Open Access Article
Open Access ArticleLight-triggered hydrophilic drug release from liposomes through removal of a photolabile protecting group†
Yuya Gotoa,
Masafumi Uedaab,
Kouta Sugikawa a,
Kazuma Yasuharac and
Atsushi Ikeda
a,
Kazuma Yasuharac and
Atsushi Ikeda *a
*a
aDepartment of Applied Chemistry, Graduate School of Engineering, Hiroshima University, 1-4-1 Kagamiyama, Higashi-Hiroshima 739-8527, Japan
bSchool of Science, Kitasato University, 1-15-1 Kitasato, Minami-ku, Sagamihara, Kanagawa 252-0373, Japan
cGraduate School of Materials Science, Nara Institute of Science and Technology, 8916-5 Takayama, Ikoma, Nara 630-0192, Japan
First published on 21st December 2018
Abstract
The antibiotic penicillin G cannot be completely incorporated into hydrophobic lipid-membranes owing to its hydrophilicity. Through modification with a hydrophobic and photolabile protecting group, penicillin G was effectively incorporated into liposomes and released by photoirradiation at 365 nm.
Introduction
The ability to regulate the release of an appropriate dose of drug at a specific location with the correct timing is very important for liposomal drug-delivery systems used in selective tissue targeting.1–5 Drug release can be controlled by pH, thermal, enzymatic, or photochemical triggers. Many research groups have reported liposomal drug-delivery systems that use the liposome collapse or the considerable distortion of the membrane that results from these triggers to affect release.6–10 In contrast, several groups have reported drug-delivery systems that use the introduction and elimination of a guest molecule protecting group to control release. For example, hydrophilic guest molecules can be incorporated into hydrophobic lipid membranes using hydrophobic protecting groups. In lipid membrane-incorporated guest molecules (LMIGs), the incorporation or leakage percentages of guest molecules are predicted by the hydrophobicity of the guest molecules. Schwarzenbach et al. directly determined the liposome–water distribution ratio (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Klipw) of phenols and phenoxides and reported that the log
Klipw) of phenols and phenoxides and reported that the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Klipw values of phenols are very similar to the octanol–water partition coefficient (log
Klipw values of phenols are very similar to the octanol–water partition coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pow).11–13 In contrast, we investigated the relationship between the equilibrium and the log
Pow).11–13 In contrast, we investigated the relationship between the equilibrium and the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pow for some small model guest molecules with a π-moiety.14 Our results indicated that many of the guest molecules with log
Pow for some small model guest molecules with a π-moiety.14 Our results indicated that many of the guest molecules with log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pow < 1.9 leaked from the lipid membranes, while most of the guest molecules with log
Pow < 1.9 leaked from the lipid membranes, while most of the guest molecules with log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pow > 1.9 did not. That is, the threshold for leakage of guest molecules was determined to be log
Pow > 1.9 did not. That is, the threshold for leakage of guest molecules was determined to be log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pow ≈ 1.9.14 Therefore, if the log
Pow ≈ 1.9.14 Therefore, if the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pow of the guest molecules can be controlled, the amount of drug released can be varied. For example, a dimer of a coumarin derivative (log
Pow of the guest molecules can be controlled, the amount of drug released can be varied. For example, a dimer of a coumarin derivative (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pow > 3) was reverted to monomers in the lipid membrane by photoirradiation at 254 nm and the monomer (log
Pow > 3) was reverted to monomers in the lipid membrane by photoirradiation at 254 nm and the monomer (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pow = 1.78) was released.15 In this work, we used penicillin G (1)16,17 (log
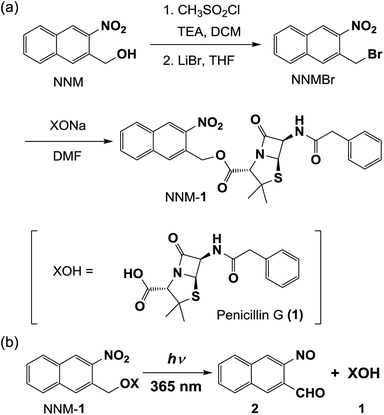
Pow = 1.78) was released.15 In this work, we used penicillin G (1)16,17 (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pow = 1.85)18 as an example of a hydrophilic drug. We report the synthesis of 1 protected with the hydrophobic photolabile protecting group 2-nitro-3-naphthalenemethanol (NNM)19–21 (Scheme 1), and the release of the penicillin G from liposomes by photocleavage of the protecting group (Scheme 2).
Pow = 1.85)18 as an example of a hydrophilic drug. We report the synthesis of 1 protected with the hydrophobic photolabile protecting group 2-nitro-3-naphthalenemethanol (NNM)19–21 (Scheme 1), and the release of the penicillin G from liposomes by photocleavage of the protecting group (Scheme 2).
Results and discussion
Photocleavage of the protecting group in NNM-1 in DMSO
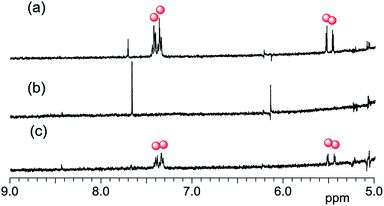
To confirm the photocleavage of the protecting group in NNM-1 (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pow = 4.13),13 we measured the 1H NMR and UV-vis absorption spectra of NNM-1 in DMSO after photoirradiation at 365 nm for 24 h. As shown in Fig. 1 and S2,† NNM moiety 1H NMR peaks in NNM-1 (Fig. 1b; blue circles) were no longer detected after photoirradiation at 365 nm, and new peaks appeared (Fig. 1c). Because these peaks (Fig. 1c; red circles) are similar to those of the sodium salt of 1 (Fig. 1a; red circles), the photocleavage of NNM-1 and generation of 1 were suggested as shown in Scheme 1b. Because of the disagreement in the chemical shifts of the 1H NMR peaks in Fig. 1a and c, we attempted to prepare 1. However, 1 decomposed when treated with an HCl solution, as it is unstable in acidic conditions.22
Pow = 4.13),13 we measured the 1H NMR and UV-vis absorption spectra of NNM-1 in DMSO after photoirradiation at 365 nm for 24 h. As shown in Fig. 1 and S2,† NNM moiety 1H NMR peaks in NNM-1 (Fig. 1b; blue circles) were no longer detected after photoirradiation at 365 nm, and new peaks appeared (Fig. 1c). Because these peaks (Fig. 1c; red circles) are similar to those of the sodium salt of 1 (Fig. 1a; red circles), the photocleavage of NNM-1 and generation of 1 were suggested as shown in Scheme 1b. Because of the disagreement in the chemical shifts of the 1H NMR peaks in Fig. 1a and c, we attempted to prepare 1. However, 1 decomposed when treated with an HCl solution, as it is unstable in acidic conditions.22
 | ||
| Fig. 1 Partial 1H NMR spectra (400 MHz, DMSO-d6, 25 °C) of (a) sodium salt of 1 and (b and c) NNM-1 (b) before and (c) after photoirradiation at 365 nm for 24 h (red circles: 1, blue circles: NNM-1). | ||
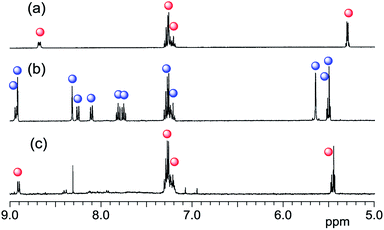
To confirm the generation of 1, we measured the electrospray ionisation mass spectroscopy (ESI-MS) (Fig. 2). Before photoirradiation, 1 was not observed in the ESI-MS spectrum of NNM-1 as a fragment (Fig. 2a). An ambiguous ion at m/z 345.09 was found to be a fragment of NNM-1 because the same peak was observed as a product ion of m/z 520 in the ESI-MS/MS for fragments generated by collision induced dissociation (Fig. S3†). However, only trace amounts of ions of 1 were observed in the sample of NNM-1 before the photoirradiation at 365 nm. In contrast, after photoirradiation, ions of 1 were detected in both the positive and negative modes, while only trace amounts of ions of NNM-1 were observed (Fig. 2c). These results clearly show that NNM-1 was almost completely cleaved after the photoirradiation. Therefore, the disagreement in the 1H NMR peaks between Fig. 1a and c, is thought to arise from Fig. 1a showing the spectrum of the sodium salt of 1.
 | ||
| Fig. 2 ESI-MS spectra of NNM-1 (a) before (positive ion mode), (b) after (positive ion mode), and (c) after (negative ion mode) photoirradiation at 365 nm for 24 h in CHCl3. | ||
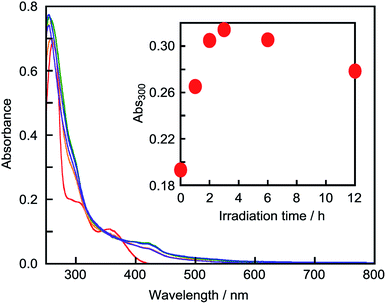
As shown in Fig. 3, the UV-vis absorption of NNM-1 changed after photoirradiation at 365 nm in DMSO. The spectrum after 12 h was similar to that of the sodium salt of 1 in H2O (Fig. S4†), and was consistent with a previous result. The absorption changes of NNM-1 were saturated after 12 h of photoirradiation. However, the absorbance decreased after 3 h of photoirradiation. This result suggests that compound 1 decomposed, and the emergence of peaks of some degradation products supported this. The structures of the degradation products could not be determined as they were only produced in small amounts.
Determination of leakage percentages of 1 and NNM-1 from liposomes
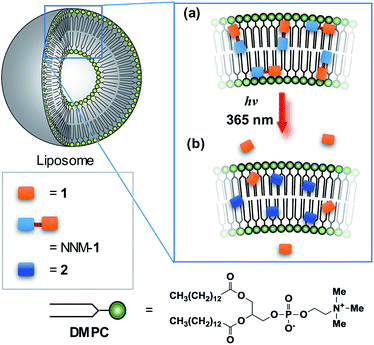
Lipid membrane-incorporated 1 and NNM-1 (LMI1 and LMI-NNM-1, respectively) were prepared using 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC, Scheme 2) and a premixing method (Scheme S1†).15,23,24 LMI1 and LMI-NNM-1 were obtained by dissolving DMPC and the guest molecules (1 or NNM-1) in chloroform, followed by concentration of the resulting mixture to give a residue that was extracted with water. Because compound 1 does not precipitate at this concentration, all 1 present existed either in the lipid membrane or in bulk solution. Therefore, the leakage percentage of 1 from liposomes was calculated to be 82%, based on the 1H NMR spectrum, which indicated that 18% of 1 was residual in the lipid membrane ([1]/[DMPC] = 10 mol%). The leakage of 1 (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pow = 1.85) is consistent with previous findings that many guest molecules with log
Pow = 1.85) is consistent with previous findings that many guest molecules with log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pow < 1.9 leak from lipid membranes.14 In contrast, NNM-1 which leaked out of the liposomes, precipitated as a result of being insoluble in water. To determine the concentration of NNM-1 in LMI-NNM-1, the residue was dissolved in CDCl3 containing ethanol (0.4 mM) as an internal standard, to measure the 1H NMR spectrum after lyophilisation of the LMI-NNM-1 solution (Fig. S5†). From the peak intensities, [NNM-1]/[DMPC] was calculated to be 3.4 mol%. Because the initial ratio of [NNM-1]/[DMPC] was 10 mol%, the residual NNM-1 ([NNM-1]/[DMPC] = 6.6 mol%) was not incorporated into the liposomes and precipitated. The precipitate was removed from the aqueous solution by filtration to give LMI-NNM-1. The incorporation of a high proportion of NNM-1 in the lipid membrane was a result of the hydrophobicity of NNM-1 (log
Pow < 1.9 leak from lipid membranes.14 In contrast, NNM-1 which leaked out of the liposomes, precipitated as a result of being insoluble in water. To determine the concentration of NNM-1 in LMI-NNM-1, the residue was dissolved in CDCl3 containing ethanol (0.4 mM) as an internal standard, to measure the 1H NMR spectrum after lyophilisation of the LMI-NNM-1 solution (Fig. S5†). From the peak intensities, [NNM-1]/[DMPC] was calculated to be 3.4 mol%. Because the initial ratio of [NNM-1]/[DMPC] was 10 mol%, the residual NNM-1 ([NNM-1]/[DMPC] = 6.6 mol%) was not incorporated into the liposomes and precipitated. The precipitate was removed from the aqueous solution by filtration to give LMI-NNM-1. The incorporation of a high proportion of NNM-1 in the lipid membrane was a result of the hydrophobicity of NNM-1 (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pow = 4.13).14 The average hydrodynamic diameter (Dhy) of LMI-NNM-1 was almost unchanged following photoirradiation, suggesting that the liposome morphology did not change after the photocleavage of NNM-1 (Table 1).
Pow = 4.13).14 The average hydrodynamic diameter (Dhy) of LMI-NNM-1 was almost unchanged following photoirradiation, suggesting that the liposome morphology did not change after the photocleavage of NNM-1 (Table 1).
Photocleavage of the protecting group in NNM-1 in liposomes
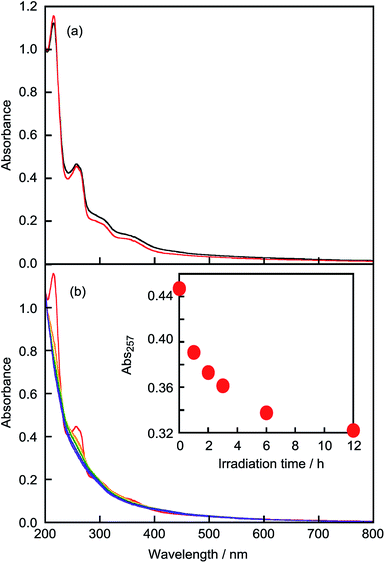
To determine the saturation time of photoirradiation, UV-vis absorption spectra were measured. The absorption of NNM-1 under both dark conditions and photoirradiation was measured for the LMI-NNM-1 solution (Fig. 4). Under dark conditions, the UV-vis absorption spectrum showed only marginal change (Fig. 4a). This result indicates that, (i) NNM-1 did not decompose in the absence of photoirradiation and (ii) the LMI-NNM-1 solution was stable for 1 d. After photoirradiation, the absorption intensity decreased and then saturated after 12 h. Peak broadening was observed in a similar manner to NNM-1 in DMSO (Fig. 3 and 4b). This result suggests that NNM-1 was almost completely photocleaved after 12 h.To confirm whether NNM-1 and cleaved 1 were present in the lipid membrane or bulk water, we measured the 1H NMR spectra of LMI-NNM-1 after photoirradiation for 12 h. The peak intensities in Fig. 5 and S6† were normalised relative to the value of DMSO (0.4 mM) as an internal standard. The leakage percentage of 1 was found to be 74%, based on the peak intensities of 1 relative to the DMSO peak. In contrast, no peaks were observed for the LMI-NNM-1 solution (Fig. 5b). It is well known that lipids peaks and those belonging to guest molecules are not detected for LMIGs as a consequence of peak broadening resulting from the formation of the liposomes.25–30 Therefore, the absence of peaks belonging to NNM-1 suggests that all of the NNM-1 was completely incorporated into the liposomes.
Antimicrobial activity of LMI-NNM-1 before and after photoirradiation
We determined the antimicrobial activity of LMI-NNM-1 before and after photoirradiation toward Staphylococcus aureus (S. aureus) ATCC 25923. The minimum inhibitory concentration (MIC) of LMI-NNM-1 after photoirradiation at 365 nm for 12 h (0.23 μg mL−1) was much lower than that measured before photoirradiation (>1.82 μg mL−1). This indicates that the antimicrobial activity of LMI-NNM-1 after photoirradiation was higher than that before photoirradiation. Consequently, the penicillin G 1 released from the lipid membranes was able to function as an antimicrobial agent. However, the MIC value was higher than that of pristine penicillin G sodium salt (0.03 μg mL−1) because the leakage percentage of 1 was found to be 74%, and 1 released to the inner aqueous phase of the vesicles did not act as an antimicrobial agent.Conclusions
The leakage percentage of pristine penicillin G 1 (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pow = 1.85) from lipid membranes was found to be 82%. In contrast, the majority of a sample of 1 was incorporated into the hydrophobic lipid membrane following modification with NNM as a hydrophobic protecting group. Consequently, the leakage of NNM-1 (log
Pow = 1.85) from lipid membranes was found to be 82%. In contrast, the majority of a sample of 1 was incorporated into the hydrophobic lipid membrane following modification with NNM as a hydrophobic protecting group. Consequently, the leakage of NNM-1 (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pow = 4.13) was minimal. After photoirradiation at 365 nm, deprotected 1 was released from the lipid membrane and the leakage percentage from NNM-1 incorporated in the lipid membrane was 74%. Because the leakage percentage of 1 from the liposomes was 82%, the release of 1 by photoirradiation was considered to have proceeded efficiently.
Pow = 4.13) was minimal. After photoirradiation at 365 nm, deprotected 1 was released from the lipid membrane and the leakage percentage from NNM-1 incorporated in the lipid membrane was 74%. Because the leakage percentage of 1 from the liposomes was 82%, the release of 1 by photoirradiation was considered to have proceeded efficiently.
Experimental
Materials
Penicillin G sodium salt and 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) were purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan) and Funakoshi Co., Ltd (Tokyo, Japan), respectively. NNM19 was synthesized according to previously reported methods.Synthesis of NNMBr
A solution of NNM (100.0 mg, 0.49 mmol) in dichloromethane (5 mL) was added to triethylamine (101.2 mg, 1.00 mmol) and methylsulfonyl chloride (85.9 mg, 0.75 mmol) in an ice bath. The reaction mixture was then stirred in the ice bath for 30 min. After 30 min, the reaction mixture was extracted with dichloromethane and washed with saturated sodium hydrogen carbonate solution and saturated sodium chloride solution. The organic layer was then dried with MgSO4 and filtered. After evaporating in vacuo, the crude product was dissolved in THF (5 mL) and lithium bromide (338 mg, 3.9 mmol) was added. The reaction mixture was stirred at room temperature for 16 h. After evaporating in vacuo, the crude product was extracted with dichloromethane and washed with saturated sodium hydrogen carbonate solution and saturated sodium chloride solution. The organic layer was dried with MgSO4 and filtered. The product was dried under vacuum. Yellow solid, 55.3 mg (42%). 1H NMR (400 MHz, CDCl3) δ 8.65 (s, 1H), 8.01–7.90 (m, 3H), 7.71–7.65 (tt, J = 4.6 Hz, 2H) 5.04 (s, 2H) ppm.Preparation of lipid-membrane-incorporated 1 and NNM-1 using the premixing method
An appropriate amount of a mixture of DMPC (6.00 μmol) and 1 or NNM-1 ([DMPC]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [1 or NNM-1] = 10
[1 or NNM-1] = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mol mol−1) was dissolved in chloroform. The solvent was evaporated under a gentle stream of nitrogen, followed by a period under vacuum to remove trace solvent. The resulting thin lipid films were hydrated on the wall of the vial above the phase transition temperature with an appropriate amount of water or D2O (1.5 mL). The hydrated materials were subjected to eight freeze–thaw cycles (−195 and +50 °C) to give unilamellar vesicles, which were extruded 11 times through 0.05 μm pores using a LiposoFast™ Mini-extruder (Avestin, Ottawa, ON, USA) above the phase transition temperature.
1 mol mol−1) was dissolved in chloroform. The solvent was evaporated under a gentle stream of nitrogen, followed by a period under vacuum to remove trace solvent. The resulting thin lipid films were hydrated on the wall of the vial above the phase transition temperature with an appropriate amount of water or D2O (1.5 mL). The hydrated materials were subjected to eight freeze–thaw cycles (−195 and +50 °C) to give unilamellar vesicles, which were extruded 11 times through 0.05 μm pores using a LiposoFast™ Mini-extruder (Avestin, Ottawa, ON, USA) above the phase transition temperature.
Photoirradiation of NNM-1 and LMI-NNM-1 solutions at 365 nm
A solution of NNM-1 in DMSO or LMI-NNM-1 in water was photoirradiated at 365 nm in a 1 mm quartz cell (1.1 mW cm−2). The photoreactions were monitored by UV-vis absorption and 1H NMR spectroscopies. The photocleavage of NNM-1 was confirmed by MS and 1H NMR spectra.Determination of the leakage percentages of guest molecules by 1H NMR spectroscopy
DMSO (0.4 mM) was added to aqueous solutions of LMIGs as an internal standard. The leakage percentages were determined by comparing the peak intensity of the guest molecules with that of DMSO.Dynamic light scattering (DLS) analysis
The hydrodynamic diameters and the zeta potentials of the liposomes were measured on an electrophoretic light scattering instrument equipped with a laser Doppler system (Zetasizer Nano ZS, Malvern Instruments Ltd, Malvern, UK).Determination of antimicrobial activity
Antimicrobial activity was evaluated by finding the minimum inhibitory concentration (MIC), which was determined in a turbidity-based microdilution assay.31 Staphylococcus aureus (S. aureus) ATCC 25923 was grown to the mid-logarithmic phase (OD600 = 0.5–0.6) in Muller-Hinton (MH) broth at pH 7.4 and then diluted to give OD600 = 0.001, which corresponds to ∼2 × 105 cfu mL−1. We mixed 90 μL of the prepared bacterial stock suspension with 10 μL of solution containing LMI-NNM-1 before and after photoirradiation at 365 nm for 12 h in a sterile polypropylene 96-well plate (#3359, Corning Life Sciences, Corning, NY, USA). The highest concentrations for NNM-1 was 1.82 μg mL−1, and the polymer was serially diluted two-fold. The OD600 of each well was measured after incubation at 37 °C for 18 hours, and any increase in turbidity was considered to be due to S. aureus growth. The minimum concentration of NNM-1 or 1 that completely inhibited the growth of S. aureus was defined as the MIC.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by a JSPS KAKENHI Grant-in-Aid for Scientific Research (B) (Grant No. JP16H04133) and a Grant-in-Aid for Challenging Exploratory Research (Grant No. JP16K13982). We thank Sarah Dodds, PhD, from Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.Notes and references
- Y. Malam, M. Loizidou and A. M. Seifalian, Trends Pharmacol. Sci., 2009, 30, 592–599 CrossRef CAS PubMed.
- W. T. Al-Jamal and K. Kostarelos, Acc. Chem. Res., 2011, 44, 1094–1104 CrossRef CAS PubMed.
- T. M. Allen and P. R. Cullis, Adv. Drug Delivery Rev., 2013, 65, 36–48 CrossRef CAS PubMed.
- R. van der Meel, M. Fens, P. Vader, W. W. van Solinge, O. Eniola-Adefeso and R. M. Schiffelers, J. Controlled Release, 2014, 195, 72–85 CrossRef CAS PubMed.
- B. S. Pattni, V. V. Chupin and V. P. Torchilin, Chem. Rev., 2015, 115, 10938–10966 CrossRef CAS PubMed.
- S. Bibi, E. Lattmann, A. R. Mohammed and Y. Perrie, J. Microencapsulation, 2012, 29, 262–276 CrossRef CAS PubMed.
- J. O. Eloy, M. C. D. Souza, R. Petrilli, J. P. A. Barcellos, R. J. Lee and J. M. Marchetti, Colloids Surf., B, 2014, 123, 345–363 CrossRef CAS PubMed.
- K. A. Carter, S. Shao, M. I. Hoopes, D. Luo, B. Ahsan, V. M. Grigoryants, W. Song, H. Huang, G. Zhang, R. K. Pandey, J. Geng, B. A. Pfeifer, C. P. Scholes, J. Ortega, M. Karttunen and J. F. Lovell, Nat. Commun., 2014, 5, 3546 CrossRef PubMed.
- S. Ghosh, R. Qi, K. A. Carter, G. Zhang, B. A. Pfeifer and J. F. Lovell, Biochem. Eng. J., 2019, 141, 43–48 CrossRef CAS.
- M. Mathiyazhakan, C. Wiraja and C. Xu, Nano-Micro Lett., 2018, 10, 10 CrossRef PubMed.
- B. I. Escher and R. P. Schwarzenbach, Environ. Sci. Technol., 1996, 30, 260–270 CrossRef CAS.
- Y. Katz and J. M. Diamond, J. Membr. Biol., 1974, 17, 69–86 CrossRef CAS PubMed.
- The values of log Pow can be calculated using several software packages. The value for NNM-1 was calculated using Advanced Chemistry Development Software V11.02, ACD/Labs Search PubMed.
- A. Ikeda, K. Ashizawa, Y. Tsuchiya, M. Ueda and K. Sugikawa, RSC Adv., 2016, 6, 78505–78513 RSC.
- R. Shimokawa, M. Ueda, K. Sugikawa and A. Ikeda, J. Photochem. Photobiol., B, 2018, 185, 235–240 CrossRef CAS PubMed.
- W. A. Velema, J. P. van der Berg, W. Szymanski, A. J. M. Driessen and B. L. Feringa, ACS Chem. Biol., 2014, 9, 1969–1974 CrossRef CAS PubMed.
- A. I. Kallenberg, F. van Rantwijk and R. A. Sheldon, Adv. Synth. Catal., 2005, 347, 905–926 CrossRef CAS.
- G. Gerebtzoff and A. Seelig, J. Chem. Inf. Model., 2006, 46, 2638–2650 CrossRef CAS PubMed.
- F. Kienzle, Helv. Chim. Acta, 1980, 63, 2364–2369 CrossRef CAS.
- R. A. Moss and X. Fu, Org. Lett., 2004, 6, 3353–3356 CrossRef CAS PubMed.
- A. K. Singh and P. K. Khade, Tetrahedron, 2005, 61, 10007–10012 CrossRef CAS.
- S. Arrowood, A. M. Hoyt Jr and M. J. Sepaniak, J. Chromatogr., 1992, 583, 105–110 CrossRef CAS PubMed.
- A. D. Bangham, Prog. Biophys. Mol. Biol., 1968, 18, 29–36 CrossRef CAS PubMed.
- M. Ueda, K. Ashizawa, K. Sugikawa, K. Koumoto, T. Nagasaki and A. Ikeda, Org. Biomol. Chem., 2017, 15, 1565–1569 RSC.
- E. Okamura and M. Nakahara, J. Phys. Chem. B, 1999, 103, 3505–3509 CrossRef CAS.
- E. Okamura, R. Kakitsubo and M. Nakahara, Langmuir, 1999, 15, 8332–8335 CrossRef CAS.
- M. Vermathen, P. Vermathen, U. Simonis and P. Bigler, Langmuir, 2008, 24, 12521–12533 CrossRef CAS PubMed.
- M. Marzorati, P. Bigler and M. Vermathen, Biochim. Biophys. Acta, 2011, 1808, 1661–1672 CrossRef CAS PubMed.
- N. Weizenmann, D. Huster and H. A. Scheidt, Biochim. Biophys. Acta, 2011, 1818, 3010–3018 CrossRef PubMed.
- T. Nakaya, Y. Tsuchiya, B. Horiguchi, K. Sugikawa, K. Komaguchi and A. Ikeda, Bull. Chem. Soc. Jpn., 2018, 91, 1337–1342 CrossRef CAS.
- CLSI, Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard–Ninth Edition, CLSI document M07-A9, Clinical and Laboratory Standards Institute, Wayne, PA, 2012 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures, 1H NMR, MS/MS and UV-vis absorption spectra. See DOI: 10.1039/c8ra08584f |
| This journal is © The Royal Society of Chemistry 2019 |