On-site visual discrimination of transgenic food by water-soluble DNA-binding AIEgens†
Zhe
Jiao
ab,
Zongning
Guo
c,
Xuelin
Huang
c,
Hongbo
Fan
*a,
Miao
Zhao
d,
Dianming
Zhou
d,
Xiaolei
Ruan
e,
Pengfei
Zhang
 *f,
Sixing
Zhou
a and
Ben Zhong
Tang
*f,
Sixing
Zhou
a and
Ben Zhong
Tang
 *b
*b
aSchool of Environment and Civil Engineering, Dongguan University of Technology, Dongguan 523808, China. E-mail: fhb666666@126.com
bDepartment of Chemistry, Hong Kong Branch of Chinese National Engineering Research Centre for Tissue Restoration and Reconstruction, Department of Chemical and Biological Engineering, The Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong. E-mail: tangbenz@ust.hk
cIntegrated Technology Center of Dongguan Entry-Exit Inspection and Quarantine Bureau, Dongguan 523073, China
dTianjin Centres for Disease Control and Prevention, Tianjin 300011, China
eSchool of Agriculture, South China Agricultural University, Guangzhou 510642, China
fGuangdong Key Laboratory of Nanomedicine, Shenzhen Engineering Laboratory of Nanomedicine and Nanoformulations, CAS Key Laboratory of Health Informatics, Institute of Biomedicine and Biotechnology, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen 518055, China. E-mail: pf.zhang@siat.ac.cn
First published on 17th July 2019
Abstract
Development of affordable, simple and effective methods for rapid on-site identification of genetically modified organisms (GMO) is of importance due to public concern for transgenic food. In this paper, a visual discrimination method for transgenic food was developed based on a portable polymerase chain reaction (PCR) instrument and water-soluble AIE luminogens (AIEgens). The transgenes were extracted and amplified by the PCR device, further visualized by DNA-binding AIEgens 1,1,2,2-tetrakis[4-(2-bromo-ethoxy)phenyl] ethene (TTAPE) with a label-free approach. The lighted-up emission could be distinctly identified by the naked eye under a UV lamp or by a portable smartphone. The results showed that only transgenes can be visually detected, which confirmed the specificity of the proposed method. Besides, the whole PCR process was not affected by the addition of AIEgens. The portable system was successfully applied and validated for on-site screening and identification of transgenic papaya, corn and soybean samples. The proposed technique may serve as a general DNA fragment on-site visual discrimination platform for environmental and clinical applications.
Introduction
The debate on the use of genetically modified organisms (GMO) in agriculture and the agro-industry is a public concern involving a combination of scientific, social, and political aspects. There is as yet little unanimity among governments and legislators regarding the foreseeable risks that GMO may pose to human health and the environment.1,2 Pressures from consumer groups and public demands have led several countries to require labeling for the presence of GM in foods. In many countries, vigorous supervision of genetically modified foods and other related documents has been promulgated. For GMO inspection and their environmental risk assessment, simple, effective and inexpensive on-field tests for GM events and/or GM contents are urgent to be developed. Various methodologies employed to analyze and/or detect the presence of GMO in food products have been reported, such as the polymerase chain reaction (PCR),3 enzyme linked immunosorbent assays (ELISA),4 microarrays,51H NMR,6 mass spectrometry,7 near infrared spectrometry (NIR), etc.8,9 The detection methods for GMO presence in food are versatile. However, most of them need to be performed in a laboratory. The consequences of relying on the slow central laboratory approach are that long time period is needed before the lab results are made available. An ideal identification technique should be rapid, easy to use and most importantly, suitable for on-site analysis.PCR methods are the most common and generally accepted detection methods for identifying the presence of GMO.10 These methods generally need to amplify the transgene with known sequences and compare the amplified fragment with that of the corresponding reference gene to obtain reliable results. The reported methods are sensitive, qualitative or quantitative, specific and precise, but there are also some disadvantages including high cost, difficulty to use, special needs, long duration, and so on.11 One solution was the application of an affordable, rapid, and easy to use portable PCR device to perform rapid DNA amplification.12 For identification of amplified products, agarose gel electrophoresis, which separates DNA fragments based on their size differences, is generally used. Some new DNA detection techniques with the advantages of high throughput, better specificity, and higher resolution have been developed in addition to agarose gel electrophoresis. However, they require expensive instruments with special software, such as a real-time PCR machine, microarray scanner, and capillary electrophoresis apparatus, etc. The visual detection of DNA amplification results is much more convenient for on-site analysis.13
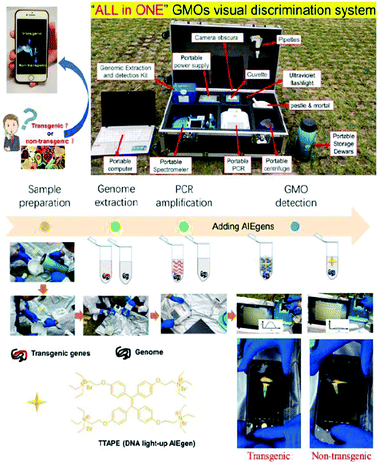
A new class of fluorescent materials with aggregation-induced emission (AIE) properties have been developed by Tang's group since 2001.14 Opposite to the photophysical phenomenon of aggregation caused quenching (ACQ) dyes, AIEgens are almost non-emissive in the molecularly dissolved state but emit intensely in the aggregated state due to the restriction of intramolecular motions and prohibition of energy dissipation via non-radiative decay channels. This unique property made easy construction of light-up/turn-on bioprobes possible by taking advantage of the AIE process. Under appropriate conditions, an AIE-based fluorescent probe luminesces dimly with very low background noise. Once it is interacted with target bioanalytes, the intramolecular motions of the AIEgen are restricted, and the lighted-up fluorescence signal can thereby be detected in a sensitive manner. Furthermore, bioprobes based on AIEgens can visualize the organelles, cells, bacteria, tissues, etc., with high contrast and superb spatial resolution and without the need for washing.15–20 However, to the best of our knowledge, there have been few reports on GMO detection based on AIEgens. A series of cationic TPE derivatives with responsive ability to light up the negatively charged nucleic acids with a complex 3D structure in solution have been developed, which inspired us to develop a visual detection approach for GMO detection. In this work, we developed an “ALL in ONE” GMO visual discrimination system through the combination of a portable device (PCR, centrifuge, spectrometer, etc.) and DNA light-up AIEgens (1,1,2,2-tetrakis[4-(2-bromo-ethoxy)phenyl]ethene, TTAPE).21–23 It is called “ALL in ONE” because all portable equipment could be packed in one bag, and all transgenic food discrimination experiments could be carried out on the field with the help of portable power. It should be pointed out that it is the first attempt using AIE materials for on-site GMO detection.
Results and discussion
Principles
As shown in Scheme 1, the “ALL in ONE” GMO visual discrimination system consists of a genomic extraction kit, detection kits, portable power supply, portable centrifuge, portable PCR device, portable spectrometer, portable storage dewar, portable computer, and a mortar and pestle. TTAPE (the mass spectrometry results could be found in Fig. S1, ESI†) showed the best performance towards a guanine-rich DNA strand (e.g., G-quadruplex structure formed by human telomeric DNA sequences) via electrostatic attraction due to the perfect structural match. Therefore, it is reasonable to anticipate that the direct visualization of transgenic food may be enabled by using TTAPE in combination with the PCR. The whole experiment could be carried out in the field. Firstly, the DNA was extracted from the samples. Different from the non-transgenic samples, there are transgene sequences in the genome of transgenic samples. The number of transgene fragments is exponentially amplified in PCR cycles upon the addition of the corresponding primer, which is further stained with TTAPE. When TTAPE is electrostatically bound to a DNA strand (G1), its intramolecular rotation is restricted, and a bright emission can be activated.Due to the large amounts of PCR products, the fluorescence from the sample can be directly observed by the naked eye or recorded by a smartphone. In contrast, the emission was very low and could not be clearly visualized for the non-transgenic sample. The UV-vis spectral changes of AIEgen before and after aggregation were also measured and the results showed only small changes (Fig. S2, ESI†).
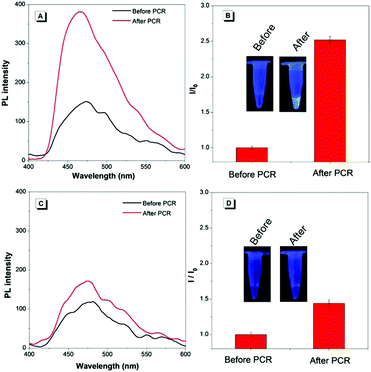
As a proof of concept, the PL (photo-luminescence) intensities of the transgenic and non-transgenic samples were obtained and compared before and after the PCR, respectively. As shown in Fig. 1A and B, for the transgenic sample, the PL intensity increased about 2.5 times after the PCR. Hence visual discrimination can be clearly made. However, the PL intensity of the non-transgenic sample showed only a small change after the PCR, which could not be discriminated visually (Fig. 1C and D). The results showed that the visual method was specific for transgenes.
Method optimization
The effect of the number of PCR cycles and TTAPE concentration on the performance of our method was studied. It's known that the amount of DNA increases exponentially during the PCR process.24 As a result, the emission was enhanced with the increase in the amount of DNA combined with TTAPE. Along with the increase in the number of PCR cycles (with 5 cycles as increments), the relative intensity increased (Fig. 2). The emission intensity was also enhanced with the increase in the amount of TTAPE. To make a compromise between analysis time and final visual results, 500 μM TTAPE was taken as the final concentration, and 25 PCR cycles were adopted. We have conducted an interference experiment. The results showed that the negatively charged compounds interfere little with detection. They can interact with positively charged TTAPE, but the emission was low (Fig. S3, ESI†). The visual discrimination can only be achieved with a large number of genes copies (long chain DNA) produced after the PCR process. As shown in Fig. S4 (ESI†), the PL intensity increased sharply from 0 to 400 μM and kept increasing between 400 μM to 1500 μM. The images taken by the camera in the Gel Imaging System were consistent with fluorescence measurement data (inset). Besides, the stability of visual discrimination results under white light was also evaluated. It showed that the discrimination results can be stable for longer than 48 h under white light (Fig. S5, ESI†). The results confirmed that our proposed method is very stable for real sample analysis. As one of the water-soluble DNA-binding AIEgens, TTAPE showed high stability and shortened the number of PCR cycles due to the typical aggregation induced emission behaviors. Compared to existing DNA-binding dyes with an ACQ effect, which could be used only in dilute solutions, TTAPE showed better performance (Fig. S6, ESI†).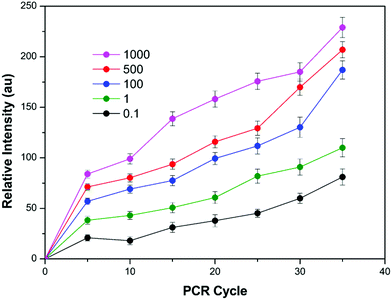 | ||
| Fig. 2 The relative intensity under different PCR cycles and different concentrations of TTAPE. The results were obtained with the portable spectrometer. | ||
We further evaluated the influence of the addition of TTAPE on the PCR process, which had little effect (Fig. S7, ESI†). It is ascribed that the restriction induced emission of TTAPE was driven by the electrostatic interaction between the positive ammonium group in TTAPE and the negative phosphate backbones of DNA. In contrast, the PCR process might be eliminated for the other nucleic acid dyes inserting themselves into the spaces between the base pairs of the double helix.25 Therefore, TTAPE showed much more potential in the real-time PCR process.
Method validation
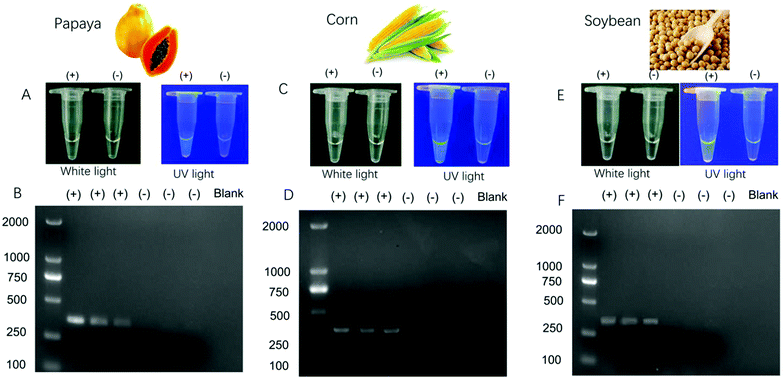
To further evaluate the versatility of our method, two more samples with different transgenic sequences were tested. As seen in Fig. 3, the three kinds of transgenic samples can be visually discriminated, which was validated by agarose gel electrophoresis. A DNA fragment of 369 bp was found in papaya, and a DNA fragment of 333 bp for transgenes in corns and soybeans. The proposed method showed great potential for in situ and on-site discrimination of transgenic samples in comparison to conventional agarose gel electrophoresis with the aid of a portable PCR device. In addition, we have conducted a limit of detection assay for contaminated seeds to define the limit of the lowest proportion of GM contamination that this method could detect. As shown in Fig. S8 (ESI†), even if only 0.01 wt% of the transgenic sample is mixed in the real sample, it could still be detected using our proposed method.Conclusions
A convenient on-site and visual discrimination method for transgenic food was developed based on water-soluble DNA-binding AIEgens. The results indicated that the visual method was specific for transgenes. The discrimination can be stable as long as 48 h under white light. With the aid of a small-scale, portable PCR instrument, it's convenient to apply this method for rapid and on-site analysis. Given its simplicity, easy operation, sensitivity, and cost effectiveness, this method can be extended to diagnosis of genetic diseases and infection monitoring in relevant studies. The ALL in ONE system could be further miniaturized through the integration of the PCR device, spectrometer and AIEgens to accomplish point-of-care detection of GMO. More importantly, it should be pointed out that this is the first attempt using AIEgens along with portable devices to develop an on-site visual discrimination approach. It is a fact that the detection time seemed to be a little long (∼2 hours); however, we believe that the detection time could be reduced by integration with a microfluidic system in which the whole process can take place. We are trying to develop more on-site visual discrimination approaches based on AIEgens with various responsive abilities and push AIEgens to real analytical applications.Experimental section
Materials and methods
Transgenic papayas were kindly provided by South China Agricultural University. Transgenic corn and beans were kindly provided by Tianjin Centres for Disease Control and Prevention. The EZ-10 Spin Column Plant Genomic DNA Purification Kit (50 PREPS, No. B518261), Taq PCR Mix (2X, without dye, No. B639293), 6X Glycerol Gel Loading Buffer II (with Xylene Cyanol, BPB, Tris–HCl, EDTA) (No. B548314) and DNA Marker D (250 PREPS, No. B600335) were purchased from Sangon Biotech (Shanghai, China). The primers for Rep genes in transgenic papaya are REP-F: 5′-CTTTGGTGCGGAAAAGTTGT-3′, REP-R: 5′-CCATAGCCCACAGTCGAATT-3′;7 the primers for CP4-epsps in transgenic corn and transgenic soybean are MCP4ES-F: 5′-ACGGTGAYCGTCTTCCMGTTAC-3′, mCP4ES-R: 5′-GAACAAGCARGGCMGCAACCA-3′.26 They were also purchased from Sangon Biotech (Shanghai, China). The PCR thermocycling process was carried out in the field using a portable thermocycling system (Leopard MINI PCR, Leopard scientific instruments Co. Ltd, China). The fluorescence of the samples was measured using a portable fluorescence measurement system (LIFS-405, Guangzhou BiaoQi Optoelectronics Technology Development Co., Ltd). The details of the portable detection system could be found in Scheme 1.Extraction of genomic DNA and PCR conditions
100 mg of the sample was ground in liquid nitrogen using a mortar and pestle. DNA was extracted following the manufacturer's specifications with minor modifications. Briefly, the buffer PCB and β-mercaptoethanol were added and vibrated for 25 min, then chloroform was added and the upper layer was transferred to a clean centrifuge tube. Then buffer and ethanol were added and transferred to the Ezup column. The column was washed twice with wash solution, and TE buffer was finally added to centrifuge the mixture for 2 min to obtain the final genomic DNA. The PCR was performed with a portable thermocycling system. For the transgenes in papaya, all PCR procedures were performed in a final volume of 30 μL with the following reagent concentrations: genomic DNA 120 ng, primer mix of REP F and REP R (6 μL, 10 μM), Taq PCR Mix 15 μL. Thermal cycler conditions were as follows: initial denaturation at 95 °C for 3 min; 25 cycles consisting of dsDNA denaturation at 94 °C for 30 s; primer annealing at 57 °C for 30 s; primer extension at 72 °C for 60 s; and final elongation at 72 °C for 7 min. For the transgenes in soybean and corn, PCR procedures were performed in a final volume of 30 μL with the following reagent concentrations: genomic DNA 120 ng, primer mix of mCP4ES-F and mCP4ES-R (6 μL, 10 μM), Taq PCR Mix 15 μL. Thermal cycler conditions were as follows: initial denaturation at 94 °C for 3 min; 25 cycles consisting of dsDNA denaturation at 94 °C for 30 s; primer annealing at 62 °C for 30 s; primer extension at 72 °C for 60 s; and final elongation at 72 °C for 5 min. TTAPE at a concentration of 500 μM was added following the PCR.Agarose gel electrophoresis
PCR products were analyzed using agarose gel electrophoresis (Beijing Liuyi biotechnology Co. Ltd, China). The gel was prepared with 2.0% of agarose in Tris Borate EDTA (TBE). 100 μM of ethidium bromide (EtBr) was used. The running conditions were constant voltage at 100 V for 50 min in TBE. The image was taken using a Tanon 1600/1600R Gel Imaging System (Shanghai, China).Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Science and Technology Planning Project of Guangdong Province of China (2018B020208005), the National Natural Science Foundation of Guangdong Province of China (2016A030313136) and the Guangdong Provincial Key Platform and Major Scientific Research Projects for Colleges and Universities (2015KCXTD029). The authors would like to acknowledge Prof. Dr Ryan T. K. Kwok for his suggestions on manuscript writing. We also acknowledge the technical support received from Thonebio. Co. Ltd, AIEgen Biotech Co., Ltd, and Mr Tianwei Chu from Guangzhou BiaoQi Optoelectronics Technology Development Co., Ltd. The authors would like to acknowledge Dr Yahui Zhang for her contribution on the figure artwork.Notes and references
- H. A. Kuiper, G. A. Kleter, H. P. J. M. Noteborn and E. J. Kok, Plant J., 2001, 27, 503 CrossRef CAS PubMed
.
- A. König, A. Cockburn, R. W. R. Crevel, E. Debruyne, R. Grafstroem, U. Hammerling, I. Kimber, I. Knudsen, H. A. Kuiper, A. A. C. M. Peijnenburg, A. H. Penninks, M. Poulsen, M. Schauzu and J. M. Wal, Food Chem. Toxicol., 2004, 42, 1047 CrossRef PubMed
.
- K. Rudi, I. Rud and A. Holck, Nucleic Acids Res., 2003, 31, e62 CrossRef PubMed
.
- G. Liu, W. Su, Q. Xu, M. Long, J. Zhou and S. Song, Food Control, 2004, 15, 303–306 CrossRef CAS
.
- R. Bordoni, A. Germini, A. Mezzelani, R. Marchelli and G. D. Bellis, J. Agric. Food Chem., 2005, 53, 912 CrossRef CAS PubMed
.
- Z. Jiao, X. X. Si, Z. M. Zhang, G. K. Li and Z. W. Cai, Food Chem., 2012, 135, 285 CrossRef CAS
.
- Z. Jiao, J. C. Deng, G. K. Li, Z. M. Zhang and Z. W. Cai, J. Food Compos. Anal., 2010, 23, 640–647 CrossRef CAS
.
- L. J. Xie, Y. B. Ying, T. J. Ying, H. Y. Yu and X. P. Fu, Anal. Chim. Acta, 2007, 584, 379 CrossRef CAS PubMed
.
- Z. Jiao, X. X. Si, G. K. Li, Z. M. Zhang and X. P. Xu, J. Agric. Food Chem., 2010, 58, 1746 CrossRef CAS PubMed
.
- Y. Zhao, F. Chen, Q. Li, L. Wang and C. Fan, Chem. Rev., 2015, 115, 12491 CrossRef CAS PubMed
.
- W. Xu, L. Xie, Z. Ye, W. Gao, Y. Yao, M. Chen, J. Qin and Y. Ying, Sci. Rep., 2015, 5, 11115 CrossRef PubMed
.
- P. Belgrader, S. Young, B. Yuan, M. Primeau, L. A. Christel, F. Pourahmadi and M. A. A. Northrup, Anal. Chem., 2001, 73, 286 CrossRef CAS PubMed
.
- Y. Cao, K. Zheng, J. Jiang, J. Wu, F. Shi, X. Song and Y. Jiang, Food Chem., 2018, 266, 73 CrossRef CAS PubMed
.
- J. D. Luo, Z. L. Xie, J. W. Y. Lam, L. Cheng, H. Y. Chen, C. F. Qiu, H. S. Kwok, X. W. Zhan, Y. Q. Liu, D. B. Zhu and B. Z. Tang, Chem. Commun., 2001, 1740 RSC
.
- J. Mei, N. L. C. Leung, R. T. K. Kwok, J. W. Y. Lam and B. Z. Tang, Chem. Rev., 2015, 115, 11718 CrossRef CAS PubMed
.
- J. Liang, B. Z. Tang and B. Liu, Chem. Soc. Rev., 2015, 44, 2798 RSC
.
- J. Mei, Y. H. Huang and H. Tian, ACS Appl. Mater. Interfaces, 2018, 10, 12217 CrossRef CAS PubMed
.
- M. Gao and B. Z. Tang, ACS Sens., 2017, 2, 1382 CrossRef CAS PubMed
.
- X. G. Gu, R. T. K. Kwok, J. W. Y. Lam and B. Z. Tang, Biomaterials, 2017, 146, 115 CrossRef CAS PubMed
.
- J. Qian and B. Z. Tang, Chem. Rev., 2017, 3, 56 CAS
.
- Y. N. Hong, M. Häußbler, J. W. Y. Lam, Z. Li, K. K. Sin, Y. Q. Dong, H. Tong, J. Z. Liu, A. J. Qin, R. Renneberg and B. Z. Tang, Chem. – Eur. J., 2008, 14, 6428 CrossRef CAS PubMed
.
- M. Wang, D. Q. Zhang, G. X. Zhang, Y. L. Tang, S. Wang and D. B. Zhu, Anal. Chem., 2008, 80, 6443 CrossRef CAS PubMed
.
- X. D. Lou, Y. Zhuang, X. L. Zuo, Y. M. Jia, Y. N. Hong, X. H. Min, Z. Y. Zhang, X. M. Xu, N. N. Liu, F. Xia and B. Z. Tang, Anal. Chem., 2015, 87, 6822 CrossRef CAS PubMed
.
- J. Li, T. Deng, X. Chu, R. Yang, J. Jiang, G. Shen and R. Yu, Anal. Chem., 2010, 82, 2811 CrossRef CAS PubMed
.
- S. Sindbert, S. Kalinin, H. Nguyen, A. Kienzler, L. Clima, W. Bannwarth and C. A. Seidel, J. Am. Chem. Soc., 2011, 133, 2463 CrossRef CAS PubMed
.
- Detection of genetically modified plants and derived products-qualitative PCR method for CP4-epsps gene. No. 1861 Announcement of the Ministry of Agriculture of the People's Republic of China.
Footnote |
| † Electronic supplementary information (ESI) available: The PL intensity of TTAPE at different concentrations, stability of visual discrimination, and the influence of TTAPE on the PCR process. See DOI: 10.1039/c9qm00279k |
| This journal is © the Partner Organisations 2019 |