Structure, properties, and biological functions of nonribosomal lipopeptides from pseudomonads†
Sebastian
Götze
 a and
Pierre
Stallforth
a and
Pierre
Stallforth
 *b
*b
aFaculty 7: Natural and Environmental Sciences, Institute for Environmental Sciences, University Koblenz Landau, Fortstraße 7, 76829 Landau, Germany
bJunior Research Group Chemistry of Microbial Communication, Leibniz Institute for Natural Product Research and Infection Biology Hans Knöll Institute (HKI), Beutenbergstraße 11a, 07745 Jena, Germany. E-mail: pierre.stallforth@leibniz-hki.de
First published on 22nd August 2019
Abstract
Bacteria of the genus Pseudomonas are ubiquitous in nature. Pseudomonads display a fascinating metabolic diversity, which correlates with their ability to colonize an extremely wide range of ecological niches. As a result, these bacteria are a prolific source of natural products. Biosynthesis of the latter is often orchestrated by arrays of chemical signals arising from intraspecies communication or interspecies relationships with bacteria, fungi, amoebae, plants, and insects. Especially nonribosomal lipopeptides, which have diverse biological activities, play important roles in the lifestyle of pseudomonads. In this review, we will focus on the molecular structures, properties, biosynthetic pathways, and biological functions of pseudomonal lipopeptides. This review is not only addressed to bio/chemists rather it serves as a comprehensive guide for all researchers (micro/biologists, ecologists, and environmental scientists) working in this multidisciplinary field.
1. Introduction
1.1 Pseudomonads
Pseudomonads are aerobic, motile, rod-shaped Gammaproteobacteria, which predominantly inhabit soil and aquatic environments.1 In their natural habitats they form mutualistic and antagonistic relationships with other prokaryotes, plants, fungi, protozoa, nematodes, or insects2 and fulfill many ecological functions that are beneficial for the environment3 and agriculture4 in general. Amongst others, pseudomonads are involved in bioremediation,5 plant growth promotion,6 and biocontrol.7 Pseudomonads, however, also represent important and often multi-drug resistant opportunistic human pathogens; the most important example being Pseudomonas aeruginosa.8,9Pseudomonas species are metabolically flexible and can process plant litter as well as a wide range of organic compounds for energy supply.10 This flexibility stems from their core genome that can adapt quickly through horizontal gene transfer or genome rearrangements with the corresponding accessory genome.11–13 Therefore, individual Pseudomonas strains can adapt to ecological niches and host organisms in a short amount of time.14 Until now, over 3000 draft genomes of Pseudomonas strains have been deposited in the Pseudomonas Genome Database, which represents a useful resource for bioinformaticians to identify genes that are linked to metabolism, pathogenicity, or antibiotic resistance.15 The genome size of most bacteria of the genus Pseudomonas is larger than the average bacterial genome size. For instance the genome of P. protegens Pf-5, which was fully sequenced in 2005 has a size of 7 Mbp.16 As a general rule it is known that bacterial genome size positively correlates with the ability to produce secondary metabolites. Hence, many Pseudomonas strains are prolific natural product producers, which is displayed by the outstanding diversity of secondary metabolites already identified in this bacterial genus.17 Pseudomonads not only biosynthesize a wide range of antimicrobials to impede competitors and protect against predators.18 They also produce chemical signal molecules to maintain intra- and interspecies interactions. For example, N-acyl-homoserine lactones synchronize the behavior in pseudomonal populations via quorum sensing (QS)19 (e.g. when changing the lifestyle from planktonic cells to biofilms) whereas phytohormones (such as indole-3-acetic acid) produced by P. syringae, a common plant pathogen, impair plant defense mechanisms to ensure successful infection of plants.20
1.2 Nonribosomal lipopeptides
Pseudomonads also biosynthesize nonribosomal lipopeptides (NRLPs), which consist of oligopeptides that are N-terminally acylated with a fatty acid. These bacterial natural products are produced by nonribosomal peptide synthetases (NRPSs) that are able to incorporate non-canonical amino acids into NRLP peptide backbones, which can range from hexa- to pentacosapeptides in size.21 In addition, NRPSs have the ability to cyclize the terminal amino acid with functional groups of the lipopeptide backbone generating macrolactones, macrolactams, or even macrocycles containing a biaryl moiety.22 Hence, NRLPs of bacterial origin are structurally diverse and have intriguing biological activities, which enable their use in medicine and industrial applications.For example, polymyxins bind to lipopolysaccharide of Gram-negative bacteria leading to a disruption of inner and outer membrane integrity causing cell death (Fig. 1).23,24 They represent one of the few last resort antibiotics that are active against Gram-negative bacteria. Another prominent NRLP is daptomycin, which binds to the cell membrane of Gram-positive bacteria in a Ca2+-dependent fashion causing a depolarization of the membrane potential.25,26 Resistance against this antibiotic is still uncommon, but was already observed.27 However, not all NRLPs are known for their antibiotic properties. The combination of a hydrophobic and a hydrophilic moiety renders NRLPs amphiphilic, which classifies most members of this natural product class as surface-active compounds or biosurfactants. The most active biosurfactant known to date is surfactin, which can lower the surface tension of water from 72 to 27 mN m−1 at concentrations in the low μM range.28 The physicochemical properties of surfactin enable its producer to easily spread over surfaces, form biofilms, and protect itself against competitors.29 Therefore, NRLPs are crucial for sustaining specific bacterial lifestyles.18
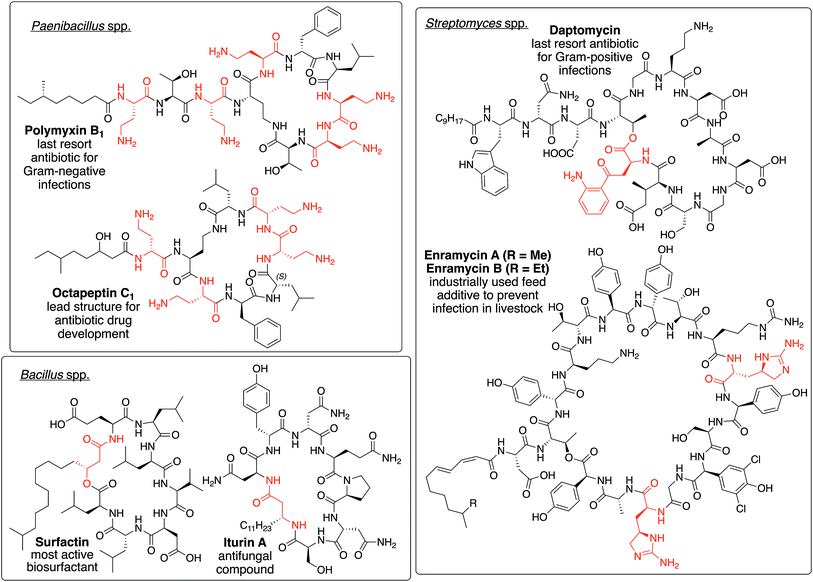 | ||
| Fig. 1 Depiction of polymxin B1,34 octapeptin C1,30,35 daptomycin,25 enramycin A/B,36,37 surfactin,38 and iturine A,39 which serve as representatives of NRLPs from different genera bearing unique structural motifs (highlighted in red). | ||
NRPLs are mainly produced by four families/genera, namely paenibacillaceae, Bacillus, Pseudomonas, and Streptomyces. Each of them produces at least one group of NRLPs that bear unique structural motifs not found in other genera or families. Polymyxins and octapeptins, which are produced by several paenibacillaceae, contain a high density of positively charged amino acids.30 The cyclization reactions during the biosynthesis of surfactins and iturins, which are of Bacillus origin, employ the β-hydroxy or β-amino functionality of the fatty acid to form the macrolactone and macrolactam ring, respectively.31Streptomyces spp. usually incorporate unique amino acids into the peptide backbone of NLPRs, such as kynurenine32 in daptomycin or enduracididine33 in enramycin.
Although more than 100 NRLPs of pseudomonal origin have been described (see ESI† for a full list), this natural product class is usually underrated, when compared to the biosynthetic variety of other genera. Thus, this comprehensive review article will highlight the incredible structural and functional diversity of Pseudomonas-derived NRLPs. A detailed account on their structure, biosynthesis, regulation, biological activities and biophysical properties will shed light on how, when, and why these natural products are produced within their ecological context. Furthermore, we will discuss the roles of pseudomonal NRLPs in interspecies interactions and their contribution to the soil ecosystem.
2. Structure of pseudomonal lipopeptides
The basic structure of pseudomonal NRLPs consists of an oligopeptide (8–25 amino acids) that is N-terminally acylated with a linear fatty acid (C5–C16). The lipid moiety is usually a β-hydroxy acid with an R-configuration of the corresponding stereocenter of all lipopeptides analyzed so far.40 In addition, the fatty acids characterized until now can also be bishydroxylated,41 unsaturated,42,43 or can contain a second carboxyl group44 (see ESI for a detailed list).The oligopeptide part can consist of a mixture of proteinogenic, modified, and non-proteinogenic amino acids. These include dehydrated (2,3-dehydroaminobutyric acid), chlorinated (4-chloro-threonine), D-/allo-configured, acylated (N5-acetyl-N5-hydroxy-ornithine) or hydroxylated (3-hydroxyaspartic acid or the rare α-hydroxy ornithine) amino acids (Fig. 2 and 3). Furthermore, biosynthetic intermediates of proteinogenic amino acids (e.g. ornithine, homoserine, 2,4-diaminobutyric acid) can also be present in the peptide chain.45 Despite their structural diversity, pseudomonal lipopeptides, with the exception of the syringomycin41,46–53 (Fig. 2) and corrugatin54–56 group (Fig. 4) as well as the ferrocins42,43 (Fig. 2), share the feature that at least half of all amino acids, which are used to construct the oligopeptide, are hydrophobic (Ala, Val, Leu, Ile, Phe).
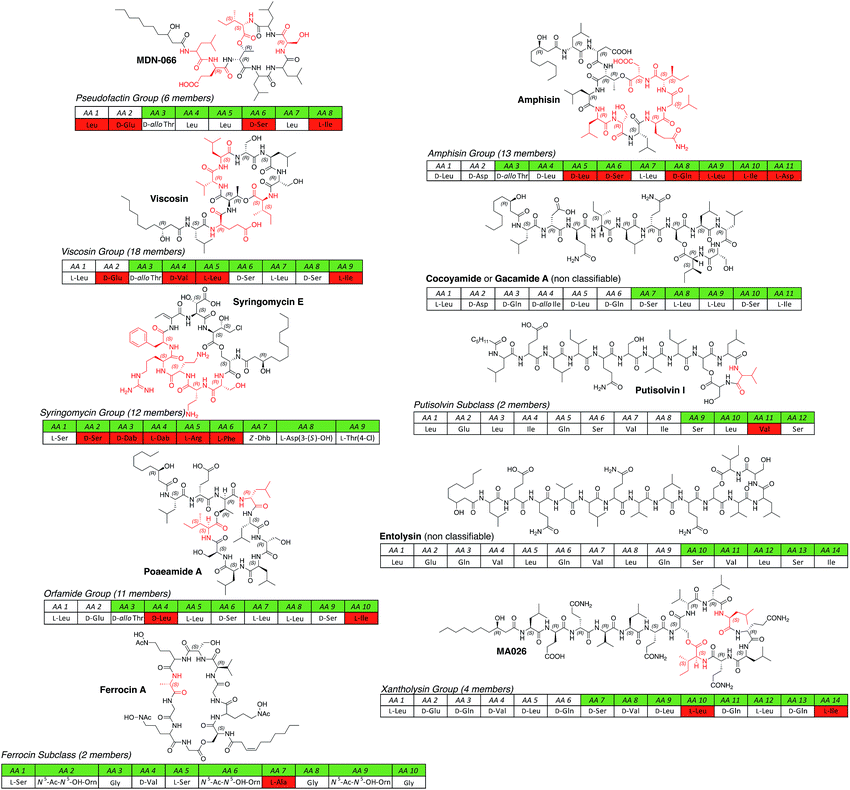 | ||
| Fig. 2 All known short CLP groups (pseudofactin,57–59 viscosin,60–67 syringomycin,41,46–53 orfamide,58,68–72 amphisin,73–79 xantholysin80,81) subgroups (ferrocin,42,43 putisolvin82) and non-classifiable individual compounds (gacamide/cocoyamide,83,84 entolysin85,86) isolated from different Pseudomonas spp. arranged according to the length of the oligopeptide and the size of the macrolactone ring. For each group either the name-giving CLP or, if more structural information is known about a related compound, one of its congeners is depicted. For a full table of all known lipopeptides please see the ESI.† In general, amino acids (AA) are written in three-letter code and AA 1 represents the N-terminus of the peptide. If the configuration of an amino acid is not indicated, no stereochemical information is known (putative assignment of stereochemical information based on bioinformatics analysis is not included). The macrolactone is indicated by the highlighting in green. Amino acids highlighted in red are not conserved in the respective CLP groups. Abbreviations: N5-Ac-N5-OH-Orn = N5-acetyl-N5-hydroxy-ornithine; Dab = 2,4-diaminobutyric acid; Dhb = 2,3-dehydroaminobutyric acid; Hse = homoserine; Orn = ornithine; WLIP = white line inducing principle. Plusbacins were not considered CLPs of pseudomonal origin, because their structure is strongly related to CLPs from Bacillus spp. and the strain was erroneously assigned.87 The structures of the ecomycins88 and hodersin89 were never elucidated. | ||
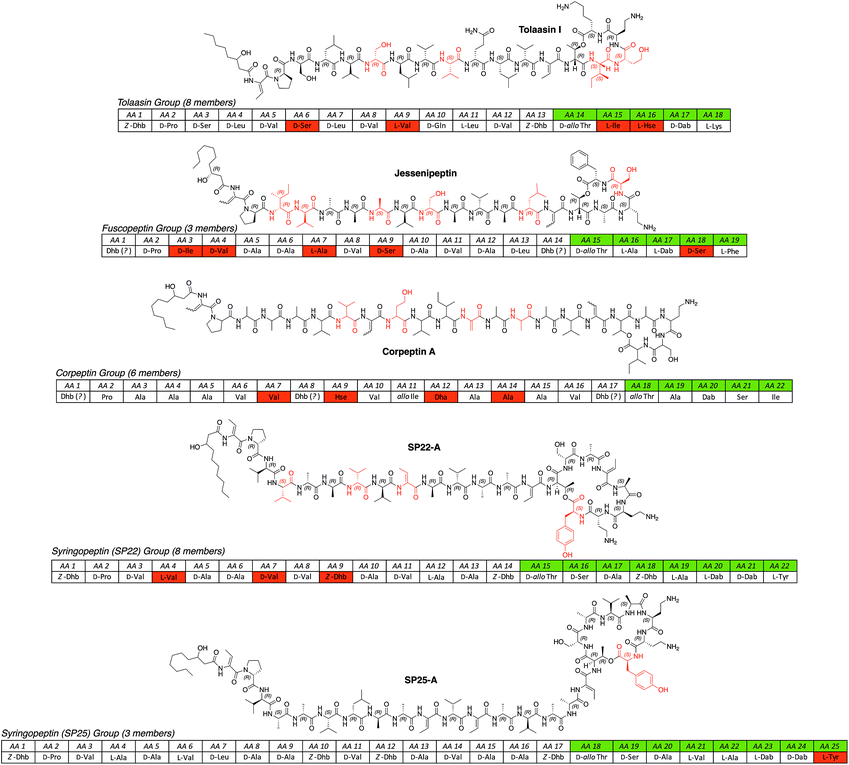 | ||
| Fig. 3 All known long CLP groups (tolaasin,44,70,90,91 fuscopeptin,92,93 corpeptin,94–97 SP22,98–102 SP25 (ref. 98,99,103) isolated from different Pseudomonas spp. arranged according to the length of the oligopeptide and the size of the macrolactone ring. For a full table of all known lipopeptides please see the ESI.† Abbreviations: Dha = 2,3-didehydroalanine; Hse = homoserine; SP = syringopeptin. | ||
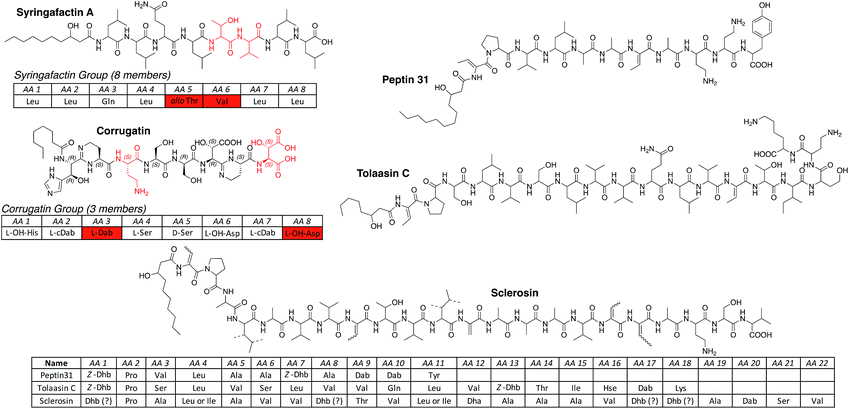 | ||
| Fig. 4 LLPs produced by Pseudomonas spp. arranged in groups according to the length and nature of the oligopeptide. For a full table of all known lipopeptides please see the ESI.† Abbreviations: cDab = cyclized Dab (cyclic amidine formation with carbonyl function of previous amino acid); Dhb = 2,3-dehydroaminobutyric acid; L-OH-His = L-threo-β-hydroxy-His; L-OH-Asp = L-threo-β-hydroxy-Asp; OH-His = β-hydroxy-His (unknown configuration); OH-Asp = β-hydroxy-Asp (unknown configuration). Ornibactins111 were not considered LLPs of pseudomonal origin, because the producing strain Burkholderia cepacia112 was falsely assigned as Pseudomonas cepacia. | ||
Interestingly, the vast majority of pseudomonal lipopeptides are not linear but display a macrolactone ring that is formed between the C-terminus of the peptide and a hydroxyl group of a serine or threonine. The size of the macrocycles ranges from 4 to 10 amino acids. All known pseudomonal cyclic lipopeptides (CLPs) can be divided into two categories, namely shorter (8–14 amino acids, Fig. 2) and longer (18–25 amino acids, Fig. 3) CLPs. With the exception of members of the syringomycin group, the ferrocins and the pseudofactins,57–59 all shorter CLPs share three common features. First, the N-terminal motif, a leucine connected to a hydrophilic amino acid (Glu/Gln/Asp), is conserved within the shorter CLP class. Second, all shorter CLPs bear acidic but no basic amino acids. Third, structural diversity of the respective groups arises mainly through fatty acid variety, configurational inversion of single amino acids, and exchange of chemically similar amino acids (Glu/Gln or Val/Leu/Ile).
In contrast, CLPs belonging to the group of syringomycins have a unique conserved structure. The lactone is formed between the C-terminus and the side chain of the N-terminal amino acid giving rise to the second largest macrocycles of all pseudomonal CLPs. They contain amino acids exclusively found in this group (4-chloro-threonine, Arg, (α-OH)Orn,52 His) and have a high content in basic amino acids. Finally, the chemical properties of amino acids occupying the same position in the peptide chain can vary significantly between members of the syringomycin group (e.g. AA 6 – hydrophobic (Phe) to hydrophilic (Thr) or AA 3 – basic (Dab) to acidic (Asp)).
Common traits of longer CLPs are the conserved motif of the first two amino acids (Dhb-Pro), the lack of acidic and the presence of basic amino acids, and the consensus-like peptide sequence indicating the threonine participating in macrolactone ring formation (hydrophobic AA-Dhb-allo-Thr).
Pseudomonal CLPs can so far be classified in eleven groups, two subclasses and the two non-classifiable compounds entolysin85 and cocoyamide A84 or gacamide A.83 Groups consist of at least two CLP members that have oligopeptides of equal length and a macrolactone ring of the same dimension and structure. In addition, at least two CLPs of one group have to be produced by different strains of a Pseudomonas sp. to ensure individual biosynthetic origins instead of one flexible or permissive biosynthetic pathway that produces congeners of one main CLP. Therefore, the putisolvins,82 as they are produced by the same strain, represent a CLP subclass whereas the fuscopeptins92,93 are classified in a group (please see the ESI† for all NRLP producing Pseudomonas strains).
When compared to CLPs, fewer linear lipopeptides (LLPs) of pseudomonal origin are described. LLPs can be divided into two groups, namely the syringafactin104,105 and corrugatin groups (Fig. 4). Members of the syringafactin group are lipidated octapeptides that show little chemical diversity in their fatty acid (3-hydroxydecanoic and 3-hydroxydodecanoic acid) and amino acid content. Only two positions of the octapeptide vary in amino acid composition, which are, however, chemically similar (AA 5 Thr/Gln – hydrophilic and AA 6 Val/Ile/Leu – hydrophobic). In contrast, representatives of the corrugatin group show unique features not found in any other group or subclass. All corrugatins contain the rare β-hydroxy histidine,106 which functions as a bidentate ligand for Fe3+ ions, and bear a cyclic amidine generated through a condensation reaction of the γ-NH2 group of Dab and the amide carbonyl of the previous amino acid. Similarly to the ferrocins, which have been isolated as their Fe3+ complexes, the corrugatins can chelate iron ions efficiently and serve as siderophores.107,108
The three additional linear lipopeptides are more closely related to the longer CLPs and are non-classifiable. Peptin31 (ref. 109) represents a truncated linear version of the syringopeptins, tolaasin C44 is most likely the hydrolysis product of tolaasin I, and sclerosin110 has a strong resemblance with nunapeptin but instead of a threonine, which could be used for a possible ring closure, contains a second Dhb at position 18.
3. Biosynthesis of pseudomonal lipopeptides
3.1 Biochemistry of CLPs and LLPs
Unlike ribosomally synthesized antimicrobial peptides113,114 or ribosomally synthesized and post-translationally modified peptides (RiPPs),115,116 NRLPs are synthesized by NRPSs,117,118 which are multi-modular megaenzymes, with molecular weights surpassing 1.0 MDa in some cases, organized in assembly lines.The biosynthetic machineries producing arthrofactin A and syringomycin E have been investigated in detail over the last decades. Therefore, this section will focus on the biosynthesis of these two short CLPs. We will point out similarities and differences between pseudomonal NRPSs and those of other genera. In addition, we also want to draw attention to enzymatic transformations unique to Pseudomonas spp. that have not been investigated yet and hold the potential to unveil new biochemistry.
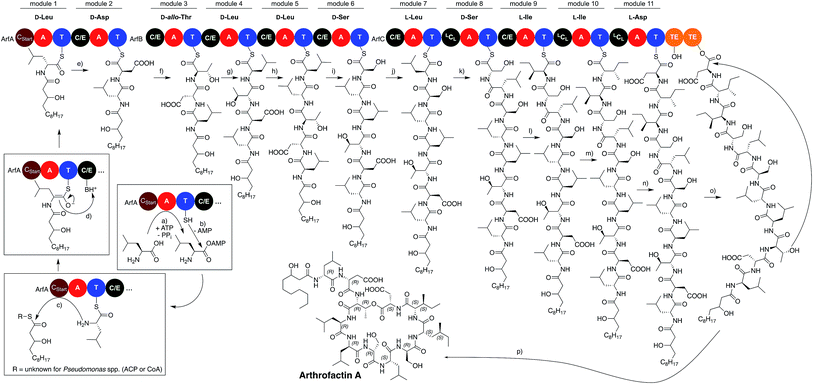
The assembly line producing arthrofactin A consist of 3 proteins (ArfA, ArfB, and ArfC) that contain multiple modules (Fig. 5).119 Each module is responsible for the incorporation of one amino acid monomer into the oligopeptide chain. Every module consists of three core domains, which are connected to each other via short flexible linker sequences.120,121 The three core domains are the condensation122 (C) and adenylation123 (A) domains, which are catalytically active, and the thiolation124 (T) domain that serves as a shuttle for biosynthetic intermediates between domains and modules.
The order of domains in all arthrofactin synthetases is C–A–T, which is typical for almost all pseudomonal NRPSs (Fig. 6). At the beginning of the biosynthesis the A domains select their corresponding L-amino acid, activate them via an adenosine monophosphate (AMP) ester, and transfer them to the T domain generating a thioester. It has to be noted that the A domains of some pseudomonal NRPSs, especially those encoding for hydrophobic amino acids, do not exclusively select for one amino acid and generate chemical diversity through their own plasticity.104,125–128 In the case of ArfC the A domain of module 11 can also activate L-Glu instead of L-Asp.76
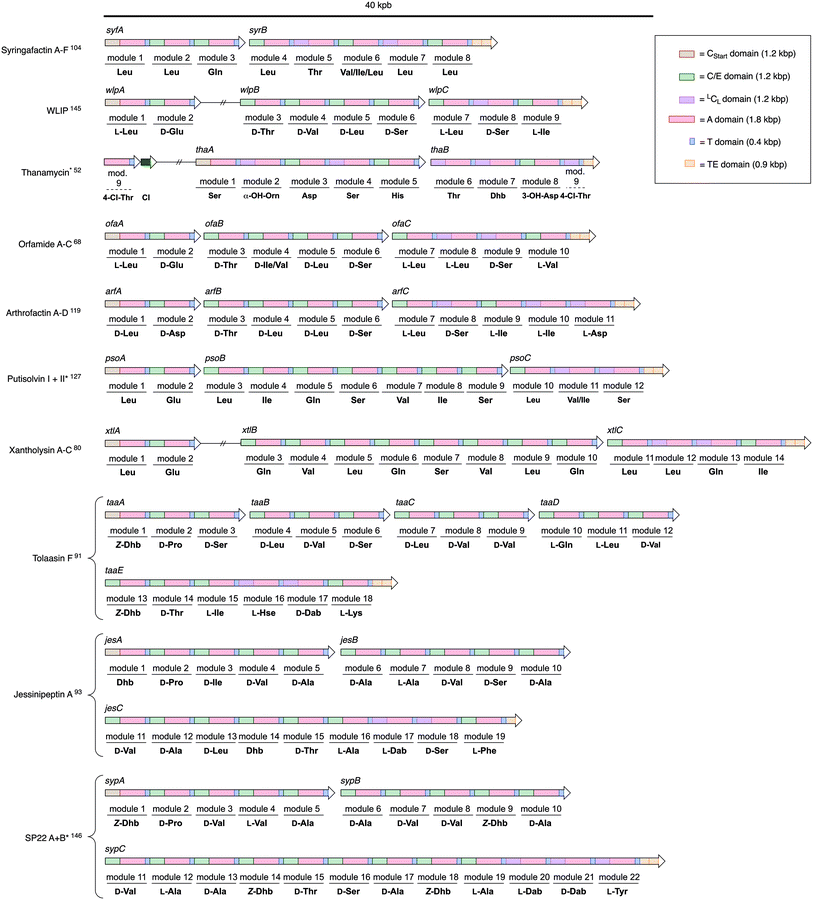 | ||
| Fig. 6 Multidomain organization of BGCs encoding different NRPSs produced by Pseudomonas spp. Gene clusters marked with an * (CLP – Genbank Accesion Number: Thanamycin – KT362216.1; Putisolvin – DQ151887.2; Syringopeptin – AF286216.2) were reanalyzed using antiSMASH Version 4.2.0 in order to assign a C domain class (see ESI†). Assignment of amino acids to the models is based on structure elucidation data and not on bioinformatics analysis. Double vertical lines represent intervening DNA. Abbreviation: Cl = chlorinase. | ||
Subsequently, module 1, which contains an N-acylation C domain subtype, called CStart domain or lipoinitiation domain, condenses an activated fatty acid thioester with the first amino acid. Surprisingly, there is no experimental evidence in which form the fatty acid is provided in Pseudomonas spp. – either tethered to an acyl carrier protein129 or linked to a single CoA130 molecule. CStart domains, like some A domains, can also be plastic and can use a wide variety of different fatty acids for the acylation reaction. Therefore, small amounts of congeners, which only differ from the major lipopeptide in their fatty acid composition, are usually produced by the same pseudomonal NRPS assembly line.
After acylation, the condensation/epimerization (C/E) domain of the second module in ArfA first epimerizes the L-leucine into the corresponding D-amino acid before it catalyzes the peptide bond formation between the amino group of the amino acid tethered to the second T domain and the lipidated amino acid thioester of the first T domain. C/E domains, which have been described first in ArfA and ArfB, are currently the only known enzymes that generate D-amino acids in pseudomonals NRPSs.131 NRPSs of Bacillus,35,132,133 Streptomyces,134–136 or other genera137–139 for example contain separate epimerization domains140,141 (E domains) that are usually located at the end of the module (C–A–T–E).142 Furthermore, they also contain C domains that only catalyze the condensation of a D- with an L-amino acid (DCL), which have yet to be identified in pseudomonal NRPSs.
The condensation/epimerization process is repeated by the following modules until the peptide backbone of the lipopeptide reaches module 8 of ArfC, which contains an LCL domain. This C domain subtype only catalyzes C–N bond formation between two L-amino acids and is not capable of generating D-amino acids. The nascent lipopeptide is further elongated by the following modules until it is covalently attached to the last T domain of module 11.
Finally, the linear arthrofactin A thioester is cyclized by a tandem thioesterase (TE) domain.143 TE domains catalyze the transesterification of the linear lipopeptide from the T domain of the last module onto a conserved Ser residue that is located in the proteins active site. This ester is then translocated again to the second TE domain where it is attacked by the hydroxyl group of the D-allo-Thr, thus forming arthofactin A. Most biosynthetic gene clusters (BGCs) that code for pseudomonal NRPSs contain a tandem TE domain, which not only synthesize CLPs but also hydrolyze the nascent thioester to yield LLPs. However, there are exceptions such as the BGCs of thanamycin52 or jessenipeptin,93 which contain only one TE domain. If only one TE domain is present, hydrolysis or cyclization reactions are performed solely by one enzyme.144
The domain and module organization of pseudomonal NRPS assembly lines is, with the general exception of the syringomycins,51–53 for the most part conserved and resemble the arthrofactin synthetase. Except for the gene cluster encoding poaeamide,71 all other gene clusters,68,80,83,93,119,127,145–148 which encode pseudomonal NRPS assembly lines, are colinear and, with the exception of the syringafactins104,105 (two) and tolaasin F91 (five), contain three open reading frames (ORF) that translate into three multidomain megaenzymes (Fig. 6). The first multidomain NRPS enzyme always contains a CStart–A–T module followed by up to four additional C–A–T modules. The second part of the assembly line consists of three to eight and the third part of three to twelve C–A–T modules. Final modules are normally followed by a tandem TE domain. External TE domains,133,149 common to NRPS gene clusters of other bacteria species, have not been identified in Pseudomonas species so far. LCL domains are only encountered in the last assembly line megaenzyme of the respective lipopeptides. However, not all C/E domains are comprised of a functional epimerization active site,91 which is why L-amino acids are not solely integrated into the peptide backbone by the last part of the assembly line (e.g. module 2 and 3 in syfA, module 7 in jesB or modules 10 and 11 in taaD).
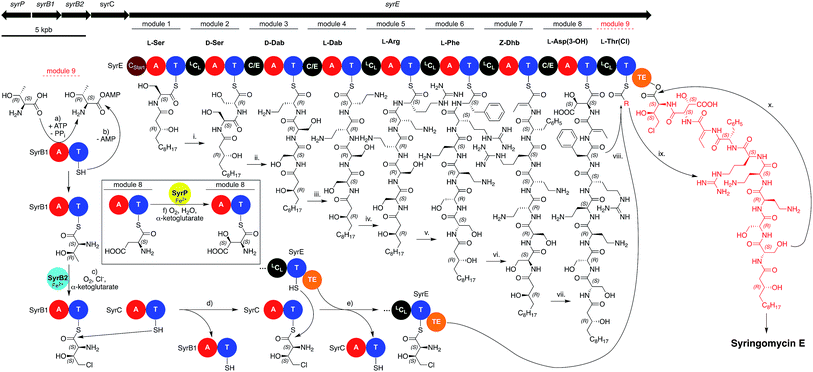
As mentioned previously, the members of the syringomycin group stand out compared to all other pseudomonal CLPs with regard to their structures. This also applies to their biosyntheses and gene cluster architecture. For example, the gene cluster encoding the syringomycin synthetases is not colinear, it contains two split modules consisting of either a C and T domain or an A and T domain, and bears more LCL than C/E domains (Fig. 7). In addition, the syringomycin synthetase machinery also contains multiple external enzymes that modify and generate amino acids unique to this CLP class. Syringomycin synthetase E (SyrE) consists of one megaenzyme comprising eight full C–A–T modules and the aforementioned ninth split module followed by a single TE domain. The biosynthesis of syringomycin E starts with the CStart domain-catalyzed acylation of L-serine, which is tethered to the T domain of the first module, with 3-hydroxy-(R)-dodecanoic acid. Then, the linear peptide backbone is elongated via a series of standard LCL and C/E domain-catalyzed C–N bond formations up to module eight. Before the hydroxylated aspartic acid can be incorporated the SyrE-bound L-Asp moiety is oxidized by the non-heme iron hydroxylase SyrP.150 After formation of the octapeptide the ninth split module of SyrE requires loading with L-4-chloro-Thr. Therefore, the external split module SyrB1,151 which consists of an A and a T domain, charges itself with L-Thr before it is chlorinated by the non-heme iron chlorinase SyrB2.152 The generated L-4-chloro-Thr is thereafter transported via the shuttle protein SyrC to the last T domain of SyrE and condensed with the nascent peptide before the TE domain finally induces cyclisation and releases syringomycin E.153
Unfortunately, only bioinformatics information is available for the putative biosynthesis of two other members of the syringomycin group, namely thanamycin52,53 and nunamycin.51 Compared to one ORF coding for an NRPS containing nine modules, the gene clusters of thanamycin and nunamycin consist of two ORFs containing five and three and a half modules (Fig. 6).
For the thanamycin gene cluster, the split module of the ninth amino acid as well as the chlorinase gene could be annotated, but genes encoding for the enzymatic machinery responsible for the hydroxylation of Asp and especially for the so far unknown α-hydroxylation of Orn are still unidentified and their products remain uncharacterized. This also applies to the biosynthesis of other unique amino acids and modifications. For example, the biochemical origin of the amino acids N5-acetyl-N5-hydroxy-ornithine (ferrocins) and L-threo-β-hydroxy-histidine (corrugatins) has not been studied. Furthermore, it is also unclear how the condensation reaction forming the cyclic amidine during the synthesis of the corrugatins is executed. Finally, the biochemical characterization of the underlying dehydration step of Thr and Ser, which most likely generates the non-canonical amino acids Dhb and Dha,154,155 has remained virtually unexplored.156 Thus, it is not clear if the corresponding C domains or external dehydratases157–159 are responsible for the elimination reaction and how the catalytic cycle operates.
Therefore, characterization of the biosynthetic pathways of extraordinary pseudomonal CLPs and LLPs could be a particularly fruitful scientific endeavor that will yield new biochemistry.
3.2 Regulation of pseudomonal lipopeptides
In Pseudomonas spp. the two-component system GacA/GacS,4 which is found in many Gammaproteobacteria,160 is one of the key elements that acts as global activator of lipopeptide production.161 GacS is a transmembrane receptor histidine sensor kinase (HSK)162,163 that, upon binding of yet unknown ligand(s),164 autophosphorylates itself. Subsequently, the phosphate residue is transferred from GacS to the acceptor domain of the response regulator GacA via a phospho-relay mechanism.165 Phosphorylated GacA is then capable of initiating the transcription of three small RNA genes called rsmX, rsmY, and rsmZ.166 The RNAs RsmX, RsmY, and RsmZ bind to the small proteins RsmA and RsmE, which serve as post-transcriptional repressors by binding to a specific site called the GacA box, and enable activation of biosynthetic regulator genes (Fig. 8).167 It was already shown for P. syringae (syringomycin),168Pseudomonas sp. DSS73 (amphisin),164P. putida PCL 1445 (putisolvin),169P. fluorescens SS101 (massetolide),170 and P. fluorescens Pf-5 (orfamide)62 that deletion or disruption of either gacA or gacS leads to mutants incapable of producing CLPs. Interestingly, some wild isolates of Pseudomonas strains also have a non-functional GacS/GacA system, which results in silenced or “cryptic” BGCs.171 In the case of the P. fluorescens Pf0-1, complementation of the strain with a functional gacA gene lead to the activation of the cryptic gene cluster encoding the gacamides, which consequently lead to the isolation and identification of a new class of pseudomonal CLPs.83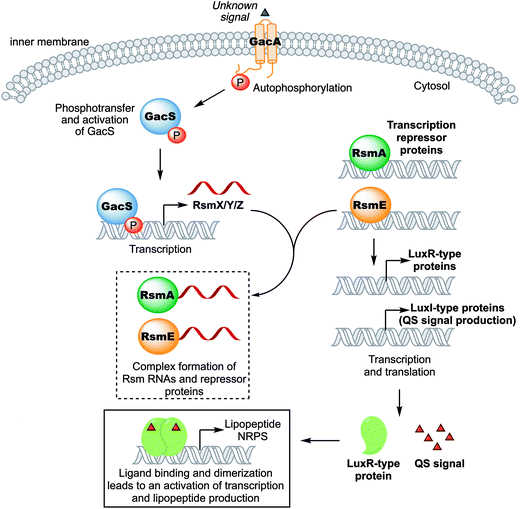 | ||
| Fig. 8 Schematic representation of the regulation of pseudomonal lipopeptide production (complex formation between RNA and RsmA/E is not stoichiometric). | ||
In Pseudomonas spp., the biosynthetic regulator genes acting downstream of the GacA/GacS system usually belong to the LuxR-type regulator family and flank CLP biosynthetic gene clusters up- or downstream (and sometimes both).172,173 LuxR-type proteins contain a helix-turn-helix DNA-binding motif and can activate174 or repress175 the expression of gene clusters. Activity of LuxR-type proteins in pseudomonal CLP production is typically regulated through QS signaling.93,176,177 QS signaling is a type of cell–cell communication used by bacteria to coordinate concerted action of a bacterial community.19,178 It involves constitutive production of a chemical signal, N-acyl L-homoserine lactones (AHLs),18N-(3-oxododecanoyl)-homoserine lactones, quinolones or the Integrated Quorum Sensing Signal (IQS)179–181 in Pseudomonas spp., which is released into the environment and accumulates as a function of bacterial cell density.182 Once a certain threshold concentration is reached the chemical signal binds to the LuxR-type regulator, which dimerizes, before it initiates transcription of, amongst others, CLP genes.183 Examples for LuxR-type regulation of CLPs in Pseudomonas spp. are divers, namely SalA (syringomycin) in P. syringae pv. syringae,184–187 PsoR (putisolvin) in P. putida,127 ViscA and ViscBC (viscosin) in P. fluorescens SBW25,173 MassA and MassBC (massetolide) in P. fluorescens SS101,170 ArfF (arthrofactin) in P. fluorescens MIS38,188 EtlR (entolysin) in P. entomophilia L48T,85 WlpR (WLIP) in P. putida RW10S2,145 XtlR (xantholysin) in P. putida BW11M1,80 PcoR and RfiA (corpeptin) in P. corrugata CFBP 5454,189 OfaR1 and OfaR2 (orfamide) in Pseudomonas sp. CMR12a,172 NunF (nunamycin) in Pseudomonas fluorescens strain In5,190 and JesR1/JesR2 (jessenipeptin) in Pseudomonas sp. QS1027.93 The protein pair JesR1/JesR2 should be highlighted here. Experiments indicated that both JesR1/JesR2 are required for the production of jessenipeptin. While further experiments are required it is conceivable that these two response regulators may form a heterodimer, which is typically associated with inactivation rather than activation of transcription.191,192
Additional genes that control CLP biosynthesis, most likely through transcriptional downregulation of LuxR-type regulators, have been identified and studied in P. fluorescens using the example of massetolide production. Genes involved include the protease complex clpA and clpP,193 the heat shock protein dnaK194 and the anti-sigma factor195prtR.194 Furthermore, a mutant lacking a functional D-3-phosphoglycerate dehydrogenase (phgdh), which is involved in the biosynthesis of L-serine, was also incapable of producing massetolide. Interestingly, production could be restored by adding L-serine to the growth medium.194 This and other results indicate that CLP biosynthesis is strongly influenced by the cellular substrate pool.196
Albeit that regulation of pseudomonal lipopeptides is frequently addressed, fundamental aspects of this research area are still unknown. For example it remains to be determined, which chemical signals bind to GacA, orphan LuxRs or LuxR-solos,197 how lipopeptide production is regulated in diverse microbial communities, and if the biochemical network orchestrating lipopeptide production in P. fluorescens also applies to other Pseudomonas spp.?
4. Properties of pseudomonal lipopeptides
4.1 Biophysical properties
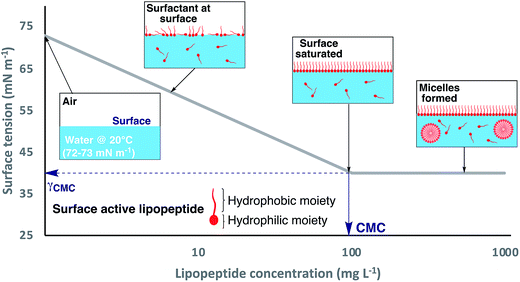
Pseudomonal lipopeptides simultaneously bear hydrophobic and hydrophilic moieties, which classifies these organic compounds as amphiphiles.198 This amphiphilicity enables NRLPs to reduce the surface tension (γ) of liquids, in which they are dissolved, in a concentration-dependent manner, thus rendering them biosurfactants (Fig. 9).199 Biosurfactants not only dissolve in water or aqueous solutions but they also adsorb to the liquid surface, with the hydrophilic moiety being immersed in the aqueous phase. The hydrophobic part associates with nonpolar molecules (air or the lipophilic part of other amphiphiles) or arranges itself on the air–water interface.200 At high surfactant concentrations, the amphiphiles form a monolayer at the liquid surface, which becomes more densely packed with increasing amphiphile concentration. The concentration at which the liquid surface is saturated with amphiphilic molecules, without surfactants aggregates, so-called micelles, having formed in the liquid phase, is called the critical micelle concentration (CMC).201 At this point, the surface tension of the corresponding liquid has reached the minimum (γCMC) and cannot be further decreased by increasing the surfactant concentration. Beyond the CMC additional surface-active molecules will form micelles in the liquid phase as they are not able to integrate into the saturated monolayer at the air–water interface. Both values (CMC and γCMC) can be determined by standard techniques of tensiometry (contact angle, Wilhelmy plate, Du Noüy Ring, pendant or spinning drop, maximum bubble pressure or capillary rise).202,203The CMC and γCMC are important physicochemical parameters that allow assessing the surface activity of a surfactant (Table 1). For example, Triton X-100 is a nonionic surfactant that is frequently used as a component of buffer solutions. Triton X-100 (in water at 25 °C) has a CMC of 216 μmol L−1 and a γCMC of ∼30 mN m−1.198 In comparison, viscosin has a CMC of 48 μmol L−1 (in water at 25 °C) and a γCMC of 28 mN m−1. Therefore, viscosin is more surface-active because it can induce a lower minimum surface tension at a concentration that is four times lower than the CMC of Triton X-100. In general, smaller CLPs, with the exception of syringomycin E, have CMCs that are an order of magnitude lower than the reported values of longer CLPs. In addition, smaller CLPs can also decrease γwater to lower values than their longer counterparts.
| Lipopeptide | CMCa | γ CMC | Swarming?c | Ref. |
|---|---|---|---|---|
| a CMC [mg L−1] in water. b Minimum surface tension between air and water at the CMC [mN m−1] at 20–25 °C. c Can the swarming ability of the producing Pseudomonas strain be attributed to the corresponding lipopeptide? | ||||
| Syringafactins | n.d. | n.d. | Yes | 104,204 |
| Cichofactins | n.d. | n.d. | Yes | 105 |
| Pseudofactin II | 72 | 32 | n.d. | 57,205 |
| Viscosin | 54 | 28 | Yes | 173,206,207 |
| WLIP | n.d. | n.d. | Yes | 145 |
| Massetolide A | n.d. | n.d. | Yes | 208 |
| Syringomycin E | 1250 | 33 | n.d. | 209,210 |
| Cormycin A | 176 | n.d. | n.d. | 50 |
| Nunamycin/nunapeptin | n.d. | n.d. | Yes | 190 |
| Orfamides | n.d. | n.d. | Yes | 68–70,211 |
| Orfamide A | ∼10 | 38 | n.d. | 212 |
| Poaeamide A | 15 | ∼40 | Yes | 71 |
| Amphisin | n.d. | n.d. | Yes | 164,213 |
| Anikasin | n.d. | n.d. | Yes | 75,214 |
| Arthrofactin A | 14 | 24 | Yes | 119,215 |
| Gacamide A | 33 | 29 | Yes | 83 |
| Putisolvins | n.d. | n.d. | Yes | 82 |
| Entolysin | n.d. | n.d. | Yes | 85 |
| Xantholysins | n.d. | n.d. | Yes | 80 |
| Tolaasin I/II (mixture) | 460 | 42 | No | 216,217 |
| Sesselins | n.d. | n.d. | No | 70 |
| SP22-A | 820 | 40 | n.d. | 218 |
| SP22-B | 800 | 41 | n.d. | 218 |
| SP25-A | 2160 | n.d. | n.d. | 210 |
Pseudomonas spp. use the surfactant properties of lipopeptides to lower the tension between the underground and the bacterial cell. In combination with their actively rotating flagella, Pseudomonas spp. can rapidly move in a multicellular fashion and spread over solid surfaces (Fig. 10a).219 This type of active bacterial movement is called swarming and has so far only been observed in three families (Firmicutes, Alpha-, and Gammaproteobacteria) of the bacterial domain.220 In order to test if a bacterial strain is able to swarm, an agar plate with a low agar content (usually around 0.5% w/w) is inoculated in the center and incubated for 24 h. A Pseudomonas strain can be classified as swarmer if after the incubation period a large area of the plate is covered with a bacterial lawn compared to a single central colony (Fig. 10b).
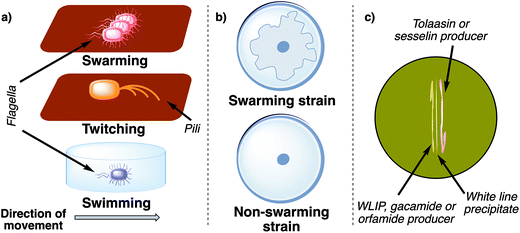 | ||
| Fig. 10 (a) Different mechanisms of active bacterial movement. Swarming is the multicellular movement of bacteria across a surface propelled by rotating flagella. Twitching is characterized by the extension of pili, which then attach to the surface and subsequently retract. Swimming is an active locomotion of individual bacteria in liquid (powered by rotating flagella). (b) Schematic comparison of a swarming and non-swarming bacterial strain on a low concentration agar plate (∼0.5% agar w/w). The swarming morphology of bacterial colonies can vary between different bacterial species and biosurfactants.227 (c) Schematic representation of a positive white line in agar test using two different strains of pseudomonads. | ||
Surprisingly, not all pseudomonal lipopeptides enable swarming. Tolaasin I for example, albeit surface active, does not induce swarming in P. tolaasii.217 Furthermore, a mutant of P. fluorescens CMR12a, which is deficient in sesselin production but generates orfamides, has enhanced swarming motility compared to the wild type. In contrast, mutants of the CMR12a strain, which were deficient in orfamide production, produced more biofilm than the wild type and were not able to swarm. Therefore, sesselins suppress the swarming ability of P. fluorescens CMR12a by inhibiting the surface active orfamides and orfamides impede biofilm formation by lowering the activity of sesselins.70
This observation can be explained by the white-line-in-agar phenomenon.221 When a tolaasin or sesselin producer is streaked out on an agar plate adjacent to a strain that produces either WLIP (White Line Inducing Principle)222,223 or orfamides70 a white line is formed between the two bacteria (Fig. 10c). This white line is the result of a complex formation between the short and long CLP, which leads to precipitation of the molecules.90,224 Thus, P. fluorescens CMR12a produces two CLPs, which mutually inhibit each other, and seem to fine tune the interplay between a motile and immotile lifestyle in this strain. Unfortunately, the structure of the complex is not known on a molecular level but the recently developed MicroED method could be an excellent tool to determine its composition.225,226
The change between a planktonic (single motile cells) and stationary (biofilm) life style is mainly regulated via QS in pseudomonads.228 In general, bacterial biofilms are comprised of mostly immobile bacterial microcolonies that are surrounded by an extracellular matrix, which consist of soluble, gel-forming polysaccharides, proteins, and DNA.229 Biofilms are highly organized matrices that are continuously remodeled and also have to be dispersed if, for example, nutrients are depleted. At this moment, single cells are released from the biofilm in order to colonize new areas.230 Probably because of their surface active properties pseudomonal CLPs have been found to be involved in biofilm regulation and surface attachment.231 Interestingly, structural similarity of lipopeptides does not indicate if they are involved in formation or degradation of biofilm. Massetolide A,208 for example, supports its producers in biofilm formation whereas the closely related CLPs WLIP145 and viscosin232 seem to inhibit biofilm production in their respective strains. Xantholysin80 also has a positive effect on biofilm formation, whereas orfamides,70 arthrofactins,119 putisolvins,82,233 and cichofactins105 seem to inhibit biofilm development. Unfortunately, the basic mechanisms of biofilm regulation through CLPs have not been deciphered yet, which can be attributed to the low number of studies that have linked lipopeptide production with biofilm formation or degradation. As a guideline for future studies concerning this topic, biofilm formation should always be investigated in a time-resolved manner232 and, if possible, molecular interaction partners234 of pseudomonal lipopeptides should be identified (using for example pull-down techniques). Furthermore, creating a temporo-spatial distribution of the investigated biosurfactant in cells and in the biofilm (for instance, by using a fluorescently labeled CLP) would help to understand the CLP controlled mechanism behind biofilm regulation.235
Pseudomonal lipopeptides not only interact with extracellular matrices. Their amphiphilic properties enable them to easily interact with and integrate into membranes. This was for example shown for a selection of members of different short CLP groups (massetolide A, orfamide A, arthrofactin, and entolysin).236 In particular, members of the viscosin group were recently investigated for their behavior in model membranes. Viscosin, viscosinamide A, pseudodesmin A, and WLIP were found to integrate and permeabilize small unilamellar vesicles consisting of a lipid composition mimicking Gram-positive bacterial cell membranes. In addition, circular dichroism (CD) spectroscopy was used to observe possible conformation changes of CLPs upon insertion into lipid vesicles.237 Interestingly, viscosin group CLPs seem to possess a very rigid α-helical conformation62,224,238 because the solution structure did not substantially differ from the membrane bound structure. Finally, the conformation and dynamics of viscosinamide A was studied in detail at the water–lipid interface using a variety of NMR methods and molecular simulations.239
Another group, whose members have been heavily studied regarding membrane activity, are the syringomycins. Syringomycin E (SR-E), for example, has ion channel-forming activity, which is affected by the sterol content, and integrates well into biomimetic membranes.240,241 It is estimated that at least six SR-E molecules are needed to form one membrane channel.242 In artificial bilayers, SR-E membrane channels also seem to exist in two distinct forms.243 Furthermore, it could be shown that a mixture of SRE, SR-A1, and SRG is able to lyse plant protoplasts at concentrations far below the CMC.218 SR-E also has hemolytic properties (with a C50 – concentration for half of total activity – of 0.41 μM) and lyses human as well as rabbit red blood cells (RBCs) by pore formation.210 In contrast to the members of the viscosin group, SR-E has at least two distinct conformations in aqueous solution, which are pH depended, and adopts an elongated structure when it is integrated into model membranes.244,245 Other members of the syringomycin group are also membrane active but less investigated. Syringotoxin forms pores in model bilayer lipid membranes and lyses erythrocytes (C50 = 1.9 μM).210,246 Pseudomycin A247 as well as cormycin A50 are hemolytic and integrate into lipid membranes, too. Interestingly, the solution structure of cormycin A is compact, L-shaped, and contains a concave hydrophobic surface that includes the fatty acid. This observation points to a possible conformational change when cormycin A is integrated into membranes because the lipid moiety of most CLPs is usually embedded in an extended conformation in the lipid bilayer.50
Long CLPs have been examined in a non-systematic fashion for their membrane activities. Tolaasin I permeabilizes model membranes248 and is hemolytic249 by forming pores on RBCs at concentrations significantly lower than its corresponding CMC. In addition, the solution structure of tolaasin I in sodium dodecyl sulfate (SDS) solutions (at a concentration above the CMC of SDS) was studied using NMR spectroscopy and molecular dynamics simulations.250 Under these conditions, tolaasin I exists as an extended left-handed α-helix (starting from AA 2 and ending at AA 14) that bears a lactone macrocycle with a “boat-like” or “seam of a tennis ball” conformation, which was first described for the lactone ring of WLIP.224 Fascinatingly, despite having differently sized rings and primary structures, the solution conformations of the lactone rings of syringotoxin,251 fuscopeptin B,252 and SP-25A99,253 are all structurally similar and closely resemble the “seam of a tennis ball”. Finally, hemolytic fuscopeptins254 and syringopeptins218,255 were studied for their ion-channel forming properties on biological and model membranes.
4.2 Biological properties
The physicochemical properties of pseudomonal lipopeptides directly influence their various biological activities. CLPs and LLPs can integrate into plasma membranes and change their microdomain membrane fluidity, although the exact mechanism for most lipopeptides is unknown and source of controversial scientific debates. This can lead to a rearrangement of plasma membrane architecture that potentially triggers depolarization and leakage or influx of ions, which can cause cell death or impaired proliferation (Fig. 11).26 This has been demonstrated by the ability of orfamide A to induce Ca2+ influx into the microalgae Chlamydomonas reinhardtii, which changed the morphology of this organism and inhibited its growth.256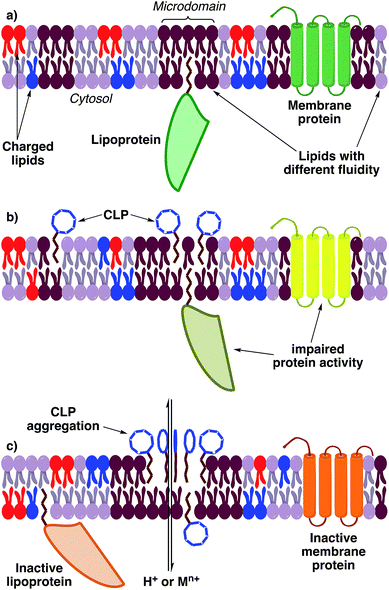 | ||
| Fig. 11 Hypothetical, general mechanism of action of lipopeptides (it cannot be ruled out that individual CLPs or LLPs also have a specific molecular targets). (a) Depiction of a normal plasma membrane. (b) At low concentrations lipopeptides integrate into specific membrane microdomains and create bilayer distortions. Distortions are counterbalanced by the attraction of lipids.257 Proteins attached or integrated into those microdomains may be impaired in their activity because the fluidity profile of the membrane changes. (c) At high concentrations of lipopeptides the distortions cannot be counterbalanced and gaps in the membrane bilayer arise. This can lead to aggregation of lipopeptides and permeabilization of the membrane.258 Permeabilization leads to an uncontrolled flux of ions into the cytosol and a depolarization of the membrane. Membrane proteins are probably inactivated by the changed membrane fluidity and environment or ion concentrations. Abbreviations: M = any metal. | ||
Therefore, most CLPs show antimicrobial, cytotoxic, and phytotoxic properties, which have been summarized in an excellent review by Geudens and Martins.2 In general, most pseudomonal CLPs tested (both short and long) show activity in the low micromolar range against Gram-positive and mycobacteria but not against Gram-negative bacteria.2 The lack of activity can be explained by the Gram-negative cell wall structure, which contains an outer and inner membrane. Lipopeptides are probably already adsorbed by the outer layer and are therefore not able to disturb the structure of the inner membrane or deregulate ion concentrations inside the bacterial cell.259 The exceptions from the rule are tolaasin I, which is active against some Pseudomonas spp.,249,260 and WLIP that has activity against different Xanthomonas strains.145
Surprisingly, ferrocins are able to inhibit the growth of Gram-negative bacteria (Escherichia coli, Salmonella typhimurium, Pseudomonas spp.) and showed strong therapeutic effects against P. aeruginosa in experimentally infected mice (at doses below 1 mg kg−1) but they have no activity against the tested Gram-positive bacteria.42 This observation can be explained by the iron-chelating properties of this CLP subgroup (Fig. 12a). Ferrocins can decrease the freely available iron concentration via complexation and thus, inhibit the growth of bacteria by limiting access to essential elements.261
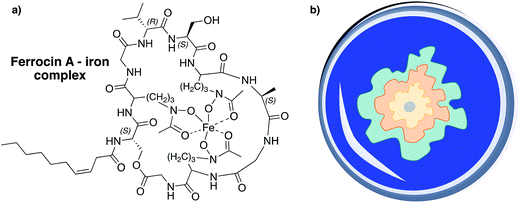 | ||
| Fig. 12 (a) Ferrocins can serve as hexadentate ligands and are able to make ferric iron (Fe3+) bioavailable to their producers.271,272 (b) Schematic illustration of a CAS assay agar plate. CAS can complex iron, which results in a blue complex. If bacteria that produce siderophores are placed on CAS-agar the iron ions are usually transferred to the better chelating agent, which results in a blue to yellow-orange color change in zones surrounding the colonies.273 The color change of the agar can be different depending on the bacterial strain and siderophore. | ||
Most CLPs have a broad antifungal activity spectrum.2 Exceptions are members of the viscosin group that show no activity against fungi belonging to the genera of Candida, Aspergillus,237 and Cryptococcus.66,249 Some smaller CLPs also have been tested for their properties as antiprotozoal agents. The CLPs, which all belong to the viscosin262,263 or amphisin75 group, showed activity but due to the fact that most protozoans are not able to grown under axenic conditions inhibitory values could not be calculated. Jessenipeptin, a long CLP belonging to the fuscopeptins, was tested against Dictyostelium discoideum, a species of soil-dwelling amoebae that can be cultured axenically,264 and showed strong amoebicidal activity (IC50 = 2 μM).93
All ever tested pseudomonal CLPs are hemolytic (at low micromolar concentrations) but showed either no or only marginal anticancer activity,2 with the exception of MDN-0066 (ref. 59) and WLIP.265 Members of the syringafactin,105 viscosin,64 tolaasin,266 fuscopeptin,92 corpeptin,96 syringomycin,50,267,268 and syringopeptin101,269 groups are also phytotoxic and induce necrosis in plants at micromolar concentrations.
One CLP has also been identified as insecticidal. Orfamide A induces high mortality rates in the green peach aphid (Myzus persicae) with an LC50 value of 34.5 μg mL−1. M. persicae is the most significant aphid pest of peach and causes decreased growth in infested trees. In addition, it serves as a vector for plant viruses. This finding is of special interest, because CLPs could be used as biodegradable pesticides that are less stressful for the environment, in comparison to conventional methods.212
Interestingly, viscosin270 and MA026 show activity against enveloped viruses, which are surrounded by a lipid bilayer. It was demonstrated that MAO26, which is produced by Pseudomonas sp. RtIB026 found in the digestive tract of rainbow trout (Oncorhynchus mykiss), protects its host from hematopoietic necrosis virus.81 The molecular basis of the antiviral properties is unclear and raises the question of whether the CLPs have a specific target or inhibit the virus by integrating into the lipid bilayer, disrupting the membrane organization of the virus.
Only limited data is available for pseudomonal LLPs as this compound class was rarely tested for their microbicidal properties. No member of the corrugatin group was tested against any microorganism. Only the iron chelating properties of ornicorrugatin and histicorrugatin were investigated using the chrome azurol S (CAS) assay (Fig. 12b).54,55 Peptin31 inhibited the growth of different fungi, the Gram-positive Bacillus megaterium and was able to lyse RBCs (C50 = 5.7 μM).109 Sclerosin is able to lyse zoospores of the causative agent of potato blight, Phytophthora infestans, and showed antimicrobial activity against multiple Bacillus strains.110
As Geudens and Martins already pointed out, pseudomonal CLPs were never tested systematically, which impedes structure–activity relationship conclusions and prevents development of anti-infectives or biocontrol agents.2,274 Therefore, a panel of standardized bioassays should be defined that can be used to test lipopeptides against selected organisms using a consistent concentration range.
Selection criteria for organisms are rapid growth, no or low pathogenicity, wide availability, and ecological or medical relevance. A useful selection of relevant microorganisms was already established by the group of Takemoto.100 Most strains are available from the American Type Culture Collection (ATCC), a global bioresource center, and fulfill the required criteria. For Gram-positive bacteria, Staphylococcus aureus (ATCC 6538, human pathogen) and B. megaterium (ATCC 14381, soil inhabitant) seem appropriate choices. E. coli (ATCC 25922, human pathogen), P. aeruginosa (ATCC 15442, human pathogen), P. fluorescens (ATCC 13525, soil inhabitant), Serratia marcescens (ATCC 8100, human pathogen and soil inhabitant), and Citrobacter freundii (ATCC 8090, soil inhabitant) are suitable organisms to test activity against Gram-negative bacteria. Mycobacterium smegmatis (ATCC 14468) is a fast-growing mycobacterium that is considered non-pathogenic, only causing diseases in rare incidents, and a standard model organism in mycobacteria research. As for fungi, Fusarium oxysporum (ATCC 36877, soil inhabitant and plant pathogen), Rhizoctonia solani (ATCC 10182, soil inhabitant and plant pathogen) and Candida albicans (ATCC 10231, opportunistic human pathogen) are excellent test organisms for evaluating antifungal properties of pseudomonal lipopeptides. Different Pythium spp., which can cause a variety of diseases, can be used to test antagonistic properties of CLPs or LLPs against plant parasites.4,275 As a preliminary test to assess the antimicrobial activities of pseudomonal lipopeptides, the standardized Kirby–Bauer disk diffusion assays using an amount of 50 μg of per test should be used.276,277 If a lipopeptide shows high activity, which is indicated by large inhibition zones that are comparable to the positive control antibiotics, minimum inhibitory concentrations (MIC) should be determined using a concentration range of 0.1–100 μM.278
In addition to the standard microorganisms, pseudomonal lipopeptides should also be tested against other relevant species. D. discoideum (strains AX2 or AX4, http://dictybase.org) could be used as a model organism to test anti-protozoal effects of CLPs and LLPs.279 This amoeba is easily cultured, a universally occurring soil inhabitant, and preys on bacteria,280,281 which makes this organism an ecologically relevant282 species. Cell lines that are useful to determine first cytotoxicity profiles, which indicate if a lipopeptide could serve as a starting point for SAR studies to develop an active pharmaceutical ingredient, are HeLa (ATCC CCL-2, cervix), MCF7 (ATCC HTB-22, mammary gland), and HUVEC (ATCC CRL-1730, umbilical vein). For cell lines and amoebae, concentration ranges of 0.1–100 μM should be adequate for testing. Tetrazolium dye-based metabolic activity assays283,284 or direct cell counting methods93,125,214 can be used to determine inhibitory concentration values (IC50).
Unfortunately, not every research team has access to a green house and is able to assess phytotoxic properties of lipopeptides in their own laboratory using potted plants under standardized conditions. Thus, as an initial experiment leafy edible vegetables, like lettuce or chicory, could be inoculated with isolated lipopeptides, dissolved in sterile water at a concentration range of 2–20 μM,50,267,268 and incubated for 24 h or longer to investigate the formation of necrotic tissue.96,105,285 In addition, the axenically growing unicellular algae C. reinhardtii (SAG 73.72) could also be used to evaluate the activity spectrum of lipopeptides.286–288 This microalgae can generally be handled in the same fashion as other microorganisms and is suitable for performing Kirby–Bauer disk diffusion and standard MIC assays (concentration range of 0.1–100 μM).256
5. The role of lipopeptides in soil and plant ecology
5.1 Soil ecology of lipopeptides
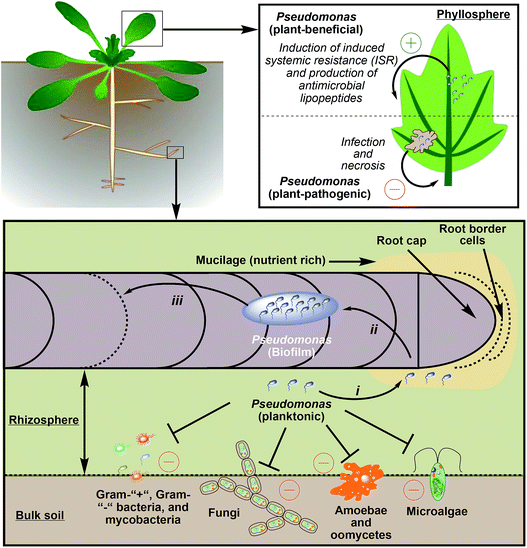
Lipopeptide-producing pseudomonads can be found in bulk soil, the rhizosphere,259 or in the phyllosphere,289,290 which is the total above-ground portion of plants (Fig. 13). When compared to bulk soil or the phyllosphere, the rhizosphere, which surrounds roots and only spans 1–3 mm in diameter, is a fertile region. In this area microbial growth is stimulated because roots secrete organic acids (e.g. citrate or succinate), monosaccharides, amino acids, and vitamins. Therefore, the density of bacteria in the rhizosphere is up to 1000 times higher than in bulk soil (rhizosphere effect).4 The area around the root cap and the border cells are especially rich in nutrients as they secrete mucilage, a matrix of high-molecular-weight compounds (e.g. polysaccharides). Mucilage not only influences structural and hydrological properties of soil,291 but also attracts plant-mutualistic292 and repels plant-antagonistic293 microorganism.The concentration gradient of carbon, nitrogen, and essential elements between bulk soil and the rhizosphere leads to a strong competition between soil organisms and a selection pressure that drives evolution.294 Especially soil pseudomonads use a diverse arsenal of chemicals to combat competitors and repel predators. In almost all ecological relevant interactions of pseudomonads with other species lipopeptides play an important role. Even nematodes, which usually prey on bacteria and can be suppressed by Pseudomonas spp. that produce 2,4-diacetylphloroglucinol and hydrogen cyanide,295–297 engage in a symbiotic relationship with pseudomonads. It was recently shown that the microbiota of Caenorhabditis elegans contains Pseudomonas strains, which protect the host against pathogenic Bacillus thuringiensis via massetolide E production.298
Pseudomonads isolated from the rhizosphere use lipopeptides to inhibit the growth of competing Gram-positive2 and Gram-negative bacteria,80 as well as fungi18 to harness scarce nutrients for their own proliferation.299 Interestingly, the antimicrobial effect of single Pseudomonas strains is sometimes related to a dual functionality of the produced CLP. For example, P. sp. DSS73 has antifungal properties because it can surround R. solani via an amphisin-dependent surface translocation and lays siege to it, which consequently leads to the death of the fungus. In this particular case, only the synergistic effects of surface motility and antifungal properties of amphisin can contain and impede the growth of R. solani.213
It was also shown that lipopeptides can inhibit, permeabilize, and immobilize microalgal cells. This enables pseudomonads to directly acquire micronutrients from algae, which are commonly found in soil, and, at the same time, to decimate the population of microbial competitors.256 In addition, lipopeptides of pseudomonal origin can also be amoebicidal75,93,263 and protect the producing organism against protists,300 which prey on and shape the rhizosphere microbiome.
In addition to antimicrobial properties, CLPs and LLPs generally confer pseudomonads a competitive advantage over other species in complex environments such as soil. Their surfactant properties allow lipopeptides to increase the wettability of water, which can lead to a better solubilization, diffusion, and uptake of water-insoluble nutrients. In addition, lipopeptide biosurfactants can also be used to entrap toxic compounds like hydrocarbons in biosurfactant micelles.7 This protects the producer organism on the one hand and enables an efficient biodegradation on the other hand.301,302 Finally, CLPs or LLPs can induce swarming in pseudomonads, which enables a fast exploration and occupation of new and potentially nutrient-rich territories such as the root tip.303
5.2 Lipopeptides in Pseudomonas-plant interactions
Pseudomonads often colonize and invade plants to form specific parasitic relationships to gain access to nutrients and sustain their growth.7 Prominent plant pathogens, which use lipopeptides as virulence factors, are for example P. syringae (induces canker disease in peach trees; syringomycin267) P. syringae pv. atrofaciens (induces basal glume rot in wheat and other cereals; syringomycin and syringopeptin),304,305P. fuscovaginae (induces sheath brown rot in rice; syringotoxin and fuscopeptin),92P. corrugata (induces tomato pit necrosis; cormycin A and corpeptin),50P. fluorescens 5064 (induces broccoli head rot; viscosin),306 and P. cichorii SF1-54 (induces lettuce midrib rot; cichofactin and chicopeptin).96,105The general infection cycle of plant-pathogenic Pseudomonas spp. starts with colonizing the leaf or fruit surfaces of a plant.307 At this stage, the biosurfactant properties of CLPs306 and LLPs105 are useful, because the pathogen can actively spread throughout the phyllosphere. This enables the efficient location of stomata or cracks in the upper epidermis of leaves and fruits. Afterwards, pseudomonads penetrate open stomata or wound sites to colonize the plant apoplast (extracellular space inside plant tissue).308 At this point pseudomonads switch from an epiphyte (organism that grows on the surface of a plant) to an endophyte (organism that grows inside a plant) life style and proliferate inside the plant tissue. This leads to an activation of the plant immune system, which can be counteracted by some pseudomonads,309,310 and to the production of phytotoxic CLPs by the pathogen, which permeabilize plant cells to access nutrients.268,311 Finally, this battle for resources leads to the formation of necrotic plant tissue and, in severe cases, to plant death (Fig. 13).
On the other side of the spectrum, lipopeptide producing Pseudomonas spp. also form plant-beneficial endophytic relationships with different plant hosts. For example, in the mutualistic relationship between sugar beets and the P. poae strain RE*1-1-14 the CLP poeamide is used by the plant-beneficial bacterium to swarm to the plant and to establish a biofilm-like colonization pattern on its roots. Thereupon, strain RE*1-1-14 penetrates the roots and establishes a stable population inside the plant. In exchange for nutrients, which are provided by the plant, the endophyte produces poeamide that has antagonistic activity against soil-borne plant pathogens such as the fungus R. solani and different oomycetes, including Phytophthora capsica and Pythium ultimum (Fig. 13).71
The massetolide A-producing strain P. fluorescens SS101 is an example of a mutualistic interaction that is only based on colonization of a plant and not on endophytic lifestyle. The strain colonizes seedlings and up-regulates plant disease resistance312 in tomato (Solanum lycopersicum), which allows the plant to withstand P. infestans infections and to reduce the expansion of existing lesions caused by the pathogen (Fig. 12).313 Colonization and activation of induced systemic resistance (ISR) are caused by massetolide A, as CLP-deficient mutants were significantly less effective in colonizing plants and in biocontrol.314
Further mutualistic interactions between epiphytic pseudomonads and plants are known. In these interactions, bacteria, in exchange for nutrients provided by the plant, produce antimicrobial lipopeptides that impede the spreading of plant pathogens in the rhizosphere. This phenomenon can be so prominent that certain types of soils are classified as disease-suppressive soils (“soils in which plants do not suffer from certain diseases or where disease severity is substantially reduced even though a virulent pathogen is present and the host plant is susceptible to the disease”).4 Usually, disease-suppressive soils contain Gammaproteobacteria that produce NRPs, which protect plants against fungal root pathogens or plant parasites.53
Many Pseudomonas spp. have been isolated from different crop rhizospheres all around the world (e.g. sugar beet,61,89,97,315 cocoyam,70,84 potato,51,94etc.), which obtain their biocontrol abilities through the production of antimicrobial lipopeptides. For example, the strain P. fluorescens In5, which was isolated from a disease-suppressive soil in southern Greenland and protects crops from potato late blight, produces two CLPs, namely nunamycin and nunapeptin. Nunamycin was demonstrated to inhibit the growth of the fungus R. solani, but not that of the oomycete Pythium aphanidermatum, which both occur in the same soil and are plant pathogens. Vice versa, nunapeptin inhibited P. aphanidermatum but not R. solani.51 In addition, a detailed analysis of fluorescent pseudomonads, found in the rhizosphere of healthy cocoyam crops in Cameroon, showed that 50% of the isolated strains produced CLPs. Most isolated and purified CLPs were also able to inhibit Pythium myriotylum, which is the causative agent of the cocoyam root rot disease.84 The ecological friendly mechanism of action of disease-suppressive soils has also lead to commercialization of different biocontrol Pseudomonas strains (e.g. Cerall® or Cedomon®), which, instead of pesticides, can be applied to soil to protect crops against microbial diseases.4
6. Conclusions
Lipopeptides are important biosurfactants and play an integral role in the life style of many Pseudomonas strains. Their physicochemical properties enable pseudomonads to efficiently swarm, establish biofilms, defend themselves against predators and competitors, form pseudomonas-plant relationships, acquire nutrients and degrade toxic compounds.Unfortunately, a universal theory that links structure of pseudomonal lipopeptides with biological activity and material properties is not available at present. This can be attributed to a lack of studies that compare the structure of CLPs and LLPs in solution with the supramolecular structure and conformation these molecules adapt in artificial and biological membranes. In addition, most biophysical studies of pseudomonal lipopeptides use a limited set of techniques (planar lipid bilayer techniques, quartz microbalance, CD-spectroscopy) to characterize the interaction between biosurfactants and model membranes, which do not reflect in vitro or in vivo conditions. In order to counteract this one-sided use of methods, we recommend the use of cell biological26 (fluorescence spectroscopy) and optical techniques316 (surface plasmon resonance, polarization interferometry, cryo-EM, atomic force microscopy) in combination with more suitable membrane mimetic systems (nanodiscs317) or live cells to generate a realistic picture of lipopeptide-biomembrane interactions.
Furthermore, evaluation of the biological properties of pseudomonal lipopeptides have focused on their antimicrobial properties. But CLPs and LLPs fulfill a more diverse set of functions and are also necessary to create and sustain symbiotic relationships; e.g. mixed bacteria communities318 and bacterial interspecies interactions.71,314 Therefore, the ecological context, such as the isolation environment, co-isolated species, and symbiotic partners should be taken into account when selecting bioassays to determine biological functionality.319 Methods that are suitable to visualize or quantify the distribution or effects of lipopeptides between pseudomonads and potential ecological interaction partners are imaging mass spectrometry (IMS)320,321 and omics techniques.322
Lastly, pseudomonal NRPS architecture and biochemistry is unique when compared to the three other genera (paenibacillaceae, Bacillus and Streptomyces) that are mainly known for lipopeptide biosynthesis.133 NRPS assembly lines in Pseudomonas spp., with the exception of the syringomycins, probably do not rely on external tailoring enzymes and modify amino acids during biosynthesis using only C domains. Therefore, experimental confirmation of these assumptions will generate insight into new biochemistry. Additionally, pseudomonal NRPS architecture follows a common genetic blueprint, which indicates common ancestors. Investigation of evolutionary processes, which caused the diversification of pseudomonal lipopeptides, could lead to an understanding of how domains in an assembly line can be duplicated, deleted, or exchanged without losing activity.323 The generated knowledge would be extremely valuable for the members of the biotechnology community, who engineer NRPSs to create peptide and lipopeptide analogs.324–326
Concerted action, from biophysical chemistry to biotechnology, is needed to understand the biological as well as the biophysical SARs of pseudomonal lipopeptides and the biochemistry behind their biosynthesis. These scientific efforts can ultimately lead to lipopeptide derivatives that represent the starting point for developing new biosurfactants, antimicrobial agents,274 anti-cancer drugs,59 antivirals,81 and even new pseudomonal biocontrol strains.4
7. Conflicts of interest
There are no conflicts to declare.8. Acknowledgments
We are grateful for financial support from the Leibniz Association and the Deutsche Forschungsgemeinschaft (collaborative research cluster ChemBioSys SFB1127). We would also like to thank the Japan Society for the Promotion of Science (Grant number: P16345) for financial support.9. Notes and references
- I. Sampedro, R. E. Parales, T. Krell and J. E. Hill, FEMS Microbiol. Rev., 2015, 39, 17–46 CAS.
- N. Geudens and J. C. Martins, Front. Microbiol., 2018, 9, 1867 CrossRef.
- R. A. Wilkes and L. Aristilde, J. Appl. Microbiol., 2017, 123, 582–593 CrossRef CAS.
- D. Haas and G. Défago, Nat. Rev. Microbiol., 2005, 3, 307–319 CrossRef CAS.
- F. L. S. de Alencar, J. A. Navoni and V. S. do Amaral, Environ. Sci. Pollut. Res., 2017, 24, 16545–16559 CrossRef.
- T. H. Mauchline and J. G. Malone, Curr. Opin. Microbiol., 2017, 37, 23–28 CrossRef.
- J. D'aes, K. De Maeyer, E. Pauwelyn and M. Höfte, Environ. Microbiol. Rep., 2010, 2, 359–372 CrossRef PubMed.
- B. S. Scales, R. P. Dickson, J. J. LiPuma and G. B. Huffnagle, Clin. Microbiol. Rev., 2014, 27, 927–948 CrossRef CAS.
- M. F. Moradali, S. Ghods and B. H. A. Rehm, Front. Cell. Infect. Microbiol., 2017, 7, 39 Search PubMed.
- F. Rojo, FEMS Microbiol. Rev., 2010, 34, 658–684 CrossRef CAS PubMed.
- C. K. Stover, X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory and M. V. Olson, Nature, 2000, 406, 959–964 CrossRef CAS.
- M. W. Silby, C. Winstanley, S. A. C. Godfrey, S. B. Levy and R. W. Jackson, FEMS Microbiol. Rev., 2011, 35, 652–680 CrossRef CAS.
- V. L. Kung, E. A. Ozer and A. R. Hauser, Microbiol. Mol. Biol. Rev., 2010, 74, 621–641 CrossRef CAS.
- R. L. Marvig, L. M. Sommer, S. Molin and H. K. Johansen, Nat. Genet., 2015, 47, 57–64 CrossRef CAS.
- G. L. Winsor, E. J. Griffiths, R. Lo, B. K. Dhillon, J. A. Shay and F. S. L. Brinkman, Nucleic Acids Res., 2016, 44, D646–D653 CrossRef CAS.
- I. T. Paulsen, C. M. Press, J. Ravel, D. Y. Kobayashi, G. S. A. Myers, D. V. Mavrodi, R. T. DeBoy, R. Seshadri, Q. Ren, R. Madupu, R. J. Dodson, A. S. Durkin, L. M. Brinkac, S. C. Daugherty, S. A. Sullivan, M. J. Rosovitz, M. L. Gwinn, L. Zhou, D. J. Schneider, S. W. Cartinhour, W. C. Nelson, J. Weidman, K. Watkins, K. Tran, H. Khouri, E. A. Pierson, L. S. P. Iii, L. S. Thomashow and J. E. Loper, Nat. Biotechnol., 2005, 23, 873–878 CrossRef CAS.
- H. Gross and J. E. Loper, Nat. Prod. Rep., 2009, 26, 1408 RSC.
- J. M. Raaijmakers, I. De Bruijn, O. Nybroe and M. Ongena, FEMS Microbiol. Rev., 2010, 34, 1037–1062 CrossRef CAS.
- K. Papenfort and B. L. Bassler, Nat. Rev. Microbiol., 2016, 14, 576–588 CrossRef CAS.
- B. N. Kunkel and C. P. Harper, J. Exp. Bot., 2018, 69, 245–254 CrossRef CAS.
- D. Schwarzer, R. Finking and M. A. Marahiel, Nat. Prod. Rep., 2003, 20, 275 RSC.
- J. Schimana, K. Gebhardt, A. Höltzel, D. G. Schmid, R. Süssmuth, J. Müller, R. Pukall and H.-P. Fiedler, J. Antibiot., 2002, 55, 565–570 CrossRef CAS.
- M. E. Evans, D. J. Feola and R. P. Rapp, Ann. Pharmacother., 1999, 33, 960–967 CrossRef CAS.
- P. G. Stansly and M. E. Schlosser, J. Bacteriol., 1947, 54, 549–556 CAS.
- R. H. Baltz, V. Miao and S. K. Wrigley, Nat. Prod. Rep., 2005, 22, 717 RSC.
- A. Müller, M. Wenzel, H. Strahl, F. Grein, T. N. V. Saaki, B. Kohl, T. Siersma, J. E. Bandow, H.-G. Sahl, T. Schneider and L. W. Hamoen, Proc. Natl. Acad. Sci. U.S.A., 2016, 113, E7077–E7086 CrossRef PubMed.
- K. Cleveland and M. Gelfand, Infect. Dis. Clin. Pract., 2013, 21(3), 213 CrossRef.
- W.-C. Chen, R.-S. Juang and Y.-H. Wei, Biochem. Eng. J., 2015, 103, 158–169 CrossRef CAS.
- H. Zeriouh, A. de Vicente, A. Pérez-García and D. Romero, Environ. Microbiol., 2014, 16, 2196–2211 CrossRef CAS.
- S. A. Cochrane and J. C. Vederas, Med. Res. Rev., 2016, 36, 4–31 CrossRef CAS.
- H. Zhao, D. Shao, C. Jiang, J. Shi, Q. Li, Q. Huang, M. S. R. Rajoka, H. Yang and M. Jin, Appl. Microbiol. Biotechnol., 2017, 101, 5951–5960 CrossRef CAS.
- O. Kurnasov, L. Jablonski, B. Polanuyer, P. Dorrestein, T. Begley and A. Osterman, FEMS Microbiol. Lett., 2003, 227, 219–227 CrossRef CAS.
- D. J. Atkinson, B. J. Naysmith, D. P. Furkert and M. A. Brimble, Beilstein J. Org. Chem., 2016, 12, 2325–2342 CrossRef CAS.
- J. A. Orwa, C. Govaerts, R. Busson, E. Roets, A. Van Schepdael and J. Hoogmartens, J. Chromatogr., A, 2001, 912, 369–373 CrossRef CAS.
- T. Velkov, A. Gallardo-Godoy, J. D. Swarbrick, M. A. T. Blaskovich, A. G. Elliott, M. Han, P. E. Thompson, K. D. Roberts, J. X. Huang, B. Becker, M. S. Butler, L. H. Lash, S. T. Henriques, R. L. Nation, S. Sivanesan, M.-A. Sani, F. Separovic, H. Mertens, D. Bulach, T. Seemann, J. Owen, J. Li and M. A. Cooper, Cell Chem. Biol., 2018, 25, 380–391 CrossRef CAS.
- F. Castiglione, A. Marazzi, M. Meli and G. Colombo, Magn. Reson. Chem., 2005, 43, 603–610 CrossRef CAS.
- M. Ngoh, C. Wrzesinski, Y. Yang, E. Zschiesche, T. Madsen, C. Stephenson, L. Newman and L. Deetz, J. Appl. Poult. Res., 2018, 27, 449–452 CrossRef CAS.
- A. Kakinuma, H. Sugino, M. Isono, G. Tamura and K. Arima, Agric. Biol. Chem., 1969, 33, 973–976 CrossRef CAS.
- F. Peypoux, M. Guinand, G. Michel, L. Delcambe, B. C. Das and E. Lederer, Biochemistry, 1978, 17, 3992–3996 CrossRef CAS.
- C. W. Moss, S. B. Samuels and R. E. Weaver, Appl. Microbiol., 1972, 24, 596–598 CAS.
- A. Ballio, F. Bossa, D. Di Giorgio, P. Ferranti, M. Paci, P. Pucci, A. Scaloni, A. Segre and G. A. Strobel, FEBS Lett., 1994, 355, 96–100 CrossRef CAS.
- N. Katayama, Y. Nozaki, K. Okonogi, S. Harada and H. Ono, J. Antibiot., 1993, 46, 65–70 CrossRef CAS.
- S. Tsubotani, N. Katayama, Y. Funabashi, H. Ono and S. Harada, J. Antibiot., 1993, 46, 287–293 CrossRef CAS.
- C. Bassarello, S. Lazzaroni, G. Bifulco, P. Lo Cantore, N. S. Iacobellis, R. Riccio, L. Gomez-Paloma and A. Evidente, J. Nat. Prod., 2004, 67, 811–816 CrossRef CAS.
- C. T. Walsh, R. V. O'Brien and C. Khosla, Angew. Chem., Int. Ed., 2013, 52, 7098–7124 CrossRef CAS.
- A. Segre, R. C. Bachmann, A. Ballio, F. Bossa, I. Grgurina, N. S. Iacobellis, G. Marino, P. Pucci, M. Simmaco and J. Y. Takemoto, FEBS Lett., 1989, 255, 27–31 CrossRef CAS.
- A. Scaloni, R. C. Bachmann, J. Y. Takemoto, D. Barra, M. Simmaco and A. Ballio, Nat. Prod. Lett., 1994, 4, 159–164 CrossRef CAS.
- N. Fukuchi, A. Isogai, J. Nakayama, S. Takayama, S. Yamashita, K. Suyama, J. Y. Takemoto and A. Suzuki, J. Chem. Soc., Perkin Trans. 1, 1992, 1149–1157 RSC.
- A. Ballio, F. Bossa, A. Collina, M. Gallo, N. S. Iacobellis, M. Paci, P. Pucci, A. Scaloni, A. Segre and M. Simmaco, FEBS Lett., 1990, 269, 377–380 CrossRef CAS.
- A. Scaloni, M. Dalla Serra, P. Amodeo, L. Mannina, R. M. Vitale, A. L. Segre, O. Cruciani, F. Lodovichetti, M. L. Greco, A. Fiore, M. Gallo, C. D'Ambrosio, M. Coraiola, G. Menestrina, A. Graniti and V. Fogliano, Biochem. J., 2004, 384, 25–36 CrossRef CAS.
- C. F. Michelsen, J. Watrous, M. A. Glaring, R. Kersten, N. Koyama, P. C. Dorrestein and P. Stougaard, mBio, 2015, 6(2), e00079 CrossRef.
- C. W. Johnston, M. A. Skinnider, M. A. Wyatt, X. Li, M. R. M. Ranieri, L. Yang, D. L. Zechel, B. Ma and N. A. Magarvey, Nat. Commun. DOI:10.1038/ncomms9421.
- R. Mendes, M. Kruijt, I. de Bruijn, E. Dekkers, M. van der Voort, J. H. M. Schneider, Y. M. Piceno, T. Z. DeSantis, G. L. Andersen, P. A. H. M. Bakker and J. M. Raaijmakers, Science, 2011, 332, 1097–1100 CrossRef CAS PubMed.
- S. Matthijs, N. Brandt, M. Ongena, W. Achouak, J.-M. Meyer and H. Budzikiewicz, BioMetals, 2016, 29, 467–485 CrossRef CAS.
- S. Matthijs, H. Budzikiewicz, M. Schäfer, B. Wathelet and P. Cornelis, Z. Naturforschung C, 2008, 63, 8–12 CAS.
- D. Risse, H. Beiderbeck, K. Taraz, H. Budzikiewicz and D. Gustine, Z. Naturforschung C, 1998, 53, 295–304 CAS.
- T. Janek, M. Łukaszewicz, T. Rezanka and A. Krasowska, Bioresour. Technol., 2010, 101, 6118–6123 CrossRef CAS.
- D. D. Nguyen, A. V. Melnik, N. Koyama, X. Lu, M. Schorn, J. Fang, K. Aguinaldo, T. L. Lincecum, M. G. K. Ghequire, V. J. Carrion, T. L. Cheng, B. M. Duggan, J. G. Malone, T. H. Mauchline, L. M. Sanchez, A. M. Kilpatrick, J. M. Raaijmakers, R. De Mot, B. S. Moore, M. H. Medema and P. C. Dorrestein, Nat. Microbiol., 2016, 2, 16197 CrossRef CAS.
- B. Cautain, N. de Pedro, C. Schulz, J. Pascual, T. da S. Sousa, J. Martin, I. Pérez-Victoria, F. Asensio, I. González, G. F. Bills, F. Reyes, O. Genilloud and F. Vicente, PLoS One, 2015, 10, e0125221 CrossRef.
- M. Hiramoto, K. Okada and S. Nagai, Tetrahedron Lett., 1970, 11, 1087–1090 CrossRef.
- T. H. Nielsen, C. Christophersen, U. Anthoni and J. Sorensen, J. Appl. Microbiol., 1999, 87, 80–90 CrossRef CAS.
- N. Geudens, M. De Vleeschouwer, K. Fehér, H. Rokni-Zadeh, M. G. K. Ghequire, A. Madder, R. De Mot, J. C. Martins and D. Sinnaeve, ChemBioChem, 2014, 15, 2736–2746 CrossRef CAS.
- F. Han, R. J. Mortishire-Smith, P. B. Rainey and D. H. Williams, Acta Crystallogr. C, 1992, 48, 1965–1968 CrossRef.
- M. S. C. Pedras, N. Ismail, J. W. Quail and S. M. Boyetchko, Phytochemistry, 2003, 62, 1105–1114 CrossRef CAS.
- J. W. Quail, N. Ismail, M. S. C. Pedras and S. M. Boyetchko, Acta Crystallogr. C, 2002, 58, o268–o271 CrossRef.
- D. Sinnaeve, C. Michaux, J. Van hemel, J. Vandenkerckhove, E. Peys, F. A. M. Borremans, B. Sas, J. Wouters and J. C. Martins, Tetrahedron, 2009, 65, 4173–4181 CrossRef CAS.
- J. Gerard, R. Lloyd, T. Barsby, P. Haden, M. T. Kelly and R. J. Andersen, J. Nat. Prod., 1997, 60, 223–229 CrossRef CAS.
- H. Gross, V. O. Stockwell, M. D. Henkels, B. Nowak-Thompson, J. E. Loper and W. H. Gerwick, Chem. Biol., 2007, 14, 53–63 CrossRef CAS.
- Z. Ma, N. Geudens, N. P. Kieu, D. Sinnaeve, M. Ongena, J. C. Martins and M. Höfte, Front. Microbiol., 2016, 7, 382 Search PubMed.
- J. D'aes, N. P. Kieu, V. Léclère, C. Tokarski, F. E. Olorunleke, K. De Maeyer, P. Jacques, M. Höfte and M. Ongena, Environ. Microbiol., 2014, 16, 2282–2300 CrossRef.
- C. Zachow, G. Jahanshah, I. de Bruijn, C. Song, F. Ianni, Z. Pataj, H. Gerhardt, I. Pianet, M. Lämmerhofer, G. Berg, H. Gross and J. M. Raaijmakers, Mol. Plant Microbe Interact., 2015, 28, 800–810 CrossRef CAS.
- H. Weisshoff, S. Hentschel, I. Zaspel, R. Jarling, E. Krause and T. L. H. Pham, Nat. Prod. Commun., 2014, 9, 989–996 CrossRef CAS.
- D. Sørensen, T. H. Nielsen, C. Christophersen, J. Sørensen and M. Gajhede, Acta Crystallogr. C, 2001, 57, 1123–1124 CrossRef.
- A. Henriksen, U. Anthoni, T. H. Nielsen, J. Sørensen, C. Christophersen and M. Gajhede, Acta Crystallogr. C, 2000, 56, 113–115 CrossRef.
- S. Götze, R. Herbst-Irmer, M. Klapper, H. Görls, K. R. A. Schneider, R. Barnett, T. Burks, U. Neu and P. Stallforth, ACS Chem. Biol., 2017, 12, 2498–2502 CrossRef.
- A. Lange, H. Sun, J. Pilger, U. M. Reinscheid and H. Gross, ChemBioChem, 2012, 13, 2671–2675 CrossRef CAS.
- H. Ui, T. Miyake, H. Iinuma, M. Imoto, H. Naganawa, S. Hattori, M. Hamada, T. Takeuchi, S. Umezawa and K. Umezawa, J. Org. Chem., 1997, 62, 103–108 CrossRef CAS.
- D. Sørensen, T. H. Nielsen, J. Sørensen and C. Christophersen, Tetrahedron Lett., 2002, 43, 4421–4423 CrossRef.
- M. Schlusselhuber, J. Godard, M. Sebban, B. Bernay, D. Garon, V. Seguin, H. Oulyadi and N. Desmasures, Front. Microbiol., 2018, 9, 1030 CrossRef.
- W. Li, H. Rokni-Zadeh, M. De Vleeschouwer, M. G. K. Ghequire, D. Sinnaeve, G.-L. Xie, J. Rozenski, A. Madder, J. C. Martins and R. De Mot, PLoS One, 2013, 8, e62946 CrossRef CAS.
- S. Shimura, M. Ishima, S. Nakajima, T. Fujii, N. Himeno, K. Ikeda, J. Izaguirre-Carbonell, H. Murata, T. Takeuchi, S. Kamisuki, T. Suzuki, K. Kuramochi, K. Watashi, S. Kobayashi and F. Sugawara, J. Am. Chem. Soc., 2013, 135, 18949–18956 CrossRef CAS.
- I. Kuiper, E. L. Lagendijk, R. Pickford, J. P. Derrick, G. E. M. Lamers, J. E. Thomas-Oates, B. J. J. Lugtenberg and G. V. Bloemberg, Mol. Microbiol., 2003, 51, 97–113 CrossRef.
- G. Jahanshah, Q. Yan, H. Gerhardt, Z. Pataj, M. Lämmerhofer, I. Pianet, M. Josten, H.-G. Sahl, M. W. Silby, J. E. Loper and H. Gross, J. Nat. Prod., 2019, 82(2), 301–308 CrossRef CAS PubMed.
- F. E. Oni, N. Geudens, O. O. Omoboye, L. Bertier, H. G. K. Hua, A. Adiobo, D. Sinnaeve, J. C. Martins and M. Höfte, Environ. Microbiol., 2019, 21(3), 1019–1034 CrossRef CAS.
- I. Vallet-Gely, A. Novikov, L. Augusto, P. Liehl, G. Bolbach, M. Pechy-Tarr, P. Cosson, C. Keel, M. Caroff and B. Lemaitre, Appl. Environ. Microbiol., 2010, 76, 910–921 CrossRef CAS.
- H. B. Bode, D. Reimer, S. W. Fuchs, F. Kirchner, C. Dauth, C. Kegler, W. Lorenzen, A. O. Brachmann and P. Grün, Chem.–Eur. J., 2012, 18, 2342–2348 CrossRef CAS.
- Y. Nakagawa, Y. Ikenishi, K. Iwatani and T. Yoshida, J. Antibiot., 1992, 45, 824–831 CrossRef.
- C. M. Miller, R. V. Miller, D. Garton-Kenny, B. Redgrave, J. Sears, M. M. Condron, D. B. Teplow and G. A. Strobel, J. Appl. Microbiol., 1998, 84, 937–944 CrossRef CAS.
- T. H. Nielsen, D. Sørensen, C. Tobiasen, J. B. Andersen, C. Christophersen, M. Givskov and J. Sørensen, Appl. Environ. Microbiol., 2002, 68, 3416–3423 CrossRef CAS.
- J. C. Nutkins, R. J. Mortishire-Smith, L. C. Packman, C. L. Brodey, P. B. Rainey, K. Johnstone and D. H. Williams, J. Am. Chem. Soc., 1991, 113, 2621–2627 CrossRef CAS.
- K. Scherlach, G. Lackner, K. Graupner, S. Pidot, T. Bretschneider and C. Hertweck, ChemBioChem, 2013, 14, 2439–2443 CrossRef CAS.
- A. Ballio, F. Bossa, L. Camoni, D. Di Giorgio, M.-C. Flamand, H. Maraite, G. Nitti, P. Pucci and A. Scaloni, FEBS Lett., 1996, 381, 213–216 CrossRef CAS.
- J. Arp, S. Götze, R. Mukherji, D. J. Mattern, M. García-Altares, M. Klapper, D. A. Brock, A. A. Brakhage, J. E. Strassmann, D. C. Queller, B. Bardl, K. Willing, G. Peschel and P. Stallforth, Proc. Natl. Acad. Sci. U.S.A., 2018, 115, 3758–3763 CrossRef CAS.
- C. F. Michelsen, H. Jensen, V. J. Venditto, R. C. Hennessy and P. Stougaard, PeerJ, 2015, 3, e1476 CrossRef.
- M. C. Emanuele, A. Scaloni, P. Lavermicocca, N. S. Jacobellis, L. Camoni, D. Di Giorgio, P. Pucci, M. Paci, A. Segre and A. Ballio, FEBS Lett., 1998, 433, 317–320 CrossRef CAS.
- C.-J. Huang, E. Pauwelyn, M. Ongena, D. Debois, V. Leclère, P. Jacques, P. Bleyaert and M. Höfte, Mol. Plant Microbe Interact., 2015, 28, 1009–1022 CrossRef CAS.
- M. Van Der Voort, H. J. G. Meijer, Y. Schmidt, J. Watrous, E. Dekkers, R. Mendes, P. C. Dorrestein, H. Gross and J. M. Raaijmakers, Front. Microbiol., 2015, 6, 693 Search PubMed.
- A. Ballio, D. Barra, F. Bossa, A. Collina, I. Grgurina, G. Marino, G. Moneti, M. Paci, P. Pucci, A. Segre and M. Simmaco, FEBS Lett., 1991, 291, 109–112 CrossRef CAS.
- A. Ballio, F. Bossa, D. Di Giorgio, A. Di Nola, C. Manetti, M. Paci, A. Scaloni and A. L. Segre, Eur. J. Biochem., 1995, 234, 747–758 CrossRef CAS.
- I. Grgurina, M. Bensaci, G. Pocsfalvi, L. Mannina, O. Cruciani, A. Fiore, V. Fogliano, K. N. Sorensen and J. Y. Takemoto, Antimicrob. Agents Chemother., 2005, 49, 5037–5045 CrossRef CAS.
- I. Grgurina, F. Mariotti, V. Fogliano, M. Gallo, A. Scaloni, N. S. Iacobellis, P. Lo Cantore, L. Mannina, V. van Axel Castelli, M. L. Greco and A. Graniti, Biochim. Biophys. Acta, 2002, 1597, 81–89 CrossRef CAS.
- A. Isogai, H. Iguchi, J. Nakayama, A. Kusai, J. Y. Takemoto and A. Suzuki, Biosc. Biotech. Biochem., 1995, 59, 1374–1376 CrossRef CAS.
- A. Scaloni, L. Camoni, D. di Giorgio, M. Scortichini, R. Cozzolino and A. Ballio, Physiol. Mol. Plant Pathol., 1997, 51, 259–264 CrossRef CAS.
- A. D. Berti, N. J. Greve, Q. H. Christensen and M. G. Thomas, J. Bacteriol., 2007, 189, 6312–6323 CrossRef CAS.
- E. Pauwelyn, C.-J. Huang, M. Ongena, V. Leclère, P. Jacques, P. Bleyaert, H. Budzikiewicz, M. Schäfer and M. Höfte, Mol. Plant Microbe Interact., 2013, 26, 585–598 CrossRef CAS.
- D. K. Hancock, B. Coxon, S.-Y. Wang, E. White V, D. J. Reeder and J. M. Bellama, J. Chem. Soc., Chem. Commun., 1993, 468–470 RSC.
- T. A. Ronnebaum and A. L. Lamb, Curr. Opin. Struct. Biol., 2018, 53, 1–11 CrossRef CAS.
- C. D. Hardy and A. Butler, J. Biol. Inorg Chem., 2018, 23(7), 957–967 CrossRef CAS.
- A. Fiore, L. Mannina, A. P. Sobolev, A. M. Salzano, A. Scaloni, I. Grgurina, M. R. Fullone, M. Gallo, C. Swasey, V. Fogliano and J. Y. Takemoto, FEMS Microbiol. Lett., 2008, 286, 158–165 CrossRef CAS.
- C. L. Berry, A. K. C. Brassinga, L. J. Donald, W. G. D. Fernando, P. C. Loewen and T. R. de Kievit, Can. J. Microbiol., 2012, 58, 1027–1034 CrossRef CAS.
- H. Stephan, S. Freund, W. Beck, G. Jung, J.-M. Meyer and G. Winkelmann, BioMetals, 1993, 6, 93–100 CrossRef CAS.
- J.-M. Meyer, T. Van Van, A. Stintzi, O. Berge and G. Winkelmann, BioMetals, 1995, 8, 309–317 CrossRef CAS.
- M. Papagianni, Biotechnol. Adv., 2003, 21, 465–499 CrossRef CAS.
- V. S. de Paula and A. P. Valente, Molecules, 2018, 23, 2040 CrossRef.
- P. G. Arnison, M. J. Bibb, G. Bierbaum, A. A. Bowers, T. S. Bugni, G. Bulaj, J. A. Camarero, D. J. Campopiano, G. L. Challis, J. Clardy, P. D. Cotter, D. J. Craik, M. Dawson, E. Dittmann, S. Donadio, P. C. Dorrestein, K.-D. Entian, M. A. Fischbach, J. S. Garavelli, U. Göransson, C. W. Gruber, D. H. Haft, T. K. Hemscheidt, C. Hertweck, C. Hill, A. R. Horswill, M. Jaspars, W. L. Kelly, J. P. Klinman, O. P. Kuipers, A. J. Link, W. Liu, M. A. Marahiel, D. A. Mitchell, G. N. Moll, B. S. Moore, R. Müller, S. K. Nair, I. F. Nes, G. E. Norris, B. M. Olivera, H. Onaka, M. L. Patchett, J. Piel, M. J. T. Reaney, S. Rebuffat, R. P. Ross, H.-G. Sahl, E. W. Schmidt, M. E. Selsted, K. Severinov, B. Shen, K. Sivonen, L. Smith, T. Stein, R. D. Süssmuth, J. R. Tagg, G.-L. Tang, A. W. Truman, J. C. Vederas, C. T. Walsh, J. D. Walton, S. C. Wenzel, J. M. Willey and W. A. van der Donk, Nat. Prod. Rep., 2013, 30, 108–160 RSC.
- M. A. Ortega and W. A. van der Donk, Cell Chem. Biol., 2016, 23, 31–44 CrossRef CAS.
- R. D. Süssmuth and A. Mainz, Angew. Chem., Int. Ed., 2017, 56, 3770–3821 CrossRef.
- M. A. Fischbach and C. T. Walsh, Chem. Rev., 2006, 106, 3468–3496 CrossRef CAS.
- N. Roongsawang, K. Hase, M. Haruki, T. Imanaka, M. Morikawa and S. Kanaya, Chem. Biol., 2003, 10, 869–880 CrossRef CAS.
- M. A. Marahiel, Nat. Prod. Rep., 2016, 33, 136–140 RSC.
- B. R. Miller, J. A. Sundlov, E. J. Drake, T. A. Makin and A. M. Gulick, Proteins, 2014, 82, 2691–2702 CrossRef CAS.
- K. Bloudoff and T. M. Schmeing, Biochim. Biophys. Acta, 2017, 1865, 1587–1604 CrossRef CAS.
- A. M. Gulick, ACS Chem. Biol., 2009, 4, 811–827 CrossRef CAS.
- T. Kittilä, A. Mollo, L. K. Charkoudian and M. J. Cryle, Angew. Chem., Int. Ed., 2016, 55, 9834–9840 CrossRef.
- M. Klapper, D. Braga, G. Lackner, R. Herbst and P. Stallforth, Cell Chem. Biol., 2018, 25, 659–665 CrossRef CAS.
- B. Wang, Q. Kang, Y. Lu, L. Bai and C. Wang, Proc. Natl. Acad. Sci. U.S.A., 2012, 109, 1287–1292 CrossRef CAS.
- J.-F. Dubern, E. R. Coppoolse, W. J. Stiekema and G. V. Bloemberg, Microbiology, 2008, 154, 2070–2083 CrossRef CAS.
- S. Meyer, J.-C. Kehr, A. Mainz, D. Dehm, D. Petras, R. D. Süssmuth and E. Dittmann, Cell Chem. Biol., 2016, 23, 462–471 CrossRef CAS.
- F. I. Kraas, T. W. Giessen and M. A. Marahiel, FEBS Lett., 2012, 586, 283–288 CrossRef CAS.
- F. I. Kraas, V. Helmetag, M. Wittmann, M. Strieker and M. A. Marahiel, Chem. Biol., 2010, 17, 872–880 CrossRef CAS.
- C. J. Balibar, F. H. Vaillancourt and C. T. Walsh, Chem. Biol., 2005, 12, 1189–1200 CrossRef CAS.
- C. Luo, X. Liu, H. Zhou, X. Wang and Z. Chen, Appl. Environ. Microbiol., 2015, 81, 422–431 CrossRef CAS.
- N. Roongsawang, K. Washio and M. Morikawa, Int. J. Mol. Sci., 2010, 12, 141–172 CrossRef.
- T. Kuranaga, K. Matsuda, A. Sano, M. Kobayashi, A. Ninomiya, K. Takada, S. Matsunaga and T. Wakimoto, Angew. Chem., Int. Ed., 2018, 57, 9447–9451 CrossRef CAS.
- H. Komaki, K. Sakurai, A. Hosoyama, A. Kimura, Y. Igarashi and T. Tamura, Sci. Rep., 2018, 8, 6888 CrossRef.
- C. Fu, L. Keller, A. Bauer, M. Brönstrup, A. Froidbise, P. Hammann, J. Herrmann, G. Mondesert, M. Kurz, M. Schiell, D. Schummer, L. Toti, J. Wink and R. Müller, J. Am. Chem. Soc., 2015, 137, 7692–7705 CrossRef CAS PubMed.
- R. Hermenau, K. Ishida, S. Gama, B. Hoffmann, M. Pfeifer-Leeg, W. Plass, J. F. Mohr, T. Wichard, H.-P. Saluz and C. Hertweck, Nat. Chem. Biol., 2018, 14, 841–843 CrossRef CAS.
- C. A. Galea, M. Han, Y. Zhu, K. Roberts, J. Wang, P. E. Thompson, J. Li and T. Velkov, J. Nat. Prod., 2017, 80, 1264–1274 CrossRef CAS.
- S. Müller, E. Garcia-Gonzalez, A. Mainz, G. Hertlein, N. C. Heid, E. Mösker, H. van den Elst, H. S. Overkleeft, E. Genersch and R. D. Süssmuth, Angew. Chem., Int. Ed., 2014, 53, 10821–10825 CrossRef.
- T. Stachelhaus and C. T. Walsh, Biochemistry, 2000, 39, 5775–5787 CrossRef CAS.
- V. De Crécy-Lagard, P. Marlière and W. Saurin, C. R. Acad. Sci., Ser. III, 1995, 318, 927–936 Search PubMed.
- W.-H. Chen, K. Li, N. S. Guntaka and S. D. Bruner, ACS Chem. Biol., 2016, 11, 2293–2303 CrossRef CAS.
- N. Roongsawang, K. Washio and M. Morikawa, ChemBioChem, 2007, 8, 501–512 CrossRef CAS.
- T. Izoré and M. J. Cryle, Nat. Prod. Rep., 2018, 35, 1120–1139 RSC.
- H. Rokni-Zadeh, W. Li, A. Sanchez-Rodriguez, D. Sinnaeve, J. Rozenski, J. C. Martins and R. De Mot, Appl. Environ. Microbiol., 2012, 78, 4826–4834 CrossRef CAS.
- B. K. Scholz-Schroeder, J. D. Soule and D. C. Gross, Mol. Plant Microbe Interact., 2003, 16, 271–280 CrossRef CAS.
- E. Guenzi, G. Galli, I. Grgurina, D. C. Gross and G. Grandi, J. Biol. Chem., 1998, 273, 32857–32863 CrossRef CAS.
- G. W. Xu and D. C. Gross, J. Bacteriol., 1988, 170, 5680–5688 CrossRef CAS.
- A. Koglin, F. Löhr, F. Bernhard, V. V. Rogov, D. P. Frueh, E. R. Strieter, M. R. Mofid, P. Güntert, G. Wagner, C. T. Walsh, M. A. Marahiel and V. Dötsch, Nature, 2008, 454, 907–911 CrossRef CAS.
- G. M. Singh, P. D. Fortin, A. Koglin and C. T. Walsh, Biochemistry, 2008, 47, 11310–11320 CrossRef CAS.
- F. H. Vaillancourt, J. Yin and C. T. Walsh, Proc. Natl. Acad. Sci. U.S.A., 2005, 102, 10111–10116 CrossRef CAS.
- L. C. Blasiak, F. H. Vaillancourt, C. T. Walsh and C. L. Drennan, Nature, 2006, 440, 368–371 CrossRef CAS.
- G. M. Singh, F. H. Vaillancourt, J. Yin and C. T. Walsh, Chem. Biol., 2007, 14, 31–40 CrossRef CAS.
- I. Grgurina and F. Mariotti, FEBS Lett., 1999, 462, 151–154 CrossRef CAS.
- O. P. Kuipers, H. S. Rollema, W. M. Yap, H. J. Boot, R. J. Siezen and W. M. de Vos, J. Biol. Chem., 1992, 267, 24340–24346 CAS.
- M. Strieker and M. A. Marahiel, ChemBioChem, 2009, 10, 607–616 CrossRef CAS.
- M. A. Ortega, Y. Hao, Q. Zhang, M. C. Walker, W. A. van der Donk and S. K. Nair, Nature, 2015, 517, 509–512 CrossRef CAS.
- C. Chatterjee, L. M. Miller, Y. L. Leung, L. Xie, M. Yi, N. L. Kelleher and W. A. van der Donk, J. Am. Chem. Soc., 2005, 127, 15332–15333 CrossRef CAS.
- N. Garg, L. M. A. Salazar-Ocampo and W. A. van der Donk, Proc. Natl. Acad. Sci. U.S.A., 2013, 110, 7258–7263 CrossRef CAS.
- M. L. Workentine, L. Chang, H. Ceri and R. J. Turner, FEMS Microbiol. Lett., 2009, 292, 50–56 CrossRef CAS PubMed.
- J. M. Raaijmakers, I. de Bruijn and M. J. D. de Kock, Mol. Plant Microbe Interact., 2006, 19, 699–710 CrossRef CAS PubMed.
- J. W. Willett and S. Crosson, Mol. Microbiol., 2017, 103, 197–202 CrossRef CAS.
- A. Ali-Ahmad, F. Fadel, C. Sebban-Kreuzer, M. Ba, G. D. Pélissier, O. Bornet, F. Guerlesquin, Y. Bourne, C. Bordi and F. Vincent, Sci. Rep., 2017, 7, 11262 CrossRef PubMed.
- B. Koch, T. H. Nielsen, D. Sørensen, J. B. Andersen, C. Christophersen, S. Molin, M. Givskov, J. Sørensen and O. Nybroe, Appl. Environ. Microbiol., 2002, 68, 4509–4516 CrossRef CAS.
- S. Zuber, F. Carruthers, C. Keel, A. Mattart, C. Blumer, G. Pessi, C. Gigot-Bonnefoy, U. Schnider-Keel, S. Heeb, C. Reimmann and D. Haas, Mol. Plant Microbe Interact., 2003, 16, 634–644 CrossRef CAS.
- A. Brencic, K. A. McFarland, H. R. McManus, S. Castang, I. Mogno, S. L. Dove and S. Lory, Mol. Microbiol., 2009, 73, 434–445 CrossRef CAS.
- C. Song, M. van der Voort, J. van de Mortel, K. A. Hassan, L. D. H. Elbourne, I. T. Paulsen, J. E. Loper and J. M. Raaijmakers, Microb. Biotechnol., 2015, 8, 296–310 CrossRef CAS.
- C. L. Bender, F. A. N-Chaidez and D. C. Gross, Microbiol. Mol. Biol. Rev., 1999, 63, 266–292 CAS.
- J.-F. Dubern, E. L. Lagendijk, B. J. J. Lugtenberg and G. V. Bloemberg, J. Bacteriol., 2005, 187, 5967–5976 CrossRef CAS.
- I. de Bruijn and J. M. Raaijmakers, J. Bacteriol., 2009, 191, 1910–1923 CrossRef CAS.
- P. Stallforth, D. A. Brock, A. M. Cantley, X. Tian, D. C. Queller, J. E. Strassmann and J. Clardy, Proc. Natl. Acad. Sci. U.S.A., 2013, 110, 14528–14533 CrossRef CAS.
- F. E. Olorunleke, N. P. Kieu, E. De Waele, M. Timmerman, M. Ongena and M. Höfte, Microbiologyopen, 2017, 6, e00499 CrossRef.
- I. de Bruijn and J. M. Raaijmakers, Appl. Environ. Microbiol., 2009, 75, 4753–4761 CrossRef CAS PubMed.
- C. Fuqua, S. C. Winans and E. P. Greenberg, Annu. Rev. Microbiol., 1996, 50, 727–751 CrossRef CAS.
- C.-S. Tsai and S. C. Winans, Mol. Microbiol., 2010, 77, 1072–1082 CrossRef CAS.
- X. Cui, R. Harling, P. Mutch and D. Darling, Eur. J. Plant Pathol., 2005, 111, 297–308 CrossRef CAS.
- J.-F. Dubern, B. J. J. Lugtenberg and G. V. Bloemberg, J. Bacteriol., 2006, 188, 2898–2906 CrossRef CAS.
- B. L. Bassler and R. Losick, Cell, 2006, 125, 237–246 CrossRef CAS PubMed.
- J. Lee and L. Zhang, Protein Cell, 2015, 6, 26–41 CrossRef CAS.
- F. Trottmann, J. Franke, K. Ishida, M. Garcia-Altares and C. Hertweck, Angew. Chem., Int. Ed. Engl., 2019, 58(1), 200–204 CrossRef CAS PubMed.
- C. Grandclément, M. Tannières, S. Moréra, Y. Dessaux and D. Faure, FEMS Microbiol. Rev., 2016, 40, 86–116 CrossRef.
- M. A. Welsh and H. E. Blackwell, FEMS Microbiol. Rev., 2016, 40, 774–794 CrossRef CAS.
- B. A. Hense and M. Schuster, Microbiol. Mol. Biol. Rev., 2015, 79, 153–169 CrossRef CAS PubMed.
- V. L. Vaughn and D. C. Gross, PLoS One, 2016, 11, e0150234 CrossRef PubMed.
- S.-E. Lu, B. K. Scholz-Schroeder and D. C. Gross, Mol. Plant Microbe Interact., 2002, 15, 43–53 CrossRef CAS PubMed.
- N. Wang, S.-E. Lu, A. R. Records and D. C. Gross, J. Bacteriol., 2006, 188, 3290–3298 CrossRef CAS.
- N. Wang, S.-E. Lu, J. Wang, Z. J. Chen and D. C. Gross, Mol. Plant Microbe Interact., 2006, 19, 257–269 CrossRef CAS PubMed.
- K. Washio, S. P. Lim, N. Roongsawang and M. Morikawa, Biosc. Biotech. Biochem., 2010, 74, 992–999 CrossRef CAS.
- C. P. Strano, P. Bella, G. Licciardello, A. Fiore, A. R. Lo Piero, V. Fogliano, V. Venturi and V. Catara, Mol. Plant Pathol., 2015, 16, 495–506 CrossRef CAS PubMed.
- R. C. Hennessy, C. B. W. Phippen, K. F. Nielsen, S. Olsson and P. Stougaard, Microbiologyopen, 2017, 6(6), e00516 CrossRef.
- C. Fuqua and E. P. Greenberg, Nat. Rev. Mol. Cell Biol., 2002, 3, 685–695 CrossRef CAS.
- J. Zhu and S. C. Winans, Proc. Natl. Acad. Sci. U.S.A., 2001, 98, 1507–1512 CrossRef CAS.
- C. Song, G. Sundqvist, E. Malm, I. de Bruijn, A. Kumar, J. van de Mortel, V. Bulone and J. M. Raaijmakers, BMC Microbiol., 2015, 15, 29 CrossRef PubMed.
- C. Song, K. Aundy, J. van de Mortel and J. M. Raaijmakers, FEMS Microbiol. Lett., 2014, 356, 166–175 CrossRef CAS PubMed.
- K. T. Hughes and K. Mathee, Annu. Rev. Microbiol., 1998, 52, 231–286 CrossRef CAS PubMed.
- S.-E. Lu, J. D. Soule and D. C. Gross, Appl. Environ. Microbiol., 2003, 69, 7273–7280 CrossRef CAS.
- R. Mukherji, S. Chowdhury and P. Stallforth, bioRxiv, 2019, 584987 Search PubMed.
- D. E. Otzen, Biochim. Biophys. Acta Biomembr., 2017, 1859, 639–649 CrossRef CAS.
- S. Cameotra and R. Makkar, Curr. Opin. Microbiol., 2004, 7, 262–266 CrossRef CAS.
- I. Langmuir, J. Am. Chem. Soc., 1917, 39, 1848–1906 CrossRef CAS.
- D. K. F. Santos, R. D. Rufino, J. M. Luna, V. A. Santos and L. A. Sarubbo, Int. J. Mol. Sci., 2016, 17(3), 401 CrossRef PubMed.
- J. D. Berry, M. J. Neeson, R. R. Dagastine, D. Y. C. Chan and R. F. Tabor, J. Colloid Interface Sci., 2015, 454, 226–237 CrossRef CAS.
- T. Kairaliyeva, E. V. Aksenenko, N. Mucic, A. V. Makievski, V. B. Fainerman and R. Miller, J. Surfactants Deterg., 2017, 20, 1225–1241 CrossRef CAS.
- A. Y. Burch, B. K. Shimada, S. W. A. Mullin, C. A. Dunlap, M. J. Bowman and S. E. Lindow, J. Bacteriol., 2012, 194, 1287–1298 CrossRef CAS.
- T. Janek, L. R. Rodrigues, E. J. Gudiña and Ż. Czyżnikowska, Colloids Surf., B, 2016, 146, 498–506 CrossRef CAS.
- I. de Bruijn, M. J. D. de Kock, M. Yang, P. de Waard, T. A. van Beek and J. M. Raaijmakers, Mol. Microbiol., 2007, 63, 417–428 CrossRef CAS PubMed.
- H. S. Saini, B. E. Barragán-Huerta, A. Lebrón-Paler, J. E. Pemberton, R. R. Vázquez, A. M. Burns, M. T. Marron, C. J. Seliga, A. A. L. Gunatilaka and R. M. Maier, J. Nat. Prod., 2008, 71, 1011–1015 CrossRef CAS PubMed.
- I. de Bruijn, M. J. D. de Kock, P. de Waard, T. A. van Beek and J. M. Raaijmakers, J. Bacteriol., 2008, 190, 2777–2789 CrossRef CAS PubMed.
- M. L. Hutchison, M. A. Tester and D. C. Gross, Mol. Plant Microbe Interact., 1995, 8, 610–620 CrossRef CAS PubMed.
- M. Dalla Serra, G. Fagiuoli, P. Nordera, I. Bernhart, C. Della Volpe, D. Di Giorgio, A. Ballio and G. Menestrina, Mol. Plant Microbe Interact., 1999, 12, 391–400 CrossRef CAS PubMed.
- P. Flury, P. Vesga, M. Péchy-Tarr, N. Aellen, F. Dennert, N. Hofer, K. P. Kupferschmied, P. Kupferschmied, Z. Metla, Z. Ma, S. Siegfried, S. de Weert, G. Bloemberg, M. Höfte, C. J. Keel and M. Maurhofer, Front. Microbiol., 2017, 8, 100 Search PubMed.
- J. Y. Jang, S. Y. Yang, Y. C. Kim, C. W. Lee, M. S. Park, J. C. Kim and I. S. Kim, J. Agric. Food Chem., 2013, 61, 6786–6791 CrossRef CAS.
- J. B. Andersen, B. Koch, T. H. Nielsen, D. Sørensen, M. Hansen, O. Nybroe, C. Christophersen, J. Sørensen, S. Molin and M. Givskov, Microbiology, 2003, 149, 37–46 CrossRef CAS.
- M. Klapper, S. Götze, R. Barnett, K. Willing and P. Stallforth, Angew. Chem., Int. Ed., 2016, 55, 8944–8947 CrossRef CAS.
- M. Morikawa, H. Daido, T. Takao, S. Murata, Y. Shimonishi and T. Imanaka, J. Bacteriol., 1993, 175, 6459–6466 CrossRef CAS.
- M. L. Hutchison and K. Johnstone, Physiol. Mol. Plant Pathol., 1993, 42, 373–384 CrossRef CAS.
- M. D. Henkels, T. A. Kidarsa, B. T. Shaffer, N. C. Goebel, P. Burlinson, D. V. Mavrodi, M. A. Bentley, L. I. Rangel, E. W. Davis, L. S. Thomashow, T. M. Zabriskie, G. M. Preston and J. E. Loper, Mol. Plant Microbe Interact., 2014, 27, 733–746 CrossRef CAS.
- M. L. Hutchison and D. C. Gross, Mol. Plant Microbe Interact., 1997, 10, 347–354 CrossRef CAS.
- J. Henrichsen, Bacteriol. Rev., 1972, 36, 478–503 CAS.
- D. B. Kearns, Nat. Rev. Microbiol., 2010, 8, 634–644 CrossRef CAS.
- W. C. Wong and T. F. Preece, J. Appl. Microbiol., 1979, 47, 401–407 CrossRef.
- P. A. Zarkower, P. J. Wuest, D. J. Royse and B. Myers, Can. J. Microbiol., 1984, 30, 360–367 CrossRef.
- P. Munsch and T. Alatossava, Microbiol. Res., 2002, 157, 7–11 CrossRef PubMed.
- R. J. Mortishire-Smith, J. C. Nutkins, L. C. Packman, C. L. Brodey, P. B. Rainey, K. Johnstone and D. H. Williams, Tetrahedron, 1991, 47, 3645–3654 CrossRef CAS.
- T. Gruene, J. T. C. Wennmacher, C. Zaubitzer, J. J. Holstein, J. Heidler, A. Fecteau-Lefebvre, S. De Carlo, E. Müller, K. N. Goldie, I. Regeni, T. Li, G. Santiso-Quinones, G. Steinfeld, S. Handschin, E. van Genderen, J. A. van Bokhoven, G. H. Clever and R. Pantelic, Angew. Chem., Int. Ed. Engl., 2018, 57, 16313–16317 CrossRef CAS.
- C. G. Jones, M. W. Martynowycz, J. Hattne, T. J. Fulton, B. M. Stoltz, J. A. Rodriguez, H. M. Nelson and T. Gonen, ACS Cent. Sci., 2018, 4, 1587–1592 CrossRef CAS PubMed.
- T. Hölscher and Á. T. Kovács, Environ. Microbiol., 2017, 19, 2537–2545 CrossRef PubMed.
- V. Lazar, Anaerobe, 2011, 17, 280–285 CrossRef.
- H.-C. Flemming, J. Wingender, U. Szewzyk, P. Steinberg, S. A. Rice and S. Kjelleberg, Nat. Rev. Microbiol., 2016, 14, 563–575 CrossRef CAS PubMed.
- O. E. Petrova and K. Sauer, Curr. Opin. Microbiol., 2016, 30, 67–78 CrossRef CAS.
- T. Janek, M. Łukaszewicz and A. Krasowska, BMC Microbiol., 2012, 12, 24 CrossRef CAS PubMed.
- L. Bonnichsen, N. Bygvraa Svenningsen, M. Rybtke, I. de Bruijn, J. M. Raaijmakers, T. Tolker-Nielsen and O. Nybroe, Microbiology, 2015, 161, 2289–2297 CrossRef CAS PubMed.
- M. Kruijt, H. Tran and J. M. Raaijmakers, J. Appl. Microbiol., 2009, 107, 546–556 CrossRef CAS.
- A. Iwasaki, O. Ohno, S. Katsuyama, M. Morita, Y. Sasazawa, S. Dan, S. Simizu, T. Yamori and K. Suenaga, Bioorg. Med. Chem. Lett., 2015, 25, 5295–5298 CrossRef CAS.
- J. Pogliano, N. Pogliano and J. A. Silverman, J. Bacteriol., 2012, 194, 4494–4504 CrossRef CAS PubMed.
- K. Reder-Christ, Y. Schmidt, M. Dörr, H.-G. Sahl, M. Josten, J. M. Raaijmakers, H. Gross and G. Bendas, Biochim. Biophys. Acta Biomembr., 2012, 1818, 566–573 CrossRef CAS PubMed.
- N. Geudens, M. N. Nasir, J.-M. Crowet, J. M. Raaijmakers, K. Fehér, T. Coenye, J. C. Martins, L. Lins, D. Sinnaeve and M. Deleu, Biochim. Biophys. Acta Biomembr., 2017, 1859, 331–339 CrossRef CAS.
- D. Sinnaeve, P. M. â. S. Hendrickx, J. Vanâhemel, E. Peys, B. Kieffer and J. Martins, Chem.–Eur. J., 2009, 15, 12653–12662 CrossRef CAS.
- Geudens, Kovács, O. Sinnaeve, Höfte and Martins, Molecules, 2019, 24, 2257 CrossRef CAS.
- L. Becucci, V. Tramonti, A. Fiore, V. Fogliano, A. Scaloni and R. Guidelli, Biochim. Biophys. Acta Biomembr., 2015, 1848, 932–941 CrossRef CAS.
- A. M. Feigin, L. V. Schagina, J. Y. Takemoto, J. H. Teeter and J. G. Brand, Biochim. Biophys. Acta Biomembr., 1997, 1324, 102–110 CrossRef CAS.
- A. M. Feigin, J. Y. Takemoto and R. Wangspa, J. Membr. Biol., 1996, 149, 41–47 CrossRef CAS.
- S. S. Efimova, A. A. Zakharova, L. V. Schagina and O. S. Ostroumova, Eur. Biophys. J., 2016, 45, 91–98 CrossRef CAS.
- E. Vaillo, A. Ballio, P. L. Luisi and R. M. Thomas, Biopolymers, 1992, 32, 1317–1326 CrossRef CAS.
- M. Anselmi, T. Eliseo, L. Zanetti-Polzi, M. R. Fullone, V. Fogliano, A. Di Nola, M. Paci and I. Grgurina, Biochim. Biophys. Acta Biomembr., 2011, 1808, 2102–2110 CrossRef CAS.
- Z. Szabó, P. Gróf, L. V. Schagina, P. A. Gurnev, J. Y. Takemoto, E. Mátyus and K. Blaskó, Biochim. Biophys. Acta Biomembr., 2002, 1567, 143–149 CrossRef.
- D. Di Giorgio, L. Camoni, C. Marchiafava and A. Ballio, Phytochemistry, 1997, 45, 1385–1391 CrossRef CAS.
- M. Coraiola, P. Lo Cantore, S. Lazzaroni, A. Evidente, N. S. Iacobellis and M. Dalla Serra, Biochim. Biophys. Acta, 2006, 1758, 1713–1722 CrossRef CAS PubMed.
- P. Lo Cantore, S. Lazzaroni, M. Coraiola, M. D. Serra, C. Cafarchia, A. Evidente and N. S. Iacobellis, Mol. Plant Microbe Interact., 2006, 19, 1113–1120 CrossRef CAS PubMed.
- F. Jourdan, S. Lazzaroni, B. L. Méndez, P. Lo Cantore, M. de Julio, P. Amodeo, N. S. Iacobellis, A. Evidente and A. Motta, Proteins, 2003, 52, 534–543 CrossRef CAS.
- A. Ballio, A. Collina, A. D. Nola, C. Manetti, M. Pad and A. Segre, Struct. Chem., 1994, 5, 43–50 CrossRef CAS.
- S. Baré, V. M. Coiro, A. Scaloni, A. Di Nola, M. Paci, A. L. Segre and A. Ballio, Eur. J. Biochem., 1999, 266, 484–492 CrossRef PubMed.
- E. Mátyus, K. Blaskó, J. Fidy and D. P. Tieleman, Eur. Biophys. J., 2008, 37, 495–502 CrossRef PubMed.
- M. Coraiola, R. Paletti, A. Fiore, V. Fogliano and M. D. Serra, J. Pept. Sci., 2008, 14, 496–502 CrossRef CAS PubMed.
- L. Becucci, A. Toppi, A. Fiore, A. Scaloni and R. Guidelli, Bioelectrochemistry, 2016, 111, 131–142 CrossRef CAS PubMed.
- P. Aiyar, D. Schaeme, M. García-Altares, D. Carrasco Flores, H. Dathe, C. Hertweck, S. Sasso and M. Mittag, Nat. Commun., 2017, 8, 1756 CrossRef PubMed.
- D. Lin and A. Grossfield, Biophys. J., 2014, 107, 1862–1872 CrossRef CAS PubMed.
- J. K. Muraih, A. Pearson, J. Silverman and M. Palmer, Biochim. Biophys. Acta, 2011, 1808, 1154–1160 CrossRef CAS PubMed.
- T. H. Nielsen and J. Sørensen, Appl. Environ. Microbiol., 2003, 69, 861–868 CrossRef CAS PubMed.
- A. Andolfi, A. Cimmino, P. L. Cantore, N. S. Iacobellis and A. Evidente, Perspect. Med. Chem., 2008, 2, 81–112 CAS.
- B. R. Wilson, A. R. Bogdan, M. Miyazawa, K. Hashimoto and Y. Tsuji, Trends Mol. Med., 2016, 22, 1077–1090 CrossRef CAS PubMed.
- K. S. Andersen and A. Winding, Biol. Fertil. Soils, 2004, 40, 230–236 CrossRef CAS.
- M. Mazzola, I. de Bruijn, M. F. Cohen and J. M. Raaijmakers, Appl. Environ. Microbiol., 2009, 75, 6804–6811 CrossRef CAS PubMed.
- L. Eichinger, J. A. Pachebat, G. Glöckner, M.-A. Rajandream, R. Sucgang, M. Berriman, J. Song, R. Olsen, K. Szafranski, Q. Xu, B. Tunggal, S. Kummerfeld, M. Madera, B. A. Konfortov, F. Rivero, A. T. Bankier, R. Lehmann, N. Hamlin, R. Davies, P. Gaudet, P. Fey, K. Pilcher, G. Chen, D. Saunders, E. Sodergren, P. Davis, A. Kerhornou, X. Nie, N. Hall, C. Anjard, L. Hemphill, N. Bason, P. Farbrother, B. Desany, E. Just, T. Morio, R. Rost, C. Churcher, J. Cooper, S. Haydock, N. van Driessche, A. Cronin, I. Goodhead, D. Muzny, T. Mourier, A. Pain, M. Lu, D. Harper, R. Lindsay, H. Hauser, K. James, M. Quiles, M. M. Babu, T. Saito, C. Buchrieser, A. Wardroper, M. Felder, M. Thangavelu, D. Johnson, A. Knights, H. Loulseged, K. Mungall, K. Oliver, C. Price, M. A. Quail, H. Urushihara, J. Hernandez, E. Rabbinowitsch, D. Steffen, M. Sanders, J. Ma, Y. Kohara, S. Sharp, M. Simmonds, S. Spiegler, A. Tivey, S. Sugano, B. White, D. Walker, J. Woodward, T. Winckler, Y. Tanaka, G. Shaulsky, M. Schleicher, G. Weinstock, A. Rosenthal, E. C. Cox, R. L. Chisholm, R. Gibbs, W. F. Loomis, M. Platzer, R. R. Kay, J. Williams, P. H. Dear, A. A. Noegel, B. Barrell and A. Kuspa, Nature, 2005, 435, 43 CrossRef CAS PubMed.
- J. Meng, Y. Fan, M. Su, C. Chen, T. Ren, J. Wang and Q. Zhao, Int. Immunopharmacol., 2014, 19, 37–44 CrossRef CAS PubMed.
- P. B. Rainey, C. L. Brodey and K. Johnstone, Physiol. Mol. Plant Pathol., 1991, 39, 57–70 CrossRef CAS.
- S. L. Sinden, J. E. DeVay and P. A. Backman, Physiol. Mol. Plant Pathol., 1971, 1, 199–213 CrossRef CAS.
- N. S. Iacobellis, P. Lavermicocca, I. Grgurina, M. Simmaco and A. Ballio, Physiol. Mol. Plant Pathol., 1992, 40, 107–116 CrossRef CAS.
- B. K. Scholz-Schroeder, M. L. Hutchison, I. Grgurina and D. C. Gross, Mol. Plant Microbe Interact., 2001, 14, 336–348 CrossRef CAS.
- V. Groupe, L. H. Pugh, D. Weiss and M. Kochi, Exp. Biol. Med., 1951, 78, 354–358 CrossRef CAS.
- B. C. Chu, A. Garcia-Herrero, T. H. Johanson, K. D. Krewulak, C. K. Lau, R. S. Peacock, Z. Slavinskaya and H. J. Vogel, BioMetals, 2010, 23, 601–611 CrossRef CAS PubMed.
- M. Miethke and M. A. Marahiel, Microbiol. Mol. Biol. Rev., 2007, 71, 413–451 CrossRef CAS PubMed.
- B. Schwyn and J. B. Neilands, Anal. Biochem., 1987, 160, 47–56 CrossRef CAS.
- N. Geudens, D. Sinnaeve and J. C. Martins, Future Med. Chem., 2018, 10, 479–481 CrossRef CAS PubMed.
- M. Mazzola, X. Zhao, M. F. Cohen and J. M. Raaijmakers, Phytopathology, 2007, 97, 1348–1355 CrossRef CAS PubMed.
- A. W. Bauer, D. M. Perry and W. M. Kirby, JAMA Intern. Med., 1959, 104, 208–216 CAS.
- A. W. Bauer, W. M. Kirby, J. C. Sherris and M. Turck, Am. J. Clin. Pathol., 1966, 45, 493–496 CrossRef CAS.
- J. M. Andrews, J. Antimicrob. Chemother., 2001, 48(Suppl. 1), 5–16 CrossRef CAS PubMed.
- J. D. Dunn, C. Bosmani, C. Barisch, L. Raykov, L. H. Lefrançois, E. Cardenal-Muñoz, A. T. López-Jiménez and T. Soldati, Front. Immunol., 2018, 8, 1906 CrossRef PubMed.
- D. A. Brock, T. E. Douglas, D. C. Queller and J. E. Strassmann, Nature, 2011, 469, 393 CrossRef CAS PubMed.
- D. A. Brock, T. S. Haselkorn, J. R. Garcia, U. Bashir, T. E. Douglas, J. Galloway, F. Brodie, D. C. Queller and J. E. Strassmann, Front. Cell. Infect. Microbiol., 2018, 8, 411 CrossRef CAS PubMed.
- R. J. Gast, R. W. Sanders and D. A. Caron, Trends Microbiol., 2009, 17, 563–569 CrossRef CAS.
- M. V. Berridge, P. M. Herst and A. S. Tan, Biotechnol. Annu. Rev., 2005, 11, 127–152 CAS.
- J. C. Stockert, R. W. Horobin, L. L. Colombo and A. Blázquez-Castro, Acta Histochem., 2018, 120, 159–167 CrossRef CAS PubMed.
- E. Pauwelyn, K. Vanhouteghem, B. Cottyn, P. De Vos, M. Maes, P. Bleyaert and M. Höfte, J. Phytopathol., 2011, 159, 298–305 CrossRef.
- I. K. Blaby, C. Blaby-Haas, N. Tourasse, E. F. Y. Hom, D. Lopez, M. Aksoy, A. Grossman, J. Umen, S. Dutcher, M. Porter, S. King, G. Witman, M. Stanke, E. H. Harris, D. Goodstein, J. Grimwood, J. Schmutz, O. Vallon, S. S. Merchant and S. Prochnik, Trends Plant Sci., 2014, 19, 672–680 CrossRef CAS PubMed.
- A. R. Grossman, E. E. Harris, C. Hauser, P. A. Lefebvre, D. Martinez, D. Rokhsar, J. Shrager, C. D. Silflow, D. Stern, O. Vallon and Z. Zhang, Eukaryotic Cell, 2003, 2, 1137–1150 CrossRef CAS.
- S. S. Merchant, S. E. Prochnik, O. Vallon, E. H. Harris, S. J. Karpowicz, G. B. Witman, A. Terry, A. Salamov, L. K. Fritz-Laylin, L. Maréchal-Drouard, W. F. Marshall, L.-H. Qu, D. R. Nelson, A. A. Sanderfoot, M. H. Spalding, V. V. Kapitonov, Q. Ren, P. Ferris, E. Lindquist, H. Shapiro, S. M. Lucas, J. Grimwood, J. Schmutz, P. Cardol, H. Cerutti, G. Chanfreau, C.-L. Chen, V. Cognat, M. T. Croft, R. Dent, S. Dutcher, E. Fernández, P. Ferris, H. Fukuzawa, D. González-Ballester, D. González-Halphen, A. Hallmann, M. Hanikenne, M. Hippler, W. Inwood, K. Jabbari, M. Kalanon, R. Kuras, P. A. Lefebvre, S. D. Lemaire, A. V. Lobanov, M. Lohr, A. Manuell, I. Meier, L. Mets, M. Mittag, T. Mittelmeier, J. V. Moroney, J. Moseley, C. Napoli, A. M. Nedelcu, K. Niyogi, S. V. Novoselov, I. T. Paulsen, G. Pazour, S. Purton, J.-P. Ral, D. M. Riaño-Pachón, W. Riekhof, L. Rymarquis, M. Schroda, D. Stern, J. Umen, R. Willows, N. Wilson, S. L. Zimmer, J. Allmer, J. Balk, K. Bisova, C.-J. Chen, M. Elias, K. Gendler, C. Hauser, M. R. Lamb, H. Ledford, J. C. Long, J. Minagawa, M. D. Page, J. Pan, W. Pootakham, S. Roje, A. Rose, E. Stahlberg, A. M. Terauchi, P. Yang, S. Ball, C. Bowler, C. L. Dieckmann, V. N. Gladyshev, P. Green, R. Jorgensen, S. Mayfield, B. Mueller-Roeber, S. Rajamani, R. T. Sayre, P. Brokstein, I. Dubchak, D. Goodstein, L. Hornick, Y. W. Huang, J. Jhaveri, Y. Luo, D. Martínez, W. C. A. Ngau, B. Otillar, A. Poliakov, A. Porter, L. Szajkowski, G. Werner, K. Zhou, I. V. Grigoriev, D. S. Rokhsar and A. R. Grossman, Science, 2007, 318, 245–250 CrossRef CAS PubMed.
- S. E. Lindow and M. T. Brandl, Appl. Environ. Microbiol., 2003, 69, 1875–1883 CrossRef CAS PubMed.
- C. Berry, W. G. D. Fernando, P. C. Loewen and T. R. de Kievit, Biol. Control, 2010, 55, 211–218 CrossRef CAS.
- M. Brax, C. Buchmann and G. E. Schaumann, J. Plant Nutr. Soil Sci., 2017, 180, 121–141 CrossRef CAS.
- J. Sasse, E. Martinoia and T. Northen, Trends Plant Sci., 2018, 23, 25–41 CrossRef CAS PubMed.
- U. Baetz and E. Martinoia, Trends Plant Sci., 2014, 19, 90–98 CrossRef CAS PubMed.
- L. Philippot, J. M. Raaijmakers, P. Lemanceau and W. H. van der Putten, Nat. Rev. Microbiol., 2013, 11, 789–799 CrossRef CAS.
- B. R. Kang, A. J. Anderson and Y. C. Kim, Plant Pathol. J., 2018, 34, 35–43 CAS.
- I. A. Siddiqui, S. S. Shaukat, I. H. Sheikh and A. Khan, World J. Microbiol. Biotechnol., 2006, 22, 641–650 CrossRef CAS.
- N. Neidig, R. J. Paul, S. Scheu and A. Jousset, Microb. Ecol., 2011, 61, 853–859 CrossRef CAS PubMed.
- K. A. B. Kissoyan, M. Drechsler, E.-L. Stange, J. Zimmermann, C. Kaleta, H. B. Bode and K. Dierking, Curr. Biol., 2019, 29, 1030–1037 CrossRef CAS PubMed.
- B. Lugtenberg, D. E. Rozen and F. Kamilova, F1000Res., 2017, 6, 343 Search PubMed.
- Z. Gao, I. Karlsson, S. Geisen, G. Kowalchuk and A. Jousset, Trends Plant Sci., 2019, 24, 165–176 CrossRef CAS PubMed.
- N. K. Bordoloi and B. K. Konwar, J. Hazard. Mater., 2009, 170, 495–505 CrossRef CAS PubMed.
- O. Zouari, D. Lecouturier, A. Rochex, G. Chataigne, P. Dhulster, P. Jacques and D. Ghribi, Biodegradation, 2019, 30(4), 259–272 CrossRef PubMed.
- A. Venieraki, P. Ch. Tsalgatidou, D. G. Georgakopoulos, M. Dimou and P. Katinakis, Hellenic Plant Protection Journal, 2016, 9, 16–27 Search PubMed.
- V. Vassilev, P. Lavermicocca, D. D. Giorgio and N. S. Iacobellis, Plant Pathol., 1996, 45, 316–322 CrossRef CAS.
- N. S. Iacobellis, G. Figliuolo, J. Janse, M. Scortichini and G. Ciuffreda, in Pseudomonas Syringae Pathovars and Related Pathogens, ed. K. Rudolph, T. J. Burr, J. W. Mansfield, D. Stead, A. Vivian and J. von Kietzell, Springer Netherlands, Dordrecht, 1997, pp. 500–504 Search PubMed.
- P. D. Hildebrand, P. G. Braun, K. B. McRae and X. Lu, Can. J. Plant Pathol., 1998, 296–303 CrossRef CAS.
- X.-F. Xin and S. Y. He, Annu. Rev. Phytopathol., 2013, 51, 473–498 CrossRef CAS PubMed.
- C. Faulkner and S. Robatzek, Curr. Opin. Plant Biol., 2012, 15, 699–707 CrossRef CAS PubMed.
- A. Block and J. R. Alfano, Curr. Opin. Microbiol., 2011, 14, 39–46 CrossRef CAS PubMed.
- X.-F. Xin, B. Kvitko and S. Y. He, Nat. Rev. Microbiol., 2018, 16, 316–328 CrossRef CAS PubMed.
- A. Carpaneto, M. Dalla Serra, G. Menestrina, V. Fogliano and F. Gambale, J. Membr. Biol., 2002, 188, 237–248 CrossRef CAS PubMed.
- C. M. J. Pieterse, C. Zamioudis, R. L. Berendsen, D. M. Weller, S. C. M. Van Wees and P. A. H. M. Bakker, Annu. Rev. Phytopathol., 2014, 52, 347–375 CrossRef CAS PubMed.
- P. A. H. M. Bakker, R. F. Doornbos, C. Zamioudis, R. L. Berendsen and C. M. J. Pieterse, Plant Pathol. J., 2013, 29, 136–143 CrossRef PubMed.
- H. Tran, A. Ficke, T. Asiimwe, M. Höfte and J. M. Raaijmakers, New Phytol., 2007, 175, 731–742 CrossRef CAS.
- M. N. Nielsen, J. Sørensen, J. Fels and H. C. Pedersen, Appl. Environ. Microbiol., 1998, 64, 3563–3569 CAS.
- T.-H. Lee, D. J. Hirst and M.-I. Aguilar, Biochim. Biophys. Acta, 2015, 1848, 1868–1885 CrossRef CAS PubMed.
- I. G. Denisov and S. G. Sligar, Chem. Rev., 2017, 117, 4669–4713 CrossRef CAS PubMed.
- T. Luzzatto-Knaan, A. V. Melnik and P. C. Dorrestein, ACS Chem. Biol., 2019, 14(3), 459–467 CrossRef CAS PubMed.
- V. V. Phelan, W.-T. Liu, K. Pogliano and P. C. Dorrestein, Nat. Chem. Biol., 2012, 8, 26–35 CrossRef CAS PubMed.
- T. Luzzatto-Knaan, A. V. Melnik and P. C. Dorrestein, Analyst, 2015, 140, 4949–4966 RSC.
- J. D. Watrous and P. C. Dorrestein, Nat. Rev. Microbiol., 2011, 9, 683–694 CrossRef CAS PubMed.
- A. E. Brunetti, F. Carnevale Neto, M. C. Vera, C. Taboada, D. P. Pavarini, A. Bauermeister and N. P. Lopes, Chem. Soc. Rev., 2018, 47, 1574–1591 RSC.
- R. A. Melnyk, S. S. Hossain and C. H. Haney, ISME J., 2019, 13(6), 1575–1588 CrossRef PubMed.
- K. A. J. Bozhüyük, F. Fleischhacker, A. Linck, F. Wesche, A. Tietze, C.-P. Niesert and H. B. Bode, Nat. Chem., 2017, 10, 275–281 CrossRef PubMed.
- F. Yan, C. Burgard, A. Popoff, N. Zaburannyi, G. Zipf, J. Maier, H. S. Bernauer, S. C. Wenzel and R. Müller, Chem. Sci., 2018, 9, 7510–7519 RSC.
- A. S. Brown, M. J. Calcott, J. G. Owen and D. F. Ackerley, Nat. Prod. Rep., 2018, 35, 1210–1228 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9np00022d |
| This journal is © The Royal Society of Chemistry 2020 |