Intracellular low-abundance microRNA imaging by a NIR-assisted entropy-driven DNA system†
Huiting
Lu‡
a,
Fan
Yang‡
b,
Benhan
Liu
b,
Kai
Zhang
b,
Yu
Cao
b,
Wenhao
Dai
b,
Wenjun
Li
*a and
Haifeng
Dong
 *b
*b
aDepartment of Chemistry, School of Chemistry and Bioengineering, University of Science and Technology Beijing, 30 Xueyuan Road, Beijing 100083, P. R. China. E-mail: wjli@sas.ustb.edu.cn
bBeijing Key Laboratory for Bioengineering and Sensing Technology, Research Center for Bioengineering and Sensing Technology, School of Chemistry & Biological Engineering University of Science & Technology Beijing, 30 Xueyuan Road, Beijing 100083, P. R. China. E-mail: hfdong@ustb.edu.cn
First published on 14th November 2018
Abstract
Intracellular microRNA (miRNA) imaging remains a key challenge due to its low abundance. Herein, we integrate a rationally designed elegant entropy-driven DNA probe with assisted DNA fuel on hollow copper sulfide nanoparticles (HCuSNPs) for intracellular miRNA imaging. The anchored assisted DNA fuel strand could be efficiently released by a NIR-II laser irradiation induced photothermal effect of the HCuSNPs. The DNA machine was activated by target miRNA binding and powered by NIR-responsive released DNA fuel through toehold-mediated strand displacement reactions, accomplished by strong fluorescence recovery. It demonstrated 2 orders of magnitude improvement in the detection sensitivity compared to molecular beacons (MBs). Reliable intracellular low-abundance miRNA imaging among different cells and monitoring of down-regulated miRNA was realized without external enzyme or fuel addition. Oncogenic miRNA imaging in vivo was also realized. The entropy-driven DNA machine system provides a facile and powerful tool for intracellular miRNA analysis and related biomedical applications.
Conceptual insightsImaging of intracellular low-abundance miRNA levels in living cells remains a great challenge. Herein, we fabricate hollow copper sulfide nanoparticles (HCuSNPs) with a good photothermal effect in the NIR-II region as a gene vector and a photothermal-responsive agent, and integrate two types of DNA probes, namely a substrate probe and a fuel probe, on it to develop a NIR-assisted elegant entropy-driven DNA system (HCuSNPs-DNA) for highly sensitive catalytic imaging of low-abundance miRNAs in living cells and in vivo. It realizes low-abundance miRNA imaging among different cells without external enzyme or fuel transfection. The change in the expression level of oncogenic miRNA is also successfully monitored. Notably, oncogenic miRNA expression in vivo is also implemented. The NIR-assisted elegant entropy-driven DNA system opens new opportunities for intracellular miRNA analysis and related biomedical applications. |
Introduction
MicroRNAs (miRNAs) are a class of noncoding small single-stranded RNAs with lengths of 18 to 23 nucleotides.1–4 They act as post-transcriptional regulators to affect an array of biological processes including cellular differentiation, proliferation and apoptosis.5–7 Importantly, the aberrant expression levels of miRNA were interrelated with diverse human diseases, including cancer,8–12 highlighting their potential as a new type of biomarker candidate for cancer classification, early disease diagnosis and prognosis.13 However, monitoring of intracellular miRNA levels in living cells remains a great challenge due to the low amount of miRNA, high sequence similarity among family members and the complex intracellular environment in vivo.14 Generally, traditional molecular imaging systems are “one-to-one” models, which cannot satisfy low-abundance intracellular miRNA imaging. Various novel intracellular enzyme-based15–17 or nanomaterial-related signal amplification strategies18–24 have been explored for miRNA detection in living cells.Currently, enzyme-free DNA toehold-mediated strand displacement amplification is widely used for intracellular nucleic acid detection due to its tunability and high amplification efficiency.25–28 Ma et al. reported a type of DNA-template gold nanoparticle-quantum dot nanoparticle assembly-based imaging system for highly sensitive imaging of intracellular miRNA.29 Yin et al. developed an elegant entropy-driven DNA nano-machine that utilized a special design of the walking leg for miRNA imaging in living cells. However, all of the cascade process requires external transfection of fuel DNA strands.30 To solve the aforementioned problems, our group previously accomplished the combination of pH or NIR-responsive DNA strands31,32 or ATP fuel33 and a DNA toehold-mediated strand displacement structure into a smart nanosystem, realizing highly sensitive intracellular miRNA detection while avoiding additional transfection. However, it is also involved in a stringent DNA modification process or complex DNA structure, and the in vivo miRNA imaging is unsatisfactory. Novel facile and reliable toehold-mediated strand displacement of intracellular miRNA detection systems is being continuously explored.
Herein, we used hollow copper sulfide nanoparticles (HCuSNPs) as a carrier and a photothermal conversion agent to integrate rationally designed elegant entropy-driven DNA probes with assisted DNA fuels for intracellular miRNA catalytic imaging (Scheme 1). The synthesized HCuSNPs present strong absorption in the second NIR (NIR-II) regions (1000–1400 nm)34–36 and good photothermal conversion efficiency, on which two types of DNA probes (HCuSNPs-DNA) including substrate probes and fuel probes were modified for miRNA detection. The substrate probes comprise BHQ2-DNA2, Cy3-DNA3 and linker DNA, in which the Cy3 fluorescence emission is almost quenched by BHQ2. The trigger target miRNA-155 could recognize toehold 1 (Toe 1) and replace Cy3-DNA to expose sequestered toehold 2 (Toe 2), leading to fluorescence recovery. Fuel DNA is attached to the HCuSNPs by hybridizing with DNA1 (12 bps), which could be released under 1064 nm laser irradiation due to the photothermal effect of the HCuSNPs. Subsequently, the fuel DNA binds to Toe 2 and liberates the target, and the released target could trigger the next cycle to produce strong fluorescence for intracellular miRNA detection. It provides a facile and competent tool for highly sensitive imaging of intracellular low-abundance miRNA without additional external transfection agents.
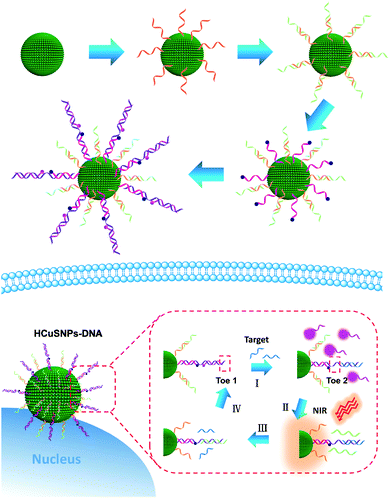 | ||
| Scheme 1 Schematic illustration of the HCuSNPs-DNA system for highly sensitive miRNA detection in living cells. | ||
Results and discussion
Characterizations of HCuSNPs and HCuSNPs-DNA
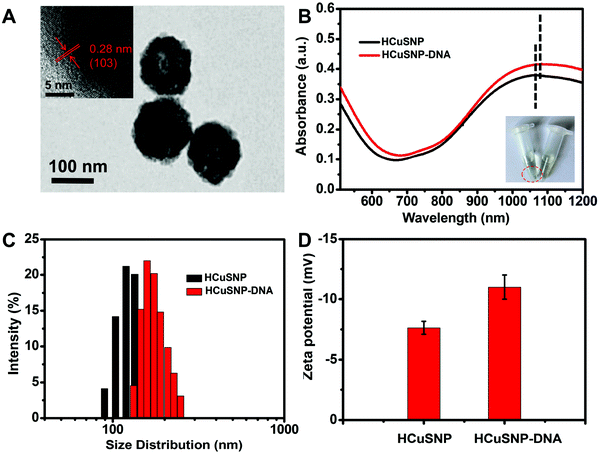
Transmission electron microscope (TEM) images presented the morphological information of HCuSNPs (Fig. 1A) that were spherical in shape with a mean diameter of about 120 nm. The high-resolution TEM (HRTEM) image revealed that the lattice constant of HCuSNPs was 0.28 nm, in agreement with the previous report (the inset in Fig. 1A).37 The functionalization of HCuSNPs with DNA probes was characterized by UV-vis spectrometry, Dynamic Light Scattering (DLS) and zeta potential analysis. The HCuSNPs showed a strong absorption peak at 1100 nm (black line), and about a 4 nm red-shift was observed after the functionalization of DNA probes (Fig. 1B, red line), and the DNA functionalization endowed the HCuSNPs with good solubility (the inset in Fig. 1B) due to the good hydrophilicity of DNA. DLS was further employed to measure the hydrodynamic size of HCuSNPs and HCuSNPs-DNA. It was found that the diameter of the HCuSNPs was about 122 nm (Fig. 1C), which was consistent with the TEM imaging. The DNA modification induced the hydrodynamic diameter increase of HCuSNPs-DNA to 158.8 nm. In addition, the surface zeta potential changed from −7.6 mV to −11 mV after the HCuSNPs were attached with negative DNA (Fig. 1D). These results confirmed that DNA probes were successfully modified on the surface of HCuSNPs.Feasibility of HCuSNPs-DNA for signal amplification
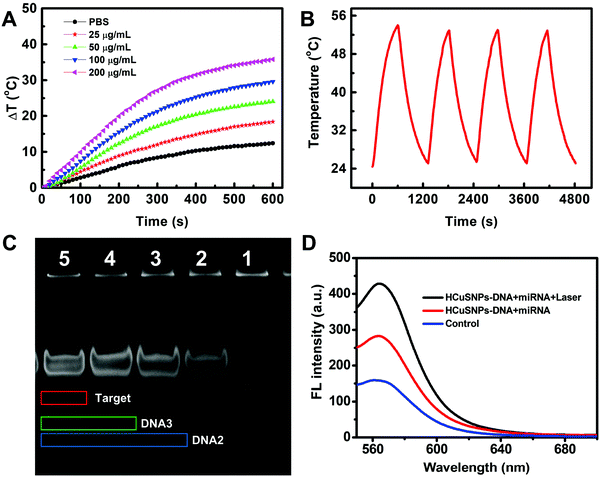
The HCuSNPs presented strong absorbance in the NIR-II region, and the photothermal effects of HCuSNPs-DNA were used for controllable release of the fuel DNA, which was first investigated. As shown in Fig. 2A, the temperature profiles indicated a concentration-dependent temperature increase pattern and good photothermal efficiency of HCuSNPs-DNA. The temperature of the HCuSNPs-DNA solution changed to 29.6 °C at 100 μg mL−1 under 1064 nm laser (1.0 W cm−2) irradiation for 10 min. The photothermal conversion efficiency was calculated to be 27.9% (Fig. S1, ESI†), suggesting the good photothermal conversion efficiency of the HCuSNPs-DNA. The photothermal stability of HCuSNPs-DNA was measured and the HCuSNPs-DNA showed a great stability and excellent reproducibility after four cycles of 1064 nm NIR laser irradiation (Fig. 2B). Polyacrylamide gel electrophoresis (PAGE) experiments were performed to verify the toehold-mediated strand displacement process (Fig. 2A). The lanes 1 to 5 were linker, linker + DNA2, linker + DNA2 + DNA3, linker + DNA2 + DNA3 + miRNA-155, and linker + DNA2 + DNA3 + fuel + miRNA-155, respectively. All the DNA probes were 10 μM and the target miRNA-155 was 5 μM. Lane 3 indicated the successful formation of the sandwich structure, while miRNA could readily displace DNA3 to form linker + DNA2 + miRNA (lane 4). Furthermore, the displacement of the target miRNA by fuel could also be observed (lane 5). These results strongly confirmed the feasibility of the entropy-driven DNA system. The feasibility was further investigated by fluorescence spectra analysis (Fig. 2B). The addition of the target miRNA caused fluorescence recovery owing to miRNA-induced displacement of DNA 3 (red curve) compared to the control, while the NIR-irradiation further induced a sharp fluorescence increase (black curve), suggesting the good feasibility of the HCuSNPs-DNA system for signal amplification.Assay performance for miRNA detection in vitro
The released temperature of the fuel was first investigated using an FAM-labeled fuel probe. As shown in Fig. S2 (ESI†), the fluorescence (FL) intensity value of the supernatant gradually increased with increasing temperature and reached a plateau when the reaction temperature increased to 46 °C, which indicated that the fuel probes could not be released at 37 °C under physiological conditions without the NIR-irradiation. Another vital experimental parameter is the molar ratio of fuel probes and substrate probes, and the FL experiments suggested that good performance could be obtained at the ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. S2, ESI†).
1 (Fig. S2, ESI†).
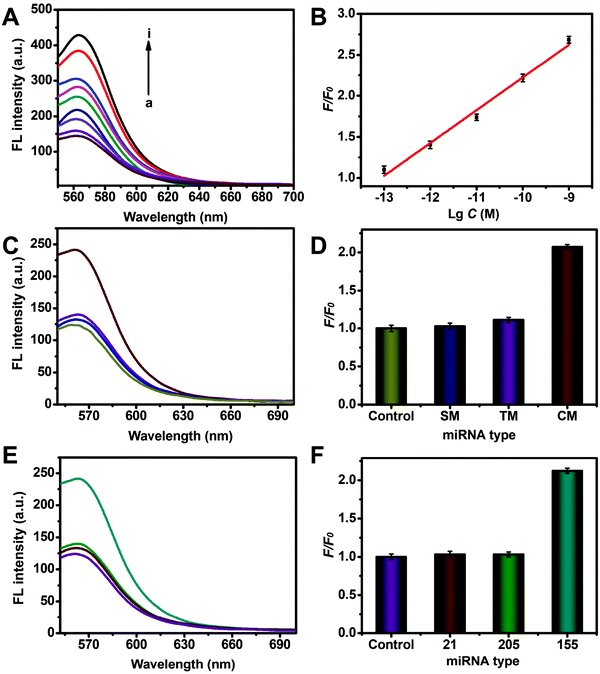
Under the optimized conditions, the performance of the assay for miRNA-155 detection was evaluated. To investigate the FL response of the proposed assay to miRNA, the fluorescence intensity was normalized as a relative fluorescence intensity change F/F0 to avoid the influence of background on the fluorescence response, where F and F0 are Cy3 FL intensity at 565 nm in the presence and absence of the target, respectively. As shown in Fig. 3A, the FL intensity increased with increasing concentration of miRNA-155. A good linear relationship exists between F/F0 and the logarithm of the miRNA-155 concentration ranging from 0.1 pM to 1 nM (Fig. 3B). The limit of detection (LOD) was calculated to be 51.4 fM by using three times the standard deviation of the control according to previous reports.38–41 It presented 2 orders of magnitude sensitivity enhancement compared to molecular beacons (MBs) without amplification (Fig. S4, ESI†).
The specificity of the proposed strategy was further investigated using the control miRNA-155 (CM) with a scrambled sequence, single-base mismatched miRNA (SM), and three-base mismatched miRNA (TM). As showed in Fig. 3C, compared with the blank control, the FL intensity in the presence of SM and TM only displayed negligible changes, while an obvious FL intensity increase was observed by adding the CM strand at the same concentration, which was 1.99-fold and 1.83-fold higher than the FL intensity of the SM and TM, respectively (Fig. 3D). Furthermore, the capability of the proposed strategy in distinguishing various types of miRNAs was also investigated in Fig. 3E. The FL signal produced by CM miRNA-155 was 2.05-fold higher compared to that produced by miRNA-21 and miRNA-205 (Fig. 3F). The results revealed the good base-mismatch discrimination capability and high specificity of the proposed assay.
Intracellular miRNA imaging via HCuSNPs-DNA probes
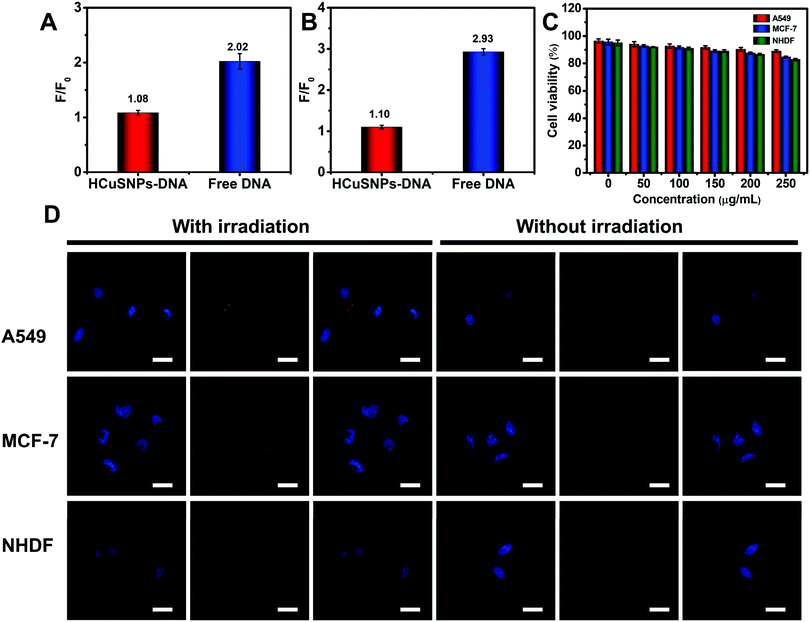
A good gene delivery system should protect the cargo from DNase degradation and protein interference during the transfection process. The protection properties of the HCuSNPs-DNA were first investigated. The HCuSNPs-DNA probes and free substrate probes were treated with 0.5 U mL−1 of human deoxyribonuclease (DNase) I at 37 °C for 1 h. As shown in Fig. 4A, no obvious fluorescence signal change was observed when Dnase I was added to the HCuSNPs-DNA probe solution during a 1 h duration and the F/F0 ratio was 1.08. In contrast, the free substrate probes displayed significant degradation after the addition of DNase I for 1 h, and the F/F0 ratio was 2.02. The serum stability of the substrate probes anchored on HCuSNPs or free substrate probes was evaluated by measuring the fluorescence recovery of Cy3 treated with 10% FBS reaction buffer over 12 h. As shown in Fig. 4B, the fluorescence leakage of HCuSNPs-DNA was much less compared to the control of free substrate probes. These results indicated that HCuSNPs could protect substrate probes from nuclease degradation and protein interference during intracellular delivery. The cytotoxicity of the HCuSNPs-DNA probe was investigated using the 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide (MTT) assay. As shown in Fig. 4C, the HCuSNPs-DNA probes displayed low cytotoxicity toward all MCF-7 cells, A549 cells and NHDF cells. The cell viabilities were above 80%, even if the concentration increases to 200 μg mL−1, which demonstrated the good biocompatibility of the HCuSNPs-DNA probes.The miRNA-155 imaging in living cells was further investigated. In the absence of NIR irradiation, both the MCF-7 cells and A549 cells displayed a weak red fluorescence related to the miRNA-155 recognition, while almost no fluorescence was observed for the NHDF cells (Fig. 4D). In contrast, under 1064 nm laser irradiation for 5 min, strong red fluorescence signals were presented for MCF-7 cells and A549 cells resulting from the entropy-driven displacement amplification, and slight red fluorescence was also observed for the NHDF cells. The corresponding fluorescence intensity was higher by 3.8-fold for A549 cells and 1.8-fold for MCF-7 cells after the NIR-laser treatment compared to that without irradiation (Fig. S5, ESI†). This demonstrated the higher expression level of oncogenic miRNA-155 in both MCF-7 cells and A549 cells compared to the NHDF normal cells. These results suggested that the HCuSNPs-DNA probe based NIR-laser assisted strand displacement amplification reaction could highly sensitively visualize low-abundance miRNAs in living cells, and provide a useful tool to discriminate cancer cells from normal cells.
The capability of the proposed nanosystem to monitor the change of oncogenic miRNA in cancer cells was further investigated using A549 cells treated with miRNA-155 inhibitors, which is pivotal to understand the bio-function of oncogenic miRNA during cancer development and explore its biomedical application. As shown in Fig. 5, the fluorescence imaging displayed an obvious decrease in fluorescence intensity after the A549 cells received miRNA-155 inhibitor treatment, and the corresponding fluorescence intensity was 0.64-fold lower than that without inhibitor treatment, which indicated a lower expression of miRNA-155. This suggested the good feasibility of the NIR-laser assisted strand displacement amplification system to monitor the expression change of low-abundance miRNA due to the high sensitivity.
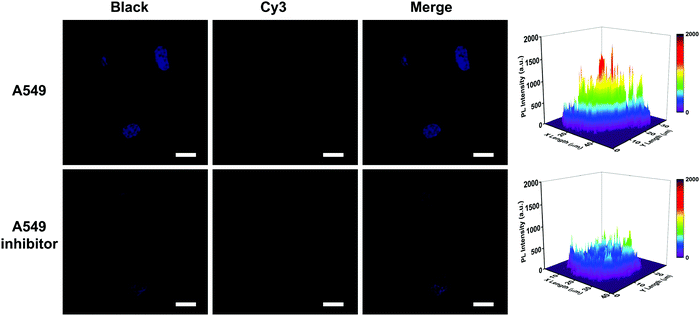 | ||
| Fig. 5 Fluorescence images for HCuSNPs-DNA-treated A549 cells without and with miRNA-155 inhibitor (300 nM) treatment. The scale bar is 40 μm. | ||
The good photothermal effect of the synthesized HCuSNPs irradiated by a NIR-II laser inspired us to further investigate the in vivo imaging performance of the system owing to the good penetration depth of the NIR-II laser. The mice that received HCuSNPs-DNA treatment displayed negligible fluorescence without NIR-laser irradiation (Fig. 6A). In contrast, obvious fluorescence associated with the miRNA-155 recognition in the tumor region was observed for the HCuSNPs-DNA treated mice receiving 1064 nm laser irradiation for 5 min (Fig. 6B), which was attributed to the good penetration capability of the NIR-II laser and high sensitivity of the entropy-driven DNA system, indicating the good feasibility of the system for in vivo low-abundance oncogenic miRNA detection.
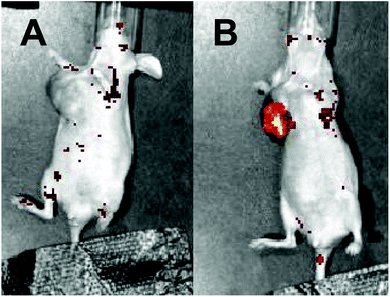 | ||
| Fig. 6 Fluorescence images of MCF-7-tumor-bearing mice preinjected with HCuSNPs and irradiated (A) without and (B) with 1064 nm NIR excitation (1.0 W cm−2) for 5 min. | ||
Conclusions
In summary, we report HCuSNPs as NIR-responsive photothermal conversion agents and gene probe carriers for NIR-assisted highly sensitive catalytic imaging of low-abundance miRNA in living cells and in vivo. The substrate probes and phothothermal-responsive fuel probes were rationally designed and anchored on HCuSNPs with good photothermal effects in the NIR-II region. The target miRNA binding with the substrate probes could activate the strand displacement reaction, which was sequentially powered by the released fuel probe under NIR laser irradiation to produce strong fluorescence for miRNA detection. It displayed good sensitivity enhancement compared to molecular beacons, and superior base mismatched capability and specificity was demonstrated in vitro. For miRNA detection in living cells, HCuSNPs-DNA presented a good enzyme degradation resistance and a low cytotoxicity, and realized low-abundance miRNA imaging in different cancers and helped sensitively monitor the change of the oncogenic miRNA expression level. Notably, the good feasibility of in vivo miRNA detection of the system was also achieved. The HCuSNPs-DNA based NIR-laser assisted entropy-driven amplification system provided a new powerful tool for intracellular miRNA analysis.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
The work was supported by the National Natural Science Foundation of China (21874008 and 21475008), the Special Foundation for State Major Research Program of China (Grant No. 2016YFC0106602 and 2016YFC0106601); the Open Research Fund Program of Beijing Key Lab of Plant Resource Research and Development, Beijing Technology and Business University (PRRD-2016-YB2); the Fundamental Research Funds for the Central Universities (Grant No. FRF-BD-17-016A) and the Beijing Municipal Science and Technology Commission (Grant No. z131102002813058).Notes and references
- H. Dong, J. Lei, L. Ding, Y. Wen, H. Ju and X. Zhang, Chem. Rev., 2013, 113, 6207–6233 CrossRef CAS PubMed.
- R. Hamam, D. Hamam, K. A. Alsaleh, M. Kassem, W. Zaher, M. Alfayez, A. Aldahmash and N. M. Alajez, Cell Death Dis., 2017, 8, e3045 CrossRef PubMed.
- S. Jonas and E. Izaurralde, Nat. Rev. Genet., 2015, 16, 421–433 CrossRef CAS PubMed.
- G. Bertoli, C. Cava and I. Castiglioni, Theranostics, 2015, 5, 1122–1143 CrossRef CAS PubMed.
- J. Krol, I. Loedige and W. Filipowicz, Nat. Rev. Genet., 2010, 11, 597–610 CrossRef CAS PubMed.
- M. Ha and V. N. Kim, Nat. Rev. Mol. Cell Biol., 2014, 15, 509–524 CrossRef CAS PubMed.
- D. P. Barte, Cell, 2004, 116, 281–297 CrossRef.
- A. L. Kasinski and F. J. Slack, Nat. Rev. Cancer, 2011, 11, 849–864 CrossRef CAS PubMed.
- M. Han, J. Toli and M. Abdellatif, Curr. Opin. Cardiol., 2011, 26, 181–189 CrossRef PubMed.
- J. Lu, G. Getz, E. A. Miska, E. Alvarez-Saavedra, J. Lamb, D. Peck, A. Sweet-Cordero, B. L. Ebert, R. H. Mak, A. A. Ferrando, J. R. Downing, T. Jacks, H. R. Horvitz and T. R. Golub, Nature, 2005, 435, 834–838 CrossRef CAS PubMed.
- A. Lujambio and S. W. Lowe, Nature, 2012, 482, 347–355 CrossRef CAS PubMed.
- C. J. Cheng, R. Bahal, I. A. Babar, Z. Pincus, F. Barrera, C. Liu, A. Svoronos, D. T. Braddock, P. M. Glazer, D. M. Engelman, W. M. Saltzman and F. J. Slack, Nature, 2015, 518, 107–110 CrossRef CAS PubMed.
- A. W. Wark, H. J. Lee and R. M. Corn, Angew. Chem., Int. Ed., 2008, 47, 644–652 CrossRef CAS PubMed.
- Y. Jiao, M. Zhu, X. Wang, Z. Zhang and S. Zhang, Part. Part. Syst. Charact., 2018, 35, 1700330 CrossRef.
- C. Xue, S. X. Zhang, C. H. Ouyang, D. Chang, B. J. Salena, Y. Li and Z. S. Wu, Angew. Chem., Int. Ed., 2018, 9739–9743 CrossRef CAS PubMed.
- J. Liu, M. Cui, H. Zhou and W. Yang, ACS Sens., 2017, 2, 1847–1853 CrossRef CAS PubMed.
- L. Li, J. Feng, H. Liu, Q. Li, L. Tong and B. Tang, Chem. Sci., 2016, 7, 1940–1945 RSC.
- H. Peng, X. F. Li, H. Zhang and X. C. Le, Nat. Commun., 2017, 8, 14378 CrossRef PubMed.
- Y. Yang, J. Huang, X. Yang, X. He, K. Quan, N. Xie, M. Ou and K. Wang, Anal. Chem., 2017, 89, 5850–5856 CrossRef CAS PubMed.
- Y. Wu, J. Huang, X. Yang, Y. Yang, K. Quan, N. Xie, J. Li, C. Ma and K. Wang, Anal. Chem., 2017, 89, 8377–8383 CrossRef CAS PubMed.
- W. Ma, P. Fu, M. Sun, L. Xu, H. Kuang and C. Xu, J. Am. Chem. Soc., 2017, 139, 11752–11759 CrossRef CAS PubMed.
- H. Wang, Y. Zhang, H. Ma, X. Ren, Y. Wang, Y. Zhang and Q. Wei, Biosens. Bioelectron., 2016, 86, 907–912 CrossRef CAS PubMed.
- Y. Shi, G. Zhang, J. Li, Y. Zhang, Y. Yu and Q. Wei, Microchim. Acta, 2017, 184, 1379–1387 CrossRef CAS.
- X. Ren, T. Zhang, D. Wu, T. Yan, X. Pang, B. Du, W. Lou and Q. Wei, Biosens. Bioelectron., 2017, 94, 694–700 CrossRef CAS PubMed.
- L. He, D. Lu, H. Liang, S. Xie, X. Zhang, Q. Liu, Q. Yuan and W. Tan, J. Am. Chem. Soc., 2017, 140, 263–271 Search PubMed.
- D. Y. Zhang and G. Seelig, Nat. Chem., 2011, 3, 103–113 CrossRef CAS PubMed.
- X. Chen, J. Am. Chem. Soc., 2012, 134, 263–271 CrossRef CAS PubMed.
- F. Huang, M. You, D. Han, X. Xiong, H. Liang and W. Tan, J. Am. Chem. Soc., 2013, 135, 7967–7973 CrossRef CAS PubMed.
- T. Z. Xuewen He, Zhi Li, Ganglin Wang and Nan Ma, Angew. Chem., Int. Ed., 2015, 55, 3073–3076 CrossRef PubMed.
- C.-P. Liang, P.-Q. Ma, H. Liu., X. Guo, B.-C. Yin and B.-C. Ye, Angew. Chem., Int. Ed., 2017, 56, 9077–9081 CrossRef CAS PubMed.
- W. Dai, H. Dong, K. Guo and X. Zhang, Chem. Sci., 2018, 9, 1753–1759 RSC.
- X. Meng, K. Zhang, W. Dai, Y. Cao, F. Yang, H. Dong and X. Zhang, Chem. Sci., 2018, 9, 7419–7425 RSC.
- X. Meng, W. Dai, K. Zhang, H. Dong and X. Zhang, Chem. Sci., 2018, 9, 1184–1190 RSC.
- J. Liu, P. Wang, X. Zhang, L. Wang, D. Wang, Z. Gu, J. Tang, M. Guo, M. Cao, H. Zhou, Y. Liu and C. Chen, ACS Nano, 2016, 10, 4587–4598 CrossRef CAS PubMed.
- Z. Liu, X. Liu, Y. Du, J. Ren and X. Qu, ACS Nano, 2015, 9, 10335–10346 CrossRef CAS PubMed.
- T. Yang, Y. Tang, L. Liu, X. Lv, Q. Wang, H. Ke, Y. Deng, H. Yang, X. Yang, G. Liu, Y. Zhao and H. Chen, ACS Nano, 2017, 11, 1848–1857 CrossRef CAS PubMed.
- D. Wang, H. Dong, M. Li, Y. Cao, F. Yang, K. Zhang, W. Dai, C. Wang and X. Zhang, ACS Nano, 2018, 12, 5241–5252 CrossRef CAS PubMed.
- S. Tang, Y. Gu, H. Lu, H. Dong, K. Zhang, W. Dai, X. Meng, F. Yang and X. Zhang, Anal. Chim. Acta, 2018, 1004, 1–9 CrossRef CAS PubMed.
- F. Xu, H. Dong, Y. Cao, H. Lu, X. Meng, W. Dai, X. Zhang, K. A. Al-Ghanim and S. Mahboob, ACS Appl. Mater. Interfaces, 2016, 8, 33499–33505 CrossRef CAS PubMed.
- H. Dong, X. Meng, W. Dai, Y. Cao, H. Lu, S. Zhou and X. Zhang, Anal. Chem., 2015, 87, 4334–4340 CrossRef CAS PubMed.
- K. Ren, Y. Xu, Y. Liu, M. Yang and H. Ju, ACS Nano, 2018, 12, 263–271 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Details of experimental procedures, the photothermal effects of HCuSNPs-DNA, and optimization of experimental conditions. See DOI: 10.1039/c8nh00330k |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |