines of human TNBC
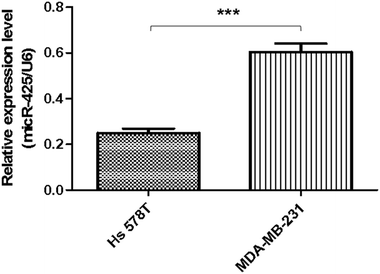
Moreover, the relative miR-425 expression levels were investigated in two cell lines of human TNBC. Compared with basal epithelial phenotype cell line Hs 578T, the relative miR-425 expression levels were significantly higher in mesenchymal phenotypic cell line MDA-MB-231 (P < 0.001) (Fig. 3).MiR-425 inhibited TGF-β1/SMAD3 pathway and mesenchymal markers expression in TNBC cell lines
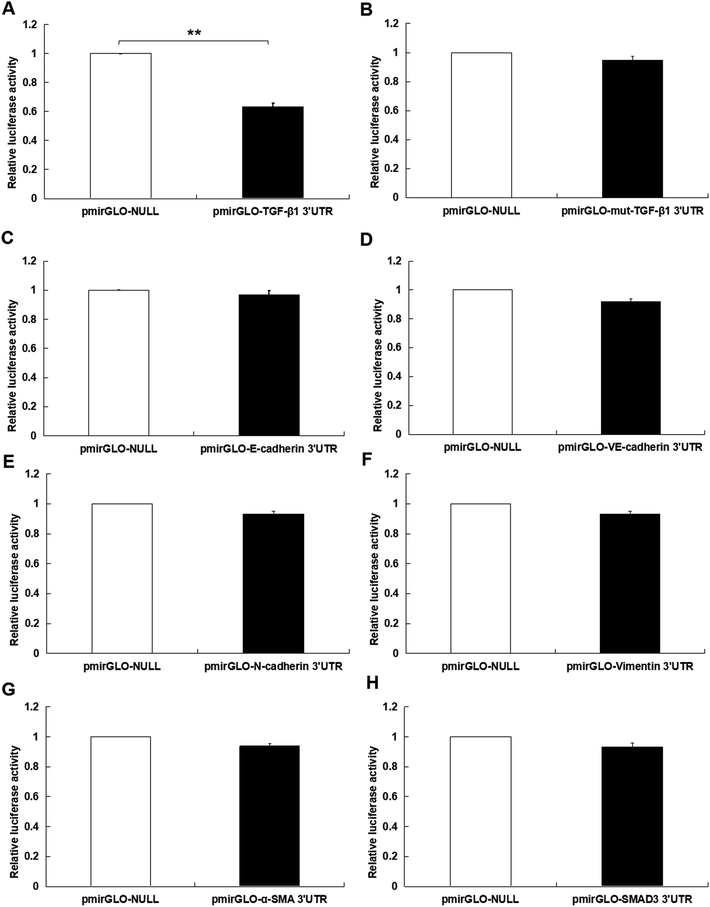
At first, the target of miR-425 were predicted with Targetscan, an online software. Therefore, miR-425 was found that it is the possibly target of 3′ UTRs untranslated region of TGF-β1 mRNA (Table 1). Further experiments were conducted using luciferase reporter gene system to identify whether miR-425 could straight bind with 3′ UTRs of TGF-β1 mRNA. The data indicated 3′ UTR luciferase activities of TGF-β1 significantly down-regulated in MDA-MB-231 cell line dealt with miR-425 (Fig. 4A). However, it did not show obviously change in the group of TGF-β1 mutation (Fig. 4B). Moreover, miR-425 did not bind directly to 3′ UTRs of E-cadherin (Fig. 4C), VE-cadherin (Fig. 4D), N-cadherin (Fig. 4E), vimentin (Fig. 4F), α-SMA (Fig. 4G), and SMAD3 (Fig. 4H), respectively. These data identified that miR-425 targeted to TGF-β1 mRNAs. The well known siRNA of TGF-β1 supported by ref. 26 had been used in our study, which had been used as a positive in our study. It could inhibit the TGF-β1/SMAD3 signaling pathway in TNBC cell lines (Fig. 5).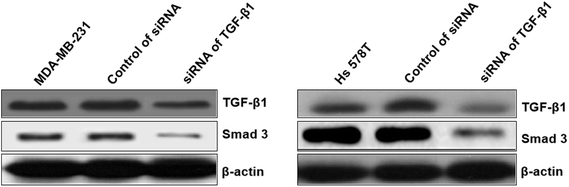 | ||
| Fig. 5 The siRNA of TGF-β1 inhibited TGF-β1/SMAD3 signaling pathway in TNBC cell lines. The well known siRNA of TGF-β1 supported by ref. 26 had been used in our study, which had been used as a positive in our study. It could inhibit the TGF-β1/SMAD3 signaling pathway. | ||
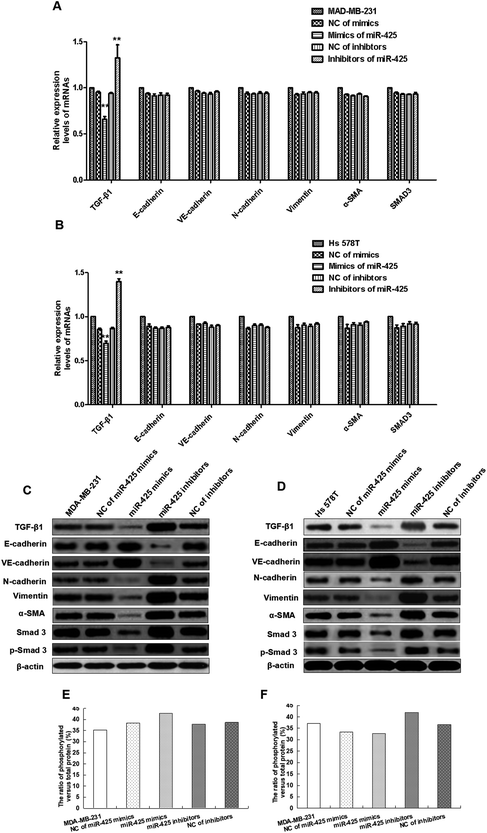
To explore the roles of miR-425 on mRNAs and proteins expression of TGF-β1, α-SMA, vimentin, N-cadherin, E-cadherin, VE-cadherin, and SMAD3 in human BC cell lines, the qRT-PCR assay and western blotting trial were performed. These results demonstrated that miR-425 restrained mRNAs and proteins expressions of TGF-β1 in MDA-MB-231 cell line. However, miR-425 could not suppress the mRNAs expression of α-SMA, vimentin, N-cadherin, VE-cadherin, E-cadherin, and SMAD3 (Fig. 6A). The western blotting result showed that miR-425 promoted proteins expression of E-cadherin and VE-cadherin, while suppressed proteins expression of α-SMA, vimentin, N-cadherin, and SMAD3 (Fig. 6C). Moreover, miR-425 inhibitors could promote expression of TGF-β1 mRNA in MDA-MB-231, but did not affect mRNAs expression of α-SMA, vimentin, N-cadherin, VE-cadherin, E-cadherin, and SMAD3 (Fig. 6A). The inhibitors of miR-425 could suppress the protein expression levels of N-cadherin, vimentin, α-SMA, and SMAD3, but promote VE-cadherin and E-cadherin protein expression level (Fig. 6C). In a word, the result indicated that, by binding with TGF-β1 3′UTR, miR-425 could straight regulate mRNA and protein expression of TGF-β1, further affect protein expression of genes associated with EndMT process and TGF-β1/SMAD3 pathway. It was same as the results in cell line Hs 578T (Fig. 6B and D).
The mesenchymal markers expression of α-SMA, vimentin, N-cadherin, SMAD3 and p-SMAD3 in TNBC cell lines were significantly inhibited, while endothelial markers expression of E-cadherin and VE-cadherin were obviously promoted by mimics of miR-425 in vitro compared with control group (Fig. 6C and D). But, the mesenchymal cell markers expression of TNBC cells were significantly promoted, while endothelial markers expression of E-cadherin and VE-cadherin were significantly suppressed by inhibitors of miR-425 in TNBC cells (Fig. 6C and D). The ratio of phosphorylated versus total protein did not show significant difference between each groups in MDA-MB-231 and Hs 578T cell lines (Fig. 6E and F). Our results identified that miR-425 suppressed mesenchymal markers expression and enhanced endothelial markers expression in TNBC cell lines, especially for the mesenchymal phenotypic cell line MDA-MB-231. The expressions of mesenchymal markers N-cadherin, vimentin, α-SMA, SMAD3 and p-SMAD3 in the mesenchymal phenotypic cell line MDA-MB-231 were higher than that in the basal phenotypic epithelial cell line Hs 578T, and all could be suppressed obviously by mimics of miR-425 in the two cell lines. Moreover, the expressions of endothelial markers E-cadherin and VE-cadherin in the mesenchymal phenotypic cell line MDA-MB-231 were less than that in the basal phenotypic epithelial cell line Hs 578T, and all two cell lines could be promoted significantly by mimics of miR-425.
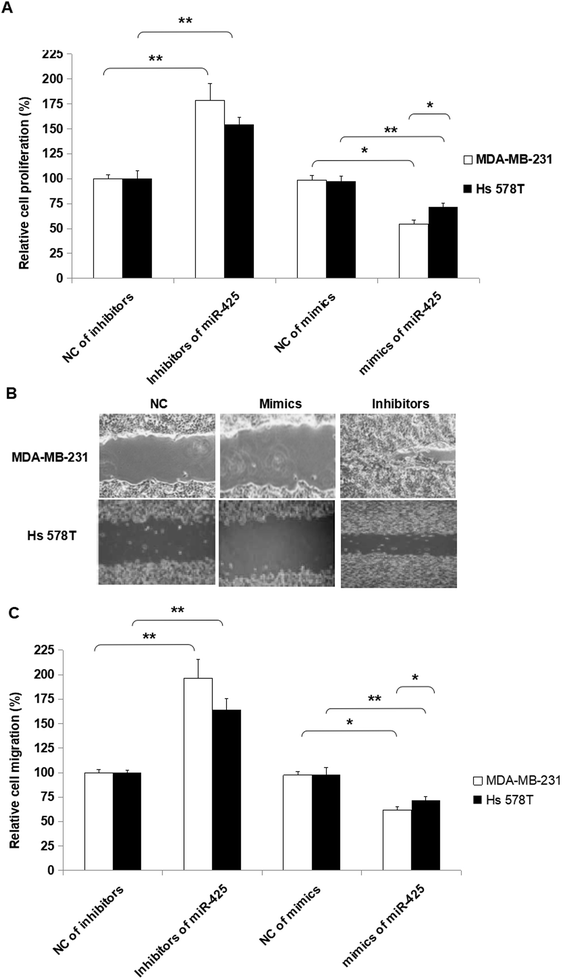
MiR-425 suppressed cell proliferation, migration, and invasion of TNBC cell lines
To assess the biofunctions of miR-425, over-expressed and down-expressed miR-425 was conducted, respectively. Furthermore, several molecular functional tests were performed in TNBC cells. The proliferation ability of TNBC cells was suppressed by miR-425 compared to that of control, and suppressed by inhibitors of miR-425 (Fig. 7A). Moreover, the change of proliferation ability induced by mimics of miR-425 in mesenchymal phenotypic cell line MDA-MB-231 was significantly more than that of basal epithelial phenotype cell line Hs 578T, while the change between the two cell lines was statistically significant difference (P < 0.05) (Fig. 7A).Additionally, the inhibitors of miR-425 promoted migration ability of TNBC cell lines, but the mimics of miR-425 suppressed the migration ability of TNBC cell lines (Fig. 7B and C). Furthermore, the change of migration ability induced by inhibitors of miR-425 in mesenchymal phenotypic cell line MDA-MB-231 was higher than that of basal epithelial phenotype cell line Hs 578T, while the change between the two type cell lines was not statistically significant difference (P > 0.05) (Fig. 7B and C). It identified that miR-425 suppressed the process of proliferation and migration in TNBC cell lines, especially for mesenchymal phenotypic cell line MDA-MB-231. The hypothesis to explain these results was that the inhibition roles induced by miR-425 in proliferation and migration of TNBC cell line, mesenchymal phenotypic cell line MDA-MB-231 were much more than the basal epithelial phenotype cell line Hs 578T (Fig. 7). Moreover, it maybe the mesenchymal characteristics of MDA-MB-231 cell line was obviously more than Hs 578T cell line.
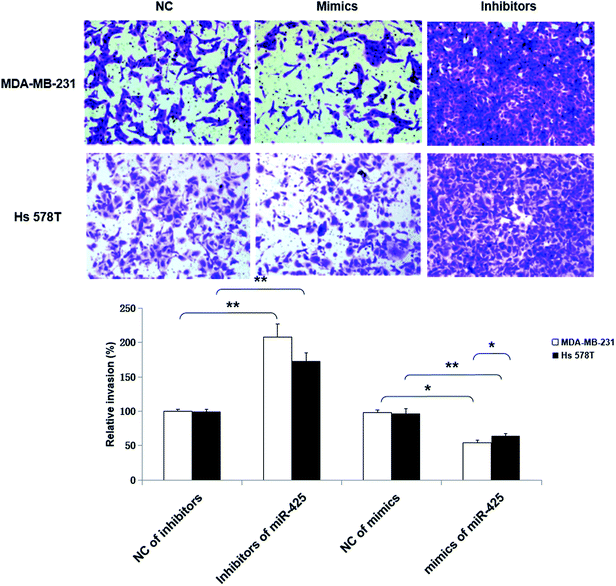
Moreover, the inhibitors of miR-425 promoted invasion ability of TNBC cell lines, but the mimics of miR-425 suppressed the invasion ability of TNBC cell lines (Fig. 8).
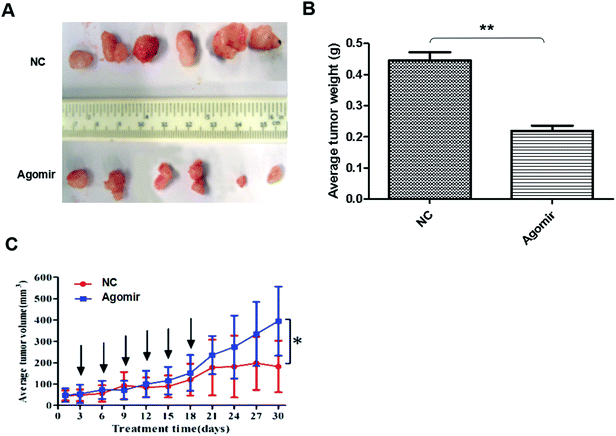
MiR-425 agomir suppressed TNBC xenografts
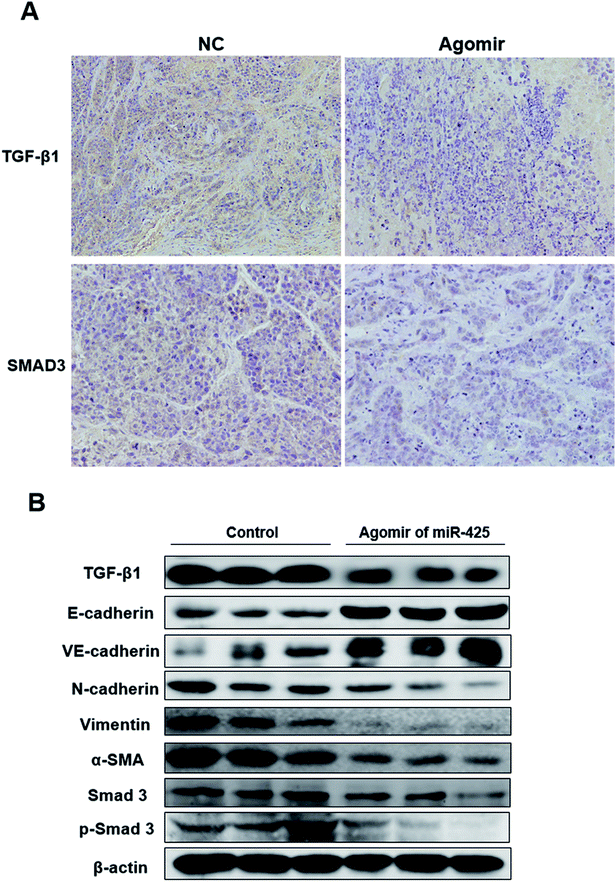
To detect anti-tumor activity induced by miR-425 agomir in vivo, human TNBC xenografts were established with MDA-MB-231 cells. Our work demonstrated that the size of tumor (Fig. 9A) and the average tumor weight (Fig. 9B) of control mice was much larger than that of experiment mice treated with agomir of miR-425, and the difference between two groups had statistical significance. Moreover, the xenografts of control groups grew rapidly, but in the experiment mice treated with agomir of miR-425, the xenografts grew slowly in vivo (Fig. 9C). The maximum diameter exhibited by a single subcutaneous tumor of control mice in our study was 1.23 cm.The expression proteins in xenografts were determined with immunohistochemical staining methods (Fig. 10A) and western blotting assay (Fig. 10B), respectively. The results revealed that when compare with control group, treatment with agomir of miR-425 could down-regulate the expression of SMAD3 and TGF-β1 with a statistically significant difference, and suppressed mesenchymal markers expression of xenograft. The mesenchymal markers expression of α-SMA, vimentin, N-cadherin, SMAD3, p-SMAD3 and endothelial markers E-cadherin, VE-cadherinin were determined in xenografts. The results indicated that compare with control group, treatment with agomir of miR-425 could obviously down-regulate the above mesenchymal markers expression in xenografts, while up-regulate expression of endothelial markers E-cadherin, VE-cadherinin. To sum up, our data confirmed that the agomir of miR-425 could inhibit development of TNBC associated with EMT through aiming TGF-β1/SMAD3 signaling pathway in vivo.
Discussion
Through EMT (epithelial–mesenchymal transition), a mesenchymal phenotype in epithelial cells can be acquire, wherein the marker expression and acquired functions of mesenchymal cell, in addition to lost the epithelial functions and markers expression of epithelia. Besides, migration and delamination of epithelial cells can be caused by EMT, then mesenchymal cells derived from epithelial cell can migrate into the underlying tissue.36 The ligands of TGF-β (transforming growth factor β) family often initiate the cellular process, EMT.37 Many investigations aim at the markers of EMT from thirty-two cancer types38,39 and the roles of TGF-β pathway.38,40 A number of elements are important factors in regulation of gene expression and EMT, while referred to development of TNBC.41–44Moreover, the transcriptional regulation mechanism of EMT and how miRNA modulated EMT or development of TNBC was unclear yet. Recent research revealed that miR-34 might serve as inhibitor in nasopharyngeal carcinoma (NPC) by targeting TNBC-derived growth factor (HDGF). It indicated that miR-34 acted as a tumor suppressor in NPC, and may be a novel therapeutic target for the treatments of patients with NPC.45 Moreover, over-expression of miR-425 is induced by IL-1β, which aim at 3′UTR of miR-425, could regulate homolog expression of tensin and phosphatase. The activation of NF-κB induced by IL-1β up-regulates miR-425 expression, while enhances transcription of miR-425 gene. Therefore, the proliferation of gastric cancer cell is promoted by miR-425 through suppressing tensin homolog and phosphatase. In summary, it confirm that over-expression of miR-425 dependent on NF-κB plays the critical role for the above process.46 EMT could induce extracellular cytokine TNF-α, TGF-β, PDGF, HGF or bFGF to bond with its receptor in BC, and then activate intracellular effectors, such as Ras, NF-κB, Smads, etc. Therefore, in this study, miRNAs were ascertained and searched after its effects on EMT and development process of TNBC.
At first, the differential miRNAs expression profiles between tumor and para-tumor tissues were assessed with miRNA-sequencing experiments. Then, we found that, compared with para-tumor tissues, 9 miRNAs were upregulated and 3 miRNAs (included miR-425) were down-regulated in tumor tissues of patients with TNBC. Furthermore, the relative expression level of miR-425 in non-tumor, para-tumor, and tumor tissues were determined using qRT-PCR method. This work identified relative levels of miR-425 expression in tumor tissues were obviously less than that in para-tumor and non-tumor tissues. However, there was no obvious difference between para-tumor and non-tumor tissues. Consequently, the relative miR-425 expression levels were investigated in two cell lines of human TNBC. The results confirmed that miR-425 expression level was significantly decreased in the basal epithelial phenotype cell lines Ts 578T and mesenchymal phenotypic cell line MDA-MB-231, which indicated that miR-425 down-expression was related to phenotype of EMT.
Therefore, the targets of miR-425 were foretasted with Targetscan, an online software, and TGF-β1 was found that it is the possibly target of 3′ UTRs of TGF-β1 mRNA. Then, further experiments were performed and identified that miR-425 could straight bind with 3′ UTRs of TGF-β1 mRNA. In a word, miR-425 directly targeted to TGF-β1 mRNAs. Hence, miR-425 could straight control mRNA and the protein expression level of TGF-β1, further affect expression levels of E-cadherin, VE-cadherin, N-cadherin, vimentin, α-SMA, and SMAD3 protein in TNBC cell lines. Taken together, our study maybe point out a novel aim at treatment for TNBC associated with EMT.
Besides, to assess the biofunctions of miR-425, several molecular functional tests were performed in TNBC cell lines transfected with mimics or inhibitors of miR-425, respectively. It identified that miR-425 suppressed proliferation process and migration or invasion ability of TNBC cell lines, significantly promoted the expressions of epithelial markers E-cadherin and VE-cadherin, suppressed mesenchymal markers expression of N-cadherin, vimentin, α-SMA, total and phosphorylated SMAD3 (p-SMAD 3) in TNBC cell lines. Our results demonstrated that miR-425 targets to TGF-β1, was a crucial mediator of EMT and development of TNBC through regulating TGF-β1/SMAD3 signaling pathway. Moreover, we did not found that it is involved in well-known molecules such as mTOR, Ki-67, etc.
Furthermore, the effects on Human TNBC xenografts and its mechanism were investigated using miR-425 agomir. Our results confirmed that the xenografts of control groups grew rapidly. But in the experiment group, which treated with agomir of miR-425, the xenografts grew slowly in vivo. Moreover, the size and weight of tumors in experiment group (treated with agomir of miR-425) was significantly less than that of control group. At the time of end, the average tumor volume of control group was much larger than that of experiment group, and the difference between the two groups had statistical significance. Therefore, the agomir of miR-425 could obviously inhibit tumor proliferation and its development process. Moreover, treatment with agomir of miR-425 could down-regulate the expression of TGF-β1 and SMAD3 with a statistically significant, while suppressed the expression of mesenchymal cell markers and promoted epithelial markers E-cadherin and VE-cadherin expression in xenografts.
We thought that our manuscript was obviously composed only one part of the whole. The assays in vivo and in vitro were all separately used to identify our conclusions. In summary, we illustrated that miR-425 targets to TGF-β1. Moreover, miR-425 was a crucial suppressor of EMT and the development of TNBC through inhibiting TGF-β1/SMAD3 signaling pathway. It suggested that aim at TGF-β1/SMAD3 signaling pathway through enhancing miR-425 expression, was a feasible therapy strategy for TNBC.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.Ethics approval and consent to participate
This investigation and the utilization of human specimens in this work had been approved by the Ethics Committee of our hospitals (Beijing Obstetrics and Gynecology Hospital, Capital Medical University, Beijing, China) on the basis of the Declaration of Helsinki. We clearly confirmed that the informed consents were obtained from all patients. Moreover, the recorded and documented of participant consents were kept in our hospital.All experiments related to animals were performed based on Laboratory Animal Center, Capital Medical University, Beijing, China, Ethical Committee Acts. The experimental mice were dealt with according to the standards supported by the Animal Protection Committee of Capital Medical University. Moreover, the ethical approval was obtained for the animal experiments conducted in your study from Animal Protection Committee of Capital Medical University.
Patient consent for publication
This investigation and the utilization of human specimens in this work had been approved by the Ethics Committee of our hospitals (Beijing Obstetrics and Gynecology Hospital, Capital Medical University, Beijing, China) on the basis of the Declaration of Helsinki. We clearly confirmed that the informed consents were obtained from all patients. Moreover, the recorded and documented of participant consents were kept in our hospital.Authors' contributions
LYP and ZY conducted the cell and animal experiments and collected the data and wrote the manuscript. LYP, YF, and CJL analyzed and interpreted the data from the experimental results. CJL and ZJQ were involved in the conception and design of the study, and revised the manuscript. All authors have read and approved the final version of the manuscript.Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 30600524 and 81341067), and the National Natural Science Foundation of Guangdong Province, China (No. 2017A030313510), Introduction of Talent Fund of Guangdong Second Provincial General Hospital (No. YY2016-006), and Capital Clinical Featured Applied Research and Results Promotion projects (Z161100000516141). The study sponsors had no involvement in the work.Conflicts of interest
The authors declare no conflict of interest.References
- J. Ferlay, I. Soerjomataram, R. Dikshit, S. Eser, C. Mathers, M. Rebelo, D. M. Parkin, D. Forman and F. Bray, Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012, Int. J. Cancer, 2015, 136, 359–386 CrossRef PubMed.
- Y. Xiao, Y. Li, H. Tao, B. Humphries, A. Li, Y. Jiang, C. Yang, R. Luo and Z. Wang, Integrin α5 down-regulation by miR-205 suppresses triple negative breast cancer stemness and metastasis by inhibiting the Src/Vav2/Rac1 pathway, Cancer Lett., 2018, 433, 199–209 CrossRef CAS PubMed.
- A. Prat and C. M. Perou, Deconstructing the molecular portraits of breast cancer, Mol. Oncol., 2011, 5, 5–23 CrossRef CAS PubMed.
- S. J. Schnitt, Classification and prognosis of invasive breast cancer: from morphology to molecular taxonomy, Mod. Pathol., 2010, 23, S60–S64 CrossRef PubMed.
- S. B. Zeichner, H. Terawaki and K. Gogineni, A review of systemic treatment in metastatic triple-negative breast cancer, Breast Cancer, 2016, 10, 25–36 CAS.
- T. K. Lee, R. T. P. Poon, A. P. Yuen, M. T. Ling, W. K. Kwok, X. H. Wang, Y. C. Wong, X. Y. Guan, K. Man, K. L. Chau and S. T. Fan, Twist Overexpression Correlates with Hepatocellular Carcinoma Metastasis through Induction of Epithelial-Mesenchymal Transition, Clin. Cancer Res., 2006, 12, 5369 CrossRef CAS PubMed.
- G. Giannelli, C. Bergamini, E. Fransvea, C. Sgarra and S. Antonaci, Laminin-5 with Transforming Growth Factor-β1 Induces Epithelial to Mesenchymal Transition in Hepatocellular Carcinoma, Gastroenterology, 2005, 129, 1375–1383 CrossRef CAS PubMed.
- Z. Y. Tang, S. L. Ye, Y. K. Liu, L. X. Qin, H. C. Sun, Q. H. Ye, L. Wang, J. Zhou, S. J. Qiu, Y. Li, X. N. Ji, H. Liu, J. L. Xia, Z. Q. Wu, J. Fan, Z. C. Ma, X. D. Zhou, Z. Y. Lin and K. D. Liu, A decade's studies on metastasis of hepatocellular carcinoma, J. Cancer Res. Clin. Oncol., 2004, 130, 187–196 CrossRef PubMed.
- M. A. Huber, N. Kraut and H. Beug, Molecular requirements for epithelial–mesenchymal transition during tumor progression, Curr. Opin. Cell Biol., 2005, 17, 548–558 CrossRef CAS PubMed.
- A. R. Lourenço and P. J. Coffer, SOX4: Joining the Master Regulators of Epithelial-to-Mesenchymal Transition?, Trends Cancer, 2017, 3, 571–582 CrossRef PubMed.
- S. Yamada, B. C. Fuchs, T. Fujii, Y. Shimoyama, H. Sugimoto, S. Nomoto, S. Takeda, K. K. Tanabe, Y. Kodera and A. Nakao, Epithelial-to-mesenchymal transition predicts prognosis of pancreatic cancer, Surgery, 2013, 154, 946–954 CrossRef PubMed.
- M. A. Huber, N. Azoitei, B. Baumann, S. Grünert, A. Sommer, H. Pehamberger, N. Kraut, H. Beug and T. Wirth, NF-κB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression, J. Clin. Invest., 2004, 114, 569–581 CrossRef CAS PubMed.
- B. De Craene and G. Berx, Regulatory networks defining EMT during cancer initiation and progression, Nat. Rev. Cancer, 2013, 13, 97–110 CrossRef CAS PubMed.
- G. Finak, et al., Stromal gene expression predicts clinical outcome in breast cancer, Nat. Med., 2008, 14, 518–527 CrossRef CAS PubMed.
- M. Sund and R. Kalluri, Tumor stroma derived biomarkers in cancer, Cancer Metastasis Rev., 2009, 28, 177–183 CrossRef PubMed.
- P. Carmeliet and R. K. Jain, Angiogenesis in cancer and other diseases, Nature, 2000, 407, 249–257 CrossRef CAS PubMed.
- D. Hanahan and R. A. Weinberg, Hallmarks of cancer: the next generation, Cell, 2011, 144, 646–674 CrossRef CAS PubMed.
- L. E. Drake and K. F. Macleod, Tumour suppressor gene function in carcinoma-associated fibroblasts: from tumour cells via EMT and back again?, J. Pathol., 2014, 232, 283–288 CrossRef CAS.
- P. Mallini, T. Lennard, J. Kirby and A. Meeson, Epithelial-to-mesenchymal transition: What is the impact on breast cancer stem cells and drug resistance, Cancer Treat. Rev., 2014, 40, 341–348 CrossRef CAS PubMed.
- R. Kalluri and M. Zeisberg, Fibroblasts in cancer, Nat. Rev. Cancer, 2006, 6, 392–401 CrossRef CAS PubMed.
- P. Mallini, T. Lennard, J. Kirby and A. Meeson, Epithelial-to-mesenchymal transition: What is the impact on breast cancer stem cells and drug resistance, Cancer Treat. Rev., 2014, 40, 341–348 CrossRef CAS PubMed.
- E. Dejana, Endothelial cell-cell junctions: happy together, Nat. Rev. Mol. Cell Biol., 2004, 5, 261–270 CrossRef CAS.
- J. M. Zarzynska, Two faces of TGF-beta1 in breast cancer, Mediators Inflammation, 2014, 2014, 141747 Search PubMed.
- L. Y. Jia, M. K. Shanmugam, G. Sethi and A. Bishayee, Potential role of targeted therapies in the treatment of triple-negative breast cancer, Anticancer Drugs, 2016, 27, 147–155 CrossRef CAS PubMed.
- O. Iriondo, Y. Liu, G. Lee, M. Elhodaky, C. Jimenez, L. Li, J. Lang, P. Wang and M. Yu, TAK1 mediates microenvironment-triggered autocrine signals and promotes triple-negative breast cancer lung metastasis, Nat. Commun., 2018, 9, 1994 CrossRef PubMed.
- N. Erin, N. Ogan and A. Yerlikaya, Correction to: Secretomes reveal several novel proteins as well as TGF-β1 as the top upstream regulator of metastatic process in breast cancer, Breast Cancer Res. Treat., 2018, 170, 251–256 CrossRef PubMed.
- N. E. Bhola, J. M. Balko, T. C. Dugger, M. G. Kuba, V. Sanchez, M. Sanders, J. Stanford, R. S. Cook and C. L. Arteaga, TGF-beta inhibition enhances chemotherapy action against triple-negative breast cancer, J. Clin. Invest., 2013, 123, 1348–1358 CrossRef CAS PubMed.
- B. Feng, Y. Cao, S. Chen, X. Chu, Y. Chu and S. Chakrabarti, miR-200b Mediates Endothelial-to-Mesenchymal Transition in Diabetic Cardiomyopathy, Diabetes, 2016, 65, 768–779 CrossRef CAS PubMed.
- L. Zhang, D. Huang, Q. Wang, D. Shen, Y. Wang, B. Chen, J. Zhang and L. Gai, MiR-132 inhibits expression of SIRT1 and induces pro-inflammatory processes of vascular endothelial inflammation through blockade of the SREBP-1c metabolic pathway, Cardiovasc. Drugs Ther., 2014, 28, 303–311 CrossRef CAS PubMed.
- A. Kozomara and S. Griffiths-Jones, miRBase: integrating microRNA annotation and deep-sequencing data, Nucleic Acids Res., 2011, 39, D152–D157 CrossRef CAS.
- M. M. Martin, Cutadapt removes adaptor sequences from high-throughput sequencing reads, EMBnet J., 2011, 17, 10–12 CrossRef.
- B. Langmead, C. Trapnell, M. Pop and S. L. Salzberg, Ultrafast and memory-efficient align-ment of short DNA sequences to the human genome, Genome Biol., 2009, 10, R25 CrossRef PubMed.
- W. Anna, S. Michal, K. Oskar, G. Wojciech, M. Monika and K. Monika, et al., Next generation sequencing reveals microRNA isoforms in liver cirrhosis and hepatocellular carcinoma, Int. J. Biochem. Cell Biol., 2014, 53, 208–217 CrossRef PubMed.
- W. Zhu, Y. Ma, X. Zhuang and X. Jin, MicroRNA-425 is downregulated in nasopharyngeal carcinoma and regulates tumor cell viability and invasion by targeting TNBC-derived growth factor, Oncol. Lett., 2018, 15, 6345–6351 Search PubMed.
- C. B. Nielsen, N. Shomron, R. Sandberg, E. Hornstein, J. Kitzman and C. B. Burge, Determinants of targeting by endogenous and exogenous microRNAs and siRNAs, RNA, 2007, 13, 1894–1910 CrossRef CAS.
- C. Foroni, M. Broggini, D. Generali and G. Damia, Epithelial–mesenchymal transition and breast cancer: role, molecular mechanisms and clinical impact, Cancer Treat. Rev., 2012, 38, 689–697 CrossRef CAS.
- M. Tu, Z. Li, X. Liu, N. Lv, C. Xi, Z. Lu, J. Wei, G. Song, J. Chen and F. Guo, et al., Vasohibin 2 promotes epithelial-mesenchymal transition in human breast cancer via activation of transforming growth factor β 1 and hypoxia dependent repression of GATA-binding factor 3, Cancer Lett., 2017, 388, 187–197 CrossRef CAS.
- R. B. Runyan and P. Savagner, Epithelial-mesenchymal transition and plasticity in the developmental basis of cancer and fibrosis, Dev. Dyn., 2018, 247, 330–331 CrossRef PubMed.
- D. L. Gibbons and C. J. Creighton, Pan-cancer survey of epithelial-mesenchymal transition markers across the Cancer Genome Atlas, Dev. Dyn., 2017, 247, 555–564 CrossRef PubMed.
- H. Ungefroren, D. Witte and H. Lehnert, The role of small GTPases of the Rho/Rac family in TGF-b-induced EMT and cell motility in cancer, Dev. Dyn., 2017, 247, 451–461 CrossRef.
- H. Xia, L. L. Ooi and K. M. Hui, MicroRNA-216a/217-induced epithelial-mesenchymal transition targets PTEN and SMAD7 to promote drug resistance and recurrence of liver cancer, Hepatology, 2013, 58, 629–641 CrossRef CAS PubMed.
- L. Zhai, C. Ma, W. Li, S. Yang and Z. Liu, miR-143 suppresses epithelial-mesenchymal transition and inhibits tumor growth of breast cancer through down-regulation of ERK5, Mol. Carcinog., 2016, 55, 1990–2000 CrossRef CAS PubMed.
- H. Škovierová, T. Okajčeková, J. Strnádel, E. Vidomanová and E. Halašová, Molecular regulation of epithelial-to-mesenchymal transition in tumorigenesis (review), Int. J. Mol. Med., 2018, 41, 1187–1200 Search PubMed.
- S. IXing, W. Yu, X. Zhang, Y. Luo, Z. Lei, D. Huang, J. Lin, Y. Huang, S. Huang, F. Nong, C. Zhou and G. Wei, Soviolanthin Extracted from Dendrobium officinale Reverses TGF-β1-Mediated Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma Cells via Deactivating the TGF-β/Smad and PI3K/Akt/mTOR Signaling Pathways, Int. J. Mol. Sci., 2018, 19(6) Search PubMed.
- G. H. Huanga, M. Y. Dua, H. M. Zhua, N. Zhanga, Z. W. Lua, L. X. Qiana, W. J. Zhanga, X. H. Tianc, H. Xia and L. Yina, MiRNA-34a reversed TGF-β-induced epithelial-mesenchymal transition via suppression of SMAD4 in NPC cells, Biomed. Pharmacother., 2018, 106, 217–224 CrossRef PubMed.
- J. Ma, J. Liu, Z. Wang, X. Gu, Y. Fan, W. Zhang, L. Xu, J. Zhang and D. Cai, NF-kappaB-dependent microRNA-425 upregulation promotes gastric cancer cell growth by targeting PTEN upon IL-1β induction, Mol. Cancer, 2014, 13, 40 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2019 |