Novel target NIR-fluorescent polymer for living tumor cell imaging†
Yanjie
Zhao
,
Yan
Wu
,
Bai
Xue
,
Xin
Jin
 * and
Xinyuan
Zhu
* and
Xinyuan
Zhu

School of Chemistry and Chemical Engineering, State Key Laboratory of Metal Matrix Composites, Shanghai Jiao Tong University, 800 Dong Chuan Road, Shanghai 200240, P. R. China. E-mail: jxcindy@sjtu.edu.cn; Tel: +86-21-34203400
First published on 6th November 2018
Abstract
As is well known, amphipathic polymers play a great role in drug delivery and bioimaging fields. However, the hydrophilic part always results in the poor internalization ability to tumor cells for drug delivery and bioimaging. Here, we designed and synthesized a new type of NIR-fluorescence polymer, PMMA-b-P(GATH-co-BOD), glucose-based hydrophilic segment conjugated with an NIR-fluorescence agent. Due to its amphiphilicity, the copolymer could self-assemble into micelles, with polyhydroxy glucose as the shell and a PMMA chain as the core. Such strategy provided high glucose density on the surface of the micelles. During cell internalization, the cluster glycoside effect will provide stronger recognition towards GLUT1 and act as an internal driving force. The copolymer PMMA-b-P(GATH-co-BOD) exhibited excellent advantages including low cytotoxicity and high targeting specificity for tumor cells. This near-infrared copolymer fluorescent probe system initiated a broad platform for drug delivery and bioimaging detection systems.
Introduction
Over the past few decades, polymer micelle systems have emerged as an attractive nanocarrier for the delivery of various types of bioimaging probe. As a promising vehicle, polymeric nanocarriers display considerable advantages, including favorable water solubility, long circulation time, and a passive enrichment ability for cancer through the EPR effect.1–9 Nevertheless, there are still some limitations of using polymeric nanocarriers for cancer detection, including inferior cancer cell targeting and poor internalization efficiency.10–13 Introducing active targeting ligands is a prominent approach with advantages of enhanced cell-uptake as well as being able to precisely locate tumor cells.14–17 Various active targeting ligands have been integrated into the surface of polymer nanoparticles, such as peptides including RGD, CPP, NLS, aptamers and small molecule folic acid and so on.18–23 These ligands usually possess specific binding abilities towards certain overexpressed receptors on the membrane of cancer cells.24–28 Nevertheless, most ligands mentioned above favor a hydrophobic environment during receptor identification, not a perfect choice for targeted ligands in polymeric nanocarriers for cancer therapy. Therefore, it is a pressing issue to find some hydrophilically adaptable active targeting ligands to construct efficient targeting polymeric nanocarriers for cancer treatment.D-Glucose is a significant essential nutrient and energy source for beings. As a strong hydrophilic molecule, D-glucose enters cells through an energy-independent active diffusion process with glucose transporters (GLUT) as mediators instead of by free diffusion.29 Furthermore, the D-glucose expenditure of the tumor cells is much greater than in normal cells, caused via the inefficient sugar degradation of tumor cells, which has been well-documented and named the “Warburg effect”.30,31 Based on the “Warburg effect” theory, in order to obtain massive D-sugars for rapid growth and proliferation, GLUT is markedly over-expressed in most tumor cells.29,32 Until now, at least 14 human GLUTs have been discovered and GLUT1 is the most crucial and extensively researched one, and is often considered a potential tumor target receptor.33–38 Thus, various cancer-targeted glycoconjugates based on the GLUT1 receptor have been designed and developed.39–51 For example, Eric Rohren et al. reported a radiolabeled glucose derivative 18F-FDG for the diagnostic therapy of cancer to detect the malignant involvement of peripheral nerves, and the tracer is frequently applied for the clinic diagnosis of cancers.40 Additionally, more cancer-targeted glycoconjugates consisting of cancer therapeutics and D-glucose have been designed and prepared.52–59
Although glycoconjugates based on the GLUT1 receptor are widely used and have led to large achievements, there are still some insufficiencies limiting their application, including complicated chemical reactions where numerous reactive sites need to be formed via organic synthesis. Difficult purification processes such as chromatography are inefficient, lead to time-wasting and allow small-scale purification of glycoconjugates with unsatisfactory targeting abilities. Herein, we developed a novel polymeric nanocarrier for cancer detection by NIR imaging via two-step RAFT polymerization. Compared with glycoconjugates, the synthetic process was simple and convenient, and the purification process was simple, efficient and achievable on a large-scale via the precipitation of diethyl ether. In this system, glucose was both a hydrophilic block as well as the target ligand of GLUT1. For this purpose, a polymerizable glucose monomer GATA and clickable monomer were designed and prepared separately. Hydrophobic block PMMA was first obtained using RAFT polymerization. Copolymerization of GATA and the clickable monomers was then conducted using PMMA as macro chain transfer agents. An NIR agent was further conjugated to the copolymer block through click chemistry. Such a strategy provided high glucose density on the surface of the nanocarrier. During cell internalization, the cluster glycoside effect provided stronger recognition towards GLUT1, thus was the internal driving force compared with single molecular glucose.60,61 It provides a capacious platform for the bioimaging of tumors.
Experimental
The experimental materials and measurement methods are shown in the ESI.†Synthetic procedures
Detailed synthetic procedures and characteristic spectra of the micromolecules and polymers are shown in the ESI.†Results and discussion
Synthesis and characterization
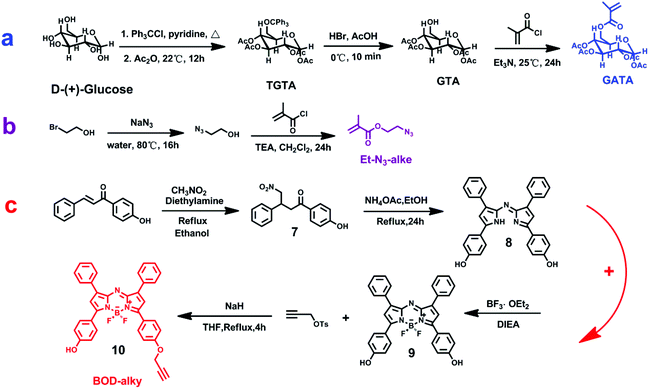
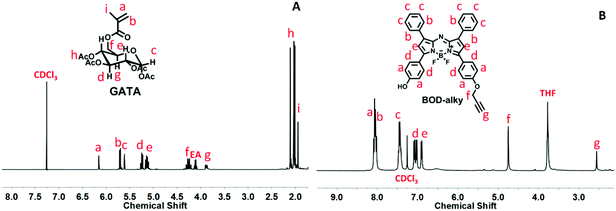
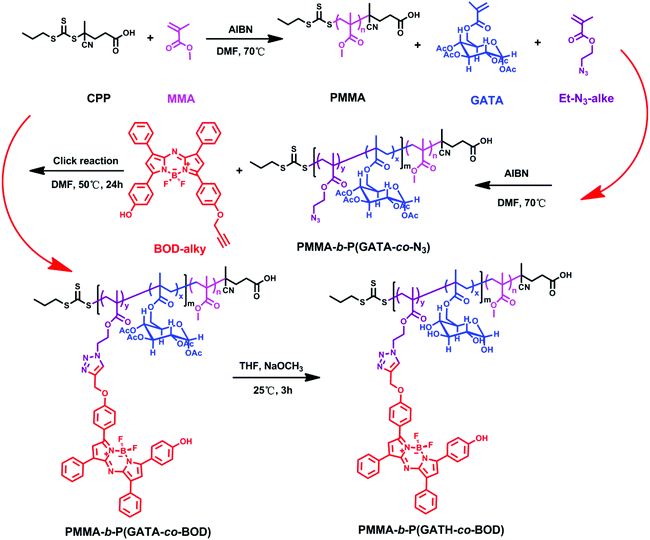
The monomers and near-infrared fluorophore in the experiment have been designed, prepared and confirmed via Chem-NMR, HRMS and FTIR. The detailed synthetic procedure is displayed in Scheme 1, and the detailed results are shown in the ESI.† The polymerizable glucosyl-monomer (GATA) was synthesized via a three-step procedure (Scheme 1a). TGA was obtained by protecting all hydroxyls in the D-(+)-glucose, and after removing triphenylmethyl protection, GTA was successfully obtained. The polymeric monomer GATA was then obtained via nucleophilic substitution with methacryloyl chloride. The clickable monomer, 2-azidoethyl methacrylate, was obtained by a two-step procedure (Scheme 1b). The 2-azidoethanol was obtained by a substitution reaction where the Br group of 2-bromoethanol was replaced by an N3 group. Then the other monomer, 2-azidoethyl methacrylate, was successfully obtained via nucleophilic substitution with methacryloyl chloride. The clickable near-infrared fluorophore BOD-alky was successfully synthesized via a complex four-step procedure (Scheme 1c). Compound 7 was obtained by an addition reaction of 4-hydroxyphenyl chalcone and nitromethane. Then compound 8, a bis-phenol-substituted azadipyrromethene, was obtained via the reflux operation of compound 7 and NH4Ac in C2H5OH for 24 h. After a chelating reaction of compound 8 with BF3-diethyletherate, the BF2-chelated derivative, compound 9, was obtained. Finally, the clickable compound 10, BOD-alky, was obtained via the nucleophilic substitution of compound 9 with propargyl tosylate under the catalysis of NaH in THF for 6 h under reflux conditions. The 1H NMR spectrum of monomer GATA is shown in Fig. 1A, the characteristic peaks of GATA are at δ (ppm) = 6.16 (m, 1H), 5.72 (d, 1H), 5.61 (t, 1H), 5.25 (t, 1H), 5.07–5.15 (m, 2H), 4.21–4.32 (m, 2H), 3.85–3.92 (m, 1H), 1.93–2.15 (m, 12H), and 1.94 (m, 3H). The 1H NMR spectrum of the near-infrared fluorophore BOD-alky is shown in Fig. 1B, the characteristic peaks of BOD-alky are at δ (ppm) = 7.95–8.10 (m, 8H), 7.38–7.48 (m, 6H), 7.01–7.09 (dd, 4H), 6.89 (d, 2H), 4.75 (s, 2H), and 2.56 (s, 1H).The polymer PMMA was prepared via RAFT polymerization, with CPP as the chain transfer agent. PMMA was obtained by adjusting the ratio of CPP, AIBN and monomer MMA. Then, PMMA was the chain transfer agent, and the polymer PMMA-b-P(GATA-co-N3) was obtained by adjusting the ratio of PMMA, AIBN and monomers GATA and 2-azidoethyl methacrylate. PMMA-b-P(GATA-co-BOD) was prepared via a click reaction with BOD-alky. Finally, polymer PMMA-b-P(GATH-co-BOD) was prepared by removing acetyl from PMMA-b-P(GATA-co-BOD). The detailed synthetic procedure is displayed in Scheme 2.
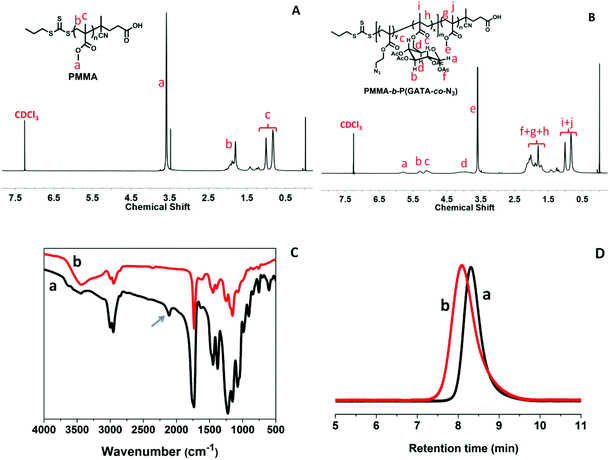
The production was determined by 1H NMR, GPC and FTIR, and the results are shown in Fig. 2. The characteristic peaks of PMMA are at δ (ppm) = 3.58 (s, –OCH3 of PMMA), 1.80 (m, –CH2– of PMMA), and 1.00 and 0.83 (s, –C(CH3)CH2 of PMMA). For PMMA-b-P(GATA-co-N3) δ = 5.7–5.9, 5.3–5.5, 5.0–5.2, 3.7–4.2 are the characteristic peaks of PGATA, and δ = 1.53–2.5 (–CH2– of PMMA and PGATA, –OAc), and 1.2–0.80 (m, –C(CH3)CH2 of PMMA and PGATA, –CH3). The signals of the BOD-alky connected to the copolymer are undetectable in the NMR spectrum due to its low content. However, as shown in Fig. 2C, the missing characteristic peak of the azide group at 2109 cm−1 in the FTIR spectrum confirms the successful connection of the BOD-alky to the copolymer. After the click reaction, the polymer PMMA-b-P(GATH-co-BOD) was obtained by removing acetyl from PGATA with NaOCH3 methanol solution. As shown in Fig. S11,† the disappearance of acetyl signals of PMMA-b-P(GATH-co-BOD) at δ = 2.27–1.58 after adding NaOCH3 methanol solution confirms the successful deprotection and removal of acetyl from polymer PMMA-b-P(GATA-co-BOD).
The polydispersity index (PDI) and molecular weight of the polymer were calculated by GPC measurements. The results are displayed in Fig. 2D and Table 1. The Mn of polymers PMMA and PMMA-b-P(GATA-co-N3) is 15.5 kDa and 19.3 kDa, and the PDI is 1.23 and 1.40. Thus, calculated by GPC data, the repeating units of MMA and GATA of the block polymers are determined to be 155 and 10 respectively.
| Polymer | M n (×103 g mol−1) | M w (×103 g mol−1) | PDI |
|---|---|---|---|
| PMMA | 15.5 | 19.0 | 1.23 |
| PMMA-b-P(GATA-co-N3) | 19.3 | 27.0 | 1.40 |
Self-assembly
The self-assembly behavior of amphiphilic polymers is greatly related to the hydrophilic segment and hydrophobic portion, which play a critical role in the polymer properties. In the designed structure, PMMA is the hydrophobic portion and the copolymer segment is the hydrophilic portion. Due to there being plenty of hydroxyls in the glucose, the diblock copolymer PMMA-b-P(GATH-co-BOD) can self-assemble into micelles, where the glucose segment is the shell and the PMMA chain is the core of the micelle. The morphology and size of the micelles were tested via TEM and DLS (Fig. 3). TEM images show that the shape of the PMMA-b-P(GATH-co-BOD) micelle is homogeneously spherical and the diameter is about 40 nm. However, when measured by DLS it was about 110 nm. The hydrodynamic diameter from DLS is usually larger than the dried size from TEM. In this case, the difference seems larger than usual, probably because glucose always shows super strong hydrogen bond formation and has a rigid structure in an aqueous system.62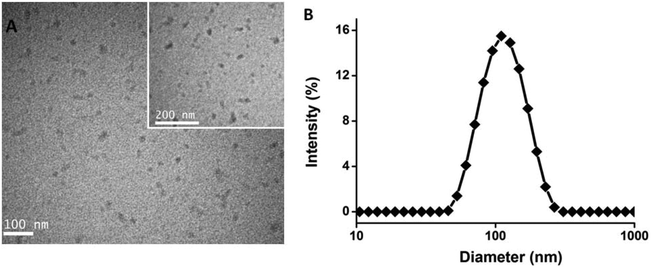 | ||
| Fig. 3 TEM pictures of the polymer micelles (A); the size distribution of polymer micelles measured via DLS (B). | ||
Fluorescence spectroscopy
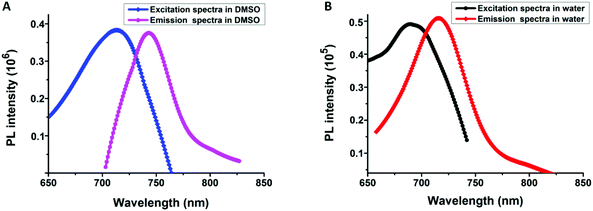
Using fluorescence detection for various kinds of biological and chemical analysis has drawn much research attention. For the fluorescence family, near-infrared fluorophores occupy a significant place in biological detection and imaging, owing to their minimal damage to cells and deeper tissue penetration.63–66The excitation and emission spectra of diblock copolymer PMMA-b-P(GATH-co-BOD) were studied in detail and are shown in Fig. 4. In a good solvent of DMSO, the maximum absorption peak of PMMA-b-P(GATH-co-BOD) was at about 713 nm, attributed to n–π* excitations of the fluorophore BOD-alky. After excitation at 713 nm, emission appears at 742 nm with a bright red color. However, in aqueous solution, the maximum absorption and emission peaks of PGATH-b-P(DPA-co-BOD) were blue-shifted to 692 nm and 716 nm, caused by the solvatochromic effect. Therefore, the PMMA-b-P(GATH-co-BOD) micelles could be used for NIR-biological imaging.
Cell cytotoxicity
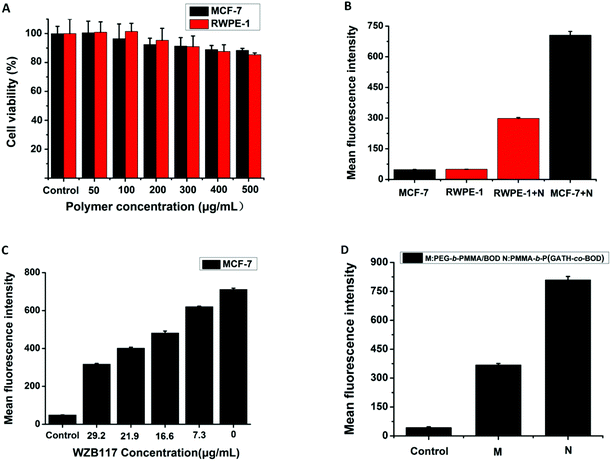
The cytotoxicity of copolymer PMMA-b-P(GATH-co-BOD) against both MCF-7 cells and RWPE-1 cells was studied by an MTT assay (Fig. 6A). The different concentrations of polymer micelles were cultured with MCF-7 cells and RWPE-1 cells, respectively for 48 h. The range changed from 50 μg mL−1 to 500 μg mL−1. Whatever the concentration conditions of the micelles, up to 80% of these two types of cell were still alive after 48 h incubation. These results showed that the copolymer PMMA-b-P(GATH-co-BOD) displayed low cytotoxicity towards both types of cell. Thus, this material exhibited good cytocompatibility, which is important for cell imaging and biological application.Cell imaging
A cell imaging experiment on two types of cell was investigated via CLSM. MCF-7 cells and RWPE-1 cells were simultaneously incubated with the same concentration of PMMA-b-P(GATH-co-BOD) micelles for 1.5 h. Then, the cell nucleus dye Hoechst 33342 was added for another 30 min. After that, PBS was employed to wash the cell dishes 3 times. After washing, cell imaging was observed, as shown in Fig. 5. The blue color is the cell nucleus dyed by Hoechst 33342 with an excitation channel of 405 nm. The red fluorescence is the copolymer PMMA-b-P(GATH-co-BOD) with an excitation channel of 633 nm. The cell imaging was conducted at room temperature. The bright red fluorescence was noticed in the MCF-7 tumor cells, and was markedly more powerful than in the normal RWPE-1 cells. The reason might be related to GLUT1. Due to the “Warburg effect”, GLUT1 is over-expressed in MCF-7 tumor cells to maintain rapid growth and proliferation compared with normal RWPE-1 cells. The copolymer PMMA-b-P(GATH-co-BOD) can self-assemble into micelles, where the polyhydroxy glucose segment is the shell. Such strategies provide a high glucose density on the surface of the micelles. During cell internalization, the cluster glycoside effect will provide stronger recognition towards GLUT1 and provide an internal driving force. Due to over-expression in the tumor cells, the uptake ability of the MCF-7 cells was obviously stronger than that of the RWPE-1 cells, thus, the red fluorescence of the MCF-7 cells was markedly brighter than the RWPE-1 cells.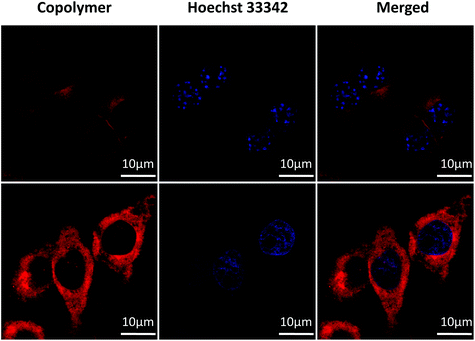 | ||
| Fig. 5 Cell images of both MCF-7 cells and RWPE-1 cells, concentration = 50 μg mL−1, blue channel: λex = 405 nm, red channel: λex = 633 nm. | ||
Flow cytometry analysis
To further confirm the cellular uptake ability of the two types of cell, flow cytometry analysis was employed to test the fluorescence intensity of PMMA-b-P(GATH-co-BOD) micelles being internalized by MCF-7 and RWPE-1 cells, respectively. The results are shown in Fig. 6B, the average fluorescence intensity after incubation with MCF-7 cells is markedly higher than with RWPE-1 cells. The results of the flow cytometry analysis show a trend which is consistent with the cell imaging.To further prove that the active targeting ability for the MCF-7 cells of PMMA-b-P(GATH-co-BOD) micelles was caused by GLUT1, the special GLUT1 inhibitor, WZB117, was incubated with MCF-7 cells for 30 min. Then, it was incubated with added PMMA-b-P(GATH-co-BOD) micelles for another 2 h. The results were evaluated by flow cytometry analysis and are shown in Fig. 6C. With the concentration of WZB117 increasing constantly, the mean fluorescence intensity of the PMMA-b-P(GATH-co-BOD) micelles internalized by MCF-7 cells decreased markedly. The reason might be that due to being inhibited by WZB117, GLUT1 could not easily play the role of especially recognizing the glucose shell of the micelles. Thus, the internalization ability of the MCF-7 cells for micelles gradually weakened. This result again proved that the active targeting ability of the PMMA-b-P(GATH-co-BOD) micelles for MCF-7 cells was mainly mediated by GLUT1.
To further prove that the active targeting ability for the MCF-7 cells of PMMA-b-P(GATH-co-BOD) micelles was caused by GLUT1, flow cytometry analysis was employed to test the fluorescence intensity of the PMMA-b-P(GATH-co-BOD) and PEG-b-PMMA/BOD micelles being internalized by MCF-7 cells, respectively. The results are shown in Fig. 6D, and the average fluorescence intensity of PMMA-b-P(GATH-co-BOD) incubated with MCF-7 cells was markedly higher than that of PEG-b-PMMA/BOD. The results of flow cytometry analysis showed that the polymer PMMA-b-P(GATH-co-BOD) micelles markedly targeted MCF-7 cells due to GLUT1.
Conclusions
In summary, a novel NIR-diblock copolymer based on a GLUTI targeting ability was successfully prepared via RAFT polymerization. Due to the inherent amphiphilicity, PMMA-b-P(GATH-co-BOD) could self-assemble into micelles with hydrophilic polyhydroxy glucose as the shell and PMMA as the core. Such a strategy provided a high glucose density on the surface of the micelles. During cell internalization, the cluster glycoside effect will provide a stronger recognition towards GLUT1 which acts as an internal driving force. Thus, PMMA-b-P(GATH-co-BOD) micelles show higher internalization efficiency against GLUT1 over-expressing cells (MCF-7 cells) than to normal cells (RWPE-1 cells). Further flow cytometry analysis results show that with an increase of GLUT1 inhibitor (special inhibitor WZB117) concentration, the amount of micelles internalized by MCF-7 cells markedly decreases. This result again proves that the active targeting ability of PMMA-b-P(GATH-co-BOD) micelles for MCF-7 cells is mainly mediated by GLUT1. A low toxicity of the micelles has been demonstrated by further cytotoxicity experiments. Overall, we believe that the novel NIR-diblock copolymer PMMA-b-P(GATH-co-BOD), with efficient cancer targeting abilities and excellent biocompatibility, synthesized in this study may pave a broad way for drug delivery and bioimaging applications.Conflicts of interest
There is no conflict to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (51690151, 51503122, 51473093), the National Basic Research Program of China (2015CB931801), the Shanghai Rising-Star Program (17QC1401100), the Shanghai Municipal Government (18JC1410800), Shanghai Jiao Tong University (15X110070008, 16X100100019, 17X100040070) and the Innovation Fund (IFPM2016B009) from the Joint Research Center for Precision Medicine set up by Shanghai Jiao Tong University & Affiliated Sixth People's Hospital South Campus (Fengxian Central Hospital).Notes and references
- K. Kataoka, A. Harada and Y. Nagasaki, Adv. Drug Delivery Rev., 2001, 47, 113–131 CrossRef CAS.
- Y. Bae, S. Fukushima, A. Harada and K. Kataoka, Angew. Chem., Int. Ed., 2003, 42, 4640–4643 CrossRef CAS.
- W. M. Zhang, J. Zhang and J. Yin, Chin. J. Polym. Sci., 2018, 36, 273–287 CrossRef CAS.
- V. Torchilin, Adv. Drug Delivery Rev., 2011, 63, 131–135 CrossRef CAS.
- A. Rösler, G. W. Vandermeulen and H.-A. Klok, Adv. Drug Delivery Rev., 2012, 64, 270–279 CrossRef.
- R. Gupta, J. Shea, C. Scafe, A. Shurlygina and N. Rapoport, J. Controlled Release, 2015, 212, 70–77 CrossRef CAS.
- C. X. Zheng, Y. Zhao and Y. Liu, Chin. J. Polym. Sci., 2018, 36, 322–346 CrossRef CAS.
- Y. Matsumura and H. Maeda, Cancer Res., 1986, 46, 6387–6392 CAS.
- J. Fang, H. Nakamura and H. Maeda, Adv. Drug Delivery Rev., 2011, 63, 136–151 CrossRef CAS.
- Y. H. Bae and H. Yin, J. Controlled Release, 2008, 131, 2–4 CrossRef CAS.
- C. Deng, Y. Jiang, R. Cheng and Z. Zhong, Nano Today, 2012, 7, 467–480 CrossRef CAS.
- K. Ulbrich and V. R. Šubr, Adv. Drug Delivery Rev., 2004, 56, 1023–1050 CrossRef CAS.
- M. Ferrari, Nat. Rev. Cancer, 2005, 5, 161–171 CrossRef CAS.
- F. Danhier, O. Feron and V. Préat, J. Controlled Release, 2010, 148, 135–146 CrossRef CAS.
- Y. Bae, W.-D. Jang, N. Nishiyama and K. Kataoka, Mol. BioSyst., 2005, 1, 242–250 RSC.
- S. Jin, J. Wan, L. Meng, L. Liu and C. Wang, ACS Appl. Mater. Interfaces, 2015, 7, 19843–19852 CrossRef CAS PubMed.
- Y. Zhuang, H. Deng and X. Y. Zhu, Biomacromolecules, 2016, 17, 2050–2062 CrossRef CAS.
- Y. Zhong, F. Meng, C. Deng and Z. Zhong, Biomacromolecules, 2014, 15, 1955–1969 CrossRef CAS PubMed.
- U. Kedar, P. Phutane, S. Shidhaye and V. Kadam, Nanomedicine, 2010, 6, 714–729 CrossRef CAS PubMed.
- E. Jin, B. Zhang, X. Sun and W. J. Murdoch, J. Am. Chem. Soc., 2013, 135, 933–940 CrossRef CAS.
- S. Kunjachan, R. Pola, F. Gremse and W. E. Hennink, Nano Lett., 2014, 14, 972–981 CrossRef CAS.
- T. Chatzisideri, S. Thysiadis, S. Katsamakas and V. Sarli, Eur. J. Med. Chem., 2017, 141, 221–231 CrossRef CAS.
- J. Yin, X. G. Meng, S. B. Zhang and C. L. Liu, Biomaterials, 2012, 33, 7884–7894 CrossRef CAS.
- Z. Xiao and O. C. Farokhzad, ACS Nano, 2012, 6, 3670–3676 CrossRef CAS.
- G. Mayer, Angew. Chem., Int. Ed., 2009, 48, 2672–2689 CrossRef CAS.
- V. Bagalkot, O. C. Farokhzad, R. Langer and S. Jon, Angew. Chem., Int. Ed., 2006, 45, 8149–8152 CrossRef CAS.
- T. M. Allen, Nat. Rev. Cancer, 2002, 2, 750–763 CrossRef CAS.
- V. P. Torchilin, A. N. Lukyanov and Z. Gao, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 6039–6044 CrossRef CAS.
- M. B. Calvo, A. Figueroa, E. G. Pulido and L. A. Aparicio, Int. J. Endocrinol., 2010, 2010, 1–14 CrossRef PubMed.
- M. G. Vander Heiden, L. C. Cantley and C. B. Thompson, Science, 2009, 324, 1029–1033 CrossRef CAS.
- O. Warburg, Science, 1956, 123, 309–314 CrossRef CAS.
- L. Szablewski, Biochim. Biophys. Acta, Rev. Cancer, 2013, 1835, 164–169 CrossRef CAS.
- M. W. Pedersen, S. Holm, E. L. Lund and P. E. Kristjansen, Neoplasia, 2001, 3, 80–87 CrossRef CAS.
- A. Krzeslak, K. Wojcik-Krowiranda, E. Forma and P. Jozwiak, Pathol. Oncol. Res., 2012, 18, 721–728 CrossRef CAS.
- P. Jozwiak, A. Krzeslak, L. Pomorski and A. Lipinska, Mol. Med. Rep., 2012, 6, 601–606 CrossRef CAS.
- K. Reinicke, P. Sotomayor, P. Cisterna and A. Godoy, J. Cell. Biochem., 2012, 113, 553–562 CrossRef CAS.
- D. Deng, P. Sun, C. Yan and N. Yan, Nature, 2015, 526, 391–396 CrossRef CAS PubMed.
- D. Deng, C. Xu, P. Sun, J. Wu and N. Yan, Nature, 2014, 510, 121–125 CrossRef CAS PubMed.
- J. R. Cantor and D. M. Sabatini, Cancer Discovery, 2012, 2, 881–898 CrossRef CAS PubMed.
- Y. Bronstein, S. Tummala and E. Rohren, Clin. Nucl. Med., 2011, 36, 96–100 CrossRef.
- M. G. Vander Heiden, Nat. Rev. Drug Discovery, 2011, 10, 671–684 CrossRef CAS PubMed.
- E. C. Calvaresi and P. J. Hergenrother, Chem. Sci., 2013, 4, 2319–2333 RSC.
- W. H. Kim, J. Lee, D.-W. Jung and D. R. Williams, Sensors, 2012, 12, 5005–5027 CrossRef CAS PubMed.
- P. Som, H. L. Atkins, D. Bandoypadhyay, J. S. Fowler and S. V. Zabinski, J. Nucl. Med., 1980, 21, 670–675 CAS.
- M. L. Bowen and C. Orvig, Chem. Commun., 2008, 41, 5077–5091 RSC.
- R. Schibli, C. Dumas, J. Petrig, L. Spadola and P. A. Schubiger, Bioconjugate Chem., 2005, 16, 105–112 CrossRef CAS PubMed.
- M. M. Welling and R. Alberto, Nucl. Med. Commun., 2010, 31, 239–248 CrossRef CAS.
- T. Storr, M. Obata, C. L. Fisher and C. Orvig, Chem. – Eur. J., 2005, 11, 195–203 CrossRef PubMed.
- J. Han, X. Gao, R. Liu and Q. Gao, Chem. Biol. Drug Des., 2016, 87, 867–877 CrossRef CAS PubMed.
- D.-K. Ji, Y. Zhang, Y. Zang and X.-P. He, J. Mater. Chem. B, 2015, 32, 6676–6689 Search PubMed.
- I. S. Wood and P. Trayhurn, Br. J. Nutr., 2003, 89, 3–9 CrossRef CAS PubMed.
- Y. Chen, M. J. Heeg, P. G. Braunschweiger and P. G. Wang, Angew. Chem., Int. Ed., 1999, 38, 1768–1769 CrossRef CAS PubMed.
- P. Liu, Y. Lu, X. Gao and Q. Gao, Chem. Commun., 2013, 49, 2421–2423 RSC.
- H. Li, X. Gao, R. Liu, Y. Wang and Q. Gao, Eur. J. Med. Chem., 2015, 101, 400–408 CrossRef CAS PubMed.
- J. Möker and J. Thiem, Eur. J. Org. Chem., 2009, 4842–4847 CrossRef.
- C. G. Hartinger, A. A. Nazarov, S. M. Ashraf and B. K. Keppler, Curr. Med. Chem., 2008, 15, 2574–2591 CrossRef CAS.
- M. Patra, T. C. Johnstone, K. Suntharalingam and S. J. Lippard, Angew. Chem., Int. Ed., 2016, 55, 2550–2554 CrossRef CAS PubMed.
- J. Pohl, B. Bertram, P. Hilgard, M. R. Nowrousian and M. Wießler, Cancer Chemother. Pharmacol., 1995, 35, 364–370 CrossRef CAS PubMed.
- M. Patra, S. G. Awuah and S. J. Lippard, J. Am. Chem. Soc., 2016, 138, 12541–12551 CrossRef CAS PubMed.
- Y. C. Lee and R. T. Lee, Acc. Chem. Res., 1995, 28, 321–327 CrossRef CAS.
- J. J. Lundquist and E. J. Toone, Chem. Rev., 2002, 102, 555–578 CrossRef CAS PubMed.
- T. Jesionowski and A. Krysztafkiewicz, Appl. Surf. Sci., 2001, 172, 18–32 CrossRef CAS.
- L. Yuan, W. Y. Lin, K. B. Zheng and W. M. Huang, Chem. Soc. Rev., 2013, 42, 622–661 RSC.
- M. Kaur and D. H. Choi, Chem. Soc. Rev., 2015, 44, 58–77 RSC.
- H. W. Liu, X. B. Zhang, J. Zhang and W. H. Tan, Anal. Chem., 2015, 87, 8896–8903 CrossRef CAS PubMed.
- W. Zhu, X. M. Huang, Z. Q. Guo and H. Tian, Chem. Commun., 2012, 48, 1784–1786 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8py01442f |
| This journal is © The Royal Society of Chemistry 2019 |