Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Elemental characterisation of the pyramidal neuron layer within the rat and mouse hippocampus†
M. J.
Hackett
 *ab,
A.
Hollings
ab,
S.
Caine
ce,
B. E.
Bewer
d,
M.
Alaverdashvili
e,
R.
Takechi
bf,
J. C. L.
Mamo
bf,
M. W. M.
Jones‡
gh,
M. D.
de Jonge
g,
P. G.
Paterson
e,
I. J.
Pickering
*ab,
A.
Hollings
ab,
S.
Caine
ce,
B. E.
Bewer
d,
M.
Alaverdashvili
e,
R.
Takechi
bf,
J. C. L.
Mamo
bf,
M. W. M.
Jones‡
gh,
M. D.
de Jonge
g,
P. G.
Paterson
e,
I. J.
Pickering
 ij and
G. N.
George
ij and
G. N.
George
 ij
ij
aCurtin Institute for Functional Molecules and Interfaces, School of Molecular and Life Sciences, Curtin University, GPOBox U1987, Bentley, WA 6845, Australia. E-mail: mark.j.hackett@curtin.edu.au
bCurtin Health Innovation Research Institute, Curtin University, Bentley, WA 6102, Australia
cDepartment of Anatomy and Cell Biology, University of Saskatchewan, 107 Wiggins Rd, Saskatoon, Saskatchewan S7N 5E5, Canada
dCanadian Light Source, 44 Innovation Boulevard, Saskatoon, Saskatchewan S7N 2V3, Canada
eCollege of Pharmacy and Nutrition, University of Saskatchewan, 107 Wiggins Rd, Saskatoon, Saskatchewan S7N 5E5, Canada
fSchool of Public Health, Curtin University, Bentley, WA 6102, Australia
gAustralian Synchrotron, ANSTO, 800 Blackburn Road, Clayton, VIC 3168, Australia
hARC Centre of Excellence for Advanced Molecular Imaging, La Trobe Institute for Molecular Science, Bundoora, Vic 3089, Australia
iMolecular and Environmental Sciences Group, Geological Sciences, University of Saskatchewan, 114 Science Place, Saskatoon, Saskatchewan S7N 5E2, Canada
jDepartment of Chemistry, University of Saskatchewan, 110 Science Place, Saskatoon, Saskatchewan S7N 5C9, Canada
First published on 29th October 2018
Abstract
A unique combination of sensitivity, resolution, and penetration make X-ray fluorescence imaging (XFI) ideally suited to investigate trace elemental distributions in the biological context. XFI has gained widespread use as an analytical technique in the biological sciences, and in particular enables exciting new avenues of research in the field of neuroscience. In this study, elemental mapping by XFI was applied to characterise the elemental content within neuronal cell layers of hippocampal sub-regions of mice and rats. Although classical histochemical methods for metal detection exist, such approaches are typically limited to qualitative analysis. Specifically, histochemical methods are not uniformly sensitive to all chemical forms of a metal, often displaying variable sensitivity to specific “pools” or chemical forms of a metal. In addition, histochemical methods require fixation and extensive chemical treatment of samples, creating the strong likelihood for metal redistribution, leaching, or contamination. Direct quantitative elemental mapping of total elemental pools, in situ within ex vivo tissue sections, without the need for chemical fixation or addition of staining reagents is not possible with traditional histochemical methods; however, such a capability, which is provided by XFI, can offer an enormous analytical advantage. The results we report herein demonstrate the analytical advantage of XFI elemental mapping for direct, label-free metal quantification, in situ within ex vivo brain tissue sections. Specifically, we definitively characterise for the first time, the abundance of Fe within the pyramidal cell layers of the hippocampus. Localisation of Fe to this cell layer is not reproducibly achieved with classical Perls histochemical Fe stains. The ability of XFI to directly quantify neuronal elemental (P, S, Cl, K, Ca, Fe, Cu, Zn) distributions, revealed unique profiles of Fe and Zn within anatomical sub-regions of the hippocampus i.e., cornu ammonis 1, 2 or 3 (CA1, CA2 or CA3) sub-regions. Interestingly, our study reveals a unique Fe gradient across neuron populations within the non-degenerating and pathology free rat hippocampus, which curiously mirrors the pattern of region-specific vulnerability of the hippocampus that has previously been established to occur in various neurodegenerative diseases.
Significance to metallomicsMetals such as Fe, Cu, and Zn are essential for brain function, and imbalance in metal levels is observed during many brain diseases and neurological conditions. The significance of the research presented herein, is that it describes a detailed characterisation of metal content within specific populations of cells in the hippocampus. The hippocampus is central to spatial memory and learning, but, is highly vulnerable to brain disease. The regional specific variation in metal content we observe, will be built upon in future studies to determine mechanistic roles of metal ions, in specific hippocampal sub-regions, during brain disease. |
Introduction
Tremendous progress in functional neuroanatomy has been made over many decades using techniques that employ staining reagents in combination with light, electron, or fluorescence microscopy.1–4 However, traditional methods typically reveal little on the chemical (molecular and elemental) composition of neurons themselves. A major reason for this is that chemical stains and chemical fixation procedures often remove, re-distribute or even add chemical content to biological samples.5–10 Consequently, it has only been through the recent advances in direct, label free elemental mapping techniques, which have provided the opportunity to examine the elemental composition of neurons, or other brain cells such as glia,11–18 with respect of their anatomical location and brain function.The hippocampus is the main centre of spatial learning and memory formation in the mammalian brain.19,20 Although elemental characterisation of neuron populations within hippocampal sub-regions has remained lacking to now, the relationships between neuron structure, biochemical composition and anatomy in relation to function have been studied extensively. The hippocampus is comprised largely of pyramidal neurons within the Cornu Ammonis sub-regions 1–3 (CA1, CA2 and CA3), while the dentate gyrus (DG) is comprised of a different type of cell, the smaller granule cells.21,22 Pyramidal neurons in different CA sub-regions (CA1, CA2, CA3) are physically different in terms of morphology and patterns of physical connection with other neurons/brain structures, however, chemical differences between pyramidal neurons located in the different CA sub-regions are only a relatively recent discovery. It is also known that differential gene and protein expression occurs across pyramidal neurons of CA1, CA2 and CA3 sub-regions.23–26 This has helped put to rest the long-standing debate regarding whether neurons within the CA2 sub-region exist as functionally and chemically distinct, or if the CA2 sub-region is simply a transition zone with a mixture of CA1 and CA3 neurons.27–29 Recent in situ-hybridization and immuno-histochemical data has demonstrated that CA2 neurons contain greater concentration of specific proteins or have specific gene expression not found in neurons of either CA1 or CA3 sub-regions.23–26,29–31 Such information has opened the door to numerous studies investigating how the distinct chemical differences between CA1, CA2 and CA3 neurons may affect function of the hippocampus during health and disease. For example, immuno-histochemistry30,31 and the use of DNA microarrays and in situ-hybridization32–34 have recently enabled examination of the differential distribution or relative concentration of specific proteins or genes between hippocampal sub-regions during normal and pathological states.23–26,35
An intriguing feature of the hippocampus is that various sub-regions of the hippocampus display selective vulnerability to damage.11,36,37 While CA1 pyramidal neurons are highly vulnerable to degeneration after ischemic insult, CA3 neurons can withstand prolonged exposure to ischemic conditions.38–40 Within the CA1 sub-region, medial CA1 neurons (those more distant from the CA2 and CA3 sub-regions) are more vulnerable to ischemic insult relative to neurons in lateral CA1 sub-regions.37,41 Further, CA1 neurons appear to be more vulnerable to damage during ageing and neurodegenerative diseases of ageing, such as Alzheimer's disease.29 In contrast, seizures experienced during epilepsy cause pronounced damage to pyramidal neurons of the CA3 sub-region but not the adjacent CA2 sub-region.31,36 It has also been established that regional vulnerability to cell death within the hippocampus occurs following traumatic brain injury.42 These disparate responses to various brain injuries, none of them particularly well understood, clearly indicate a strong requirement for increased comprehension of the “baseline” biochemical and elemental composition of neurons in the CA1, CA2 and CA3 hippocampal sub-regions in the non-degenerating brain. Such knowledge will complement current understanding of gene and protein expression, and may provide insight into the biochemical pathways that underlie the selective vulnerability to neurodegeneration within these hippocampal sub-regions.
Classical histochemical methods exist for metal detection such as Timms' and Dancher methods for Zn,43,44 Perls and Turnbull's stains for Fe,45,46 and rhodamine for Cu,47 along with a variety of modern fluorescent metal sensors (i.e. TSQ for Zn).48 Unfortunately, classical histochemical approaches are typically confined to qualitative analysis, and even so, present several drawbacks with interpretation of the staining pattern.49 Such limitations of histochemical methods include non-uniform sensitivity to all chemical forms of a metal, since often drastically increased or decreased sensitivity to specific “pools” or chemical forms of a metal exist.49 For example, heme Fe is not detected by Perls methods.45,46,49 In addition, histochemical methods require fixation and extensive chemical treatment of samples, creating the strong likelihood for metal redistribution, leaching, or contamination.45,46 Recent advances in metal-sensing has provided a range of fluorescence sensors to detect labile metal pools.14,50,51 At this stage, such approaches are typically confined to cell culture, with limited application to rodent animal models, nonetheless, the value of sensing labile metal pools in living cell culture should not be understated.14,50,51
One promising analytical technique to directly quantify elemental composition of biological samples at the cellular level, without the use of endogenous stains or sensors, is X-ray fluorescence imaging (XFI), also known as X-ray fluorescence microscopy (XFM). Application of XFI to image biological samples at third generation synchrotron facilities enables sub micro-molar detection at micron or several-micron spatial resolution, with data collection rates on the order of milli-seconds per pixel.14,52–57 For these reasons, XFI has found increasing use within the field of neuroscience, to study elemental alterations that correlate with anatomical structure,58,59 or occur as a consequence of diseases such as Alzheimer's disease,60–64 Parkinson's disease,65,66 damaging events that occur during epilepsy,67–69 traumatic brain injury,70 and stroke.17,71,72 XFI has also been used to study the elemental phenotype of individual cells and neurons within in vitro cultures,64,73–76 and to study the elemental distribution of the rat hippocampus,12,67–69,77 but detailed characterisation of elemental signatures of cells within CA1 medial, CA1 lateral, CA2 or CA3 sub-regions has not been previously undertaken.
In this investigation, we have used XFI to characterise the elemental composition of medial CA1, lateral CA1, CA2 and CA3 neuronal sub-regions of rat and mouse hippocampus. The distribution of the elements P, S, Cl, K, Ca, Fe, Cu, and Zn was quantified at 3 μm spatial resolution (semi-quantitative for lighter elements; see Methods). The measurements reveal 4 unique insights into region specific elemental distribution: (1) individual cells within CA1, CA2 and CA3 sub-regions of the rat can be differentiated based on unique Fe and Zn areal densities, providing further support that CA2 neurons are a chemically distinct subset of pyramidal neurons. In the murine hippocampus, a similar observation of elevated Fe content is observed in neurons within the CA1 sub-region, relative to CA2 and CA3 sub-regions, however, neurons within CA3 and CA2 sub-regions cannot be differentiated; (2) a lateral-to-medial trend for increasing Fe areal density occurs across the CA1 region in the non-degenerating pathology free rat and murine hippocampus. This observation parallels the trend of increasing susceptibility to delayed neurodegeneration that is known to occur within this region following ischemic insult; (3) the mossy fibres of the CA3 sub-region, which although well established to contain high Zn levels (Timm's and TSQ methods),43,48 are also enriched in Fe within the rat hippocampus, but not murine hippocampus; and (4) the unique insights into cellular Fe content provided by XFI, could not be identified by routine histochemical detection of Fe using the classic histochemical Perls stain.
Materials and methods
Animal models
Tissue was used from healthy (sham operated) male, 8-week old Sprague-Dawley rats (n = 5). These animals form part of a larger study investigating chemical pathways involved in neurodegeneration following global brain ischemia.78 The subset of animals used in this study had been subjected to the sham surgery (i.e., healthy control animal) associated with the 2-vessel occlusion model of global brain ischemia model, as previously reported from our laboratory.78 Brain tissue was obtained from the rats 2 days after sham surgery (n = 5 animals). Although these animals have undergone a sham surgery, which could potentially induce mild systemic inflammation, there is no evidence to suggest brain biochemistry is altered 48 hours after sham surgery, nor have we ever observed cell death or markers of neurodegeneration in the brain of these sham-operated animals.78,79 Many models that produce brain injury or neurodegeneration for example modelling stroke, traumatic brain injury, or vascular dementia, use a surgical model, with surgery operated sham controls. Therefore, the baseline elemental composition of hippocampal neurons from sham animals will be more readily comparable to other studies reporting on sham control animals, than would naïve animals. This work was approved by the University of Saskatchewan's Animal Research Ethics Board, and adhered to the Canadian Council on Animal Care guidelines for humane animal use.Wildtype, male, 12 week old C57 BL/6 mice (n = 5) were housed in standard cages in a temperature controlled (21 °C) colony room on a 12/12 h light/dark cycle with standard rodent maintenance chow and water available ad libitum. All experimental procedures were conducted in accordance with Curtin University Animal Ethics Guidelines.
Tissue collection and sample preparation
All rat tissue was prepared at the University of Saskatchewan and all mice tissue was prepared at Curtin University. Sample preparation procedures were identical between rats and mice. To avoid introduction of chemical artefacts that can result during sample preparation of brain tissue,5,8,10 all rats were anaesthetized with isoflurane, and humanely sacrificed through decapitation, with the head immediately frozen by dropping into liquid nitrogen, as described previously.10 The frozen brain was chiselled out from the head on dry ice. A 20 μm-thick coronal brain section (−3.3 to −2.7 mm anterior to Bregma in rats, and −1.3 to −2.2 mm anterior to Bregma in mice) was cut with a cryo-microtome at −18 °C and was melted onto a: (rat samples) silicon nitride substrate 10 × 10 mm2 silicon frame, 200 μm thick, 5 × 5 mm2 silicon nitride membrane, 200 nm thick, (Norcada, Edmonton, AB, Canada); or (mouse samples) silicon nitride substrate 10 × 10 mm2 silicon frame, 200 μm thick, 5 × 5 mm2 silicon nitride membrane, 1 μm thick, (Melbourne Centre for Nanofabrication, MCN), for imaging at the X-ray fluorescence microscopy beamline at the Australian Synchrotron. An additional two sections for each brain, 10 μm-thick, was mounted onto a regular glass microscope slide, air-dried and post-fixed with immersion for 2 minutes in 10% buffered formalin solution, for routine histology staining with cresyl violet, and Perls Fe histochemistry, as previously described.71Synchrotron imaging
Elemental mapping of was performed at the X-ray fluorescence microscopy beamline at the Australian Synchrotron.80 A monochromatic incident beam of 12.7 keV was focused to a 1 μm (1-sigma) spot with a Kirkpatrick–Baez mirror pair. X-ray emission spectra from the sample were collected in event-mode using the low-latency, 384-channel Maia detector.81 Data were collected with the sample orientated normal to the incident beam and with the detector positioned in backscatter geometry. The sample was raster scanned through the beam with an effective dwell time of 0.1 ms and an effective step (pixel) size of 1 μm. To increase signal to noise (S/N) to highlight subtle but important differences and improve chemical contrast, elemental maps presented in Fig. 4 were processed with a 3-point moving average. Elemental foils (Micromatter, Canada), were scanned in the same geometry and used as references for elemental quantification. Elemental maps were reconstructed from the full emission spectra with GeoPIXE v6.6j. (CSIRO, Australia), which uses a linear transformation matrix for spectral deconvolution.82 Quantification was performed with calibration against elemental foils of known composition, taking into account the composition and density of the silicon nitride substrate and the approximate composition of the sample. The composition of the tissue sample was approximated to equal dried organic material (C22H10N2O4), with a density of 1.42 g cm−3, as previously reported.16 The sample was approximated to have shrunk perpendicular to the section plane, by a factor of ∼3 during air-drying, to a final thickness of 7 μm. The nominal sample composition and thickness estimate is used to provide a first-order self-absorption correction, and inaccuracies have negligible impact on the quantitative concentrations of the first-row transition metals in these thin tissue sections, as reported by several groups.83–85 Lighter element such as S and P are presented for distributional purposes only; absolute quantification of these elements is difficult and subject to significant errors from the assumption of matrix composition, thickness and density. Quantitative data were extracted as tiff files of quantitative per-pixel elemental area density in ng cm−2, which were then imported into ImageJ v1.48, as described previously.16 All regions of interest and the average elemental areal density were calculated using ImageJ. The approximate radiation dose received by the samples during the experiment was calculated using the method of Jones et al.,86 and was found to be 5.39 kGy, which is more than 2 orders of magnitude below the limit at which Jones et al., reported radiation-damage-induced elemental redistribution to occur.86Data analysis
All results are represented as group means ± standard deviation (S.D.). Average elemental concentrations of P, S, Cl, K, Ca, Fe, Cu, and Zn were determined for 10 individual cells within the CA1, CA2, and CA3 sub-regions of the hippocampus, for five rats (n = 5) and five mice (n = 5). Regions of interest were drawn, using ImageJ software, for the soma of each cell body, using the K signal and the established fact that cell bodies are enriched in K.53 The elemental concentration for neurons of each hippocampal sub-region, was determined from the average of the 10 individual cells per animal. In this study we have not attempted to directly compare quantitative metal content between mice and rats, since tissue sections were cut at different times, on different cryo-microtomes, and the assumption of constant thickness between different instruments could not be made. Elemental data were considered as continuous quantitative data. Statistical analysis was performed with SPSS software, SPSS version 23. A one-way ANOVA was performed to determine if there was a significant effect of anatomical location on elemental content of neurons in each region. When the results from ANOVA indicated a significant effect, a two-tailed Tukey's post hoc test was performed. The 95% confidence limit was used (p < 0.05).Results and discussion
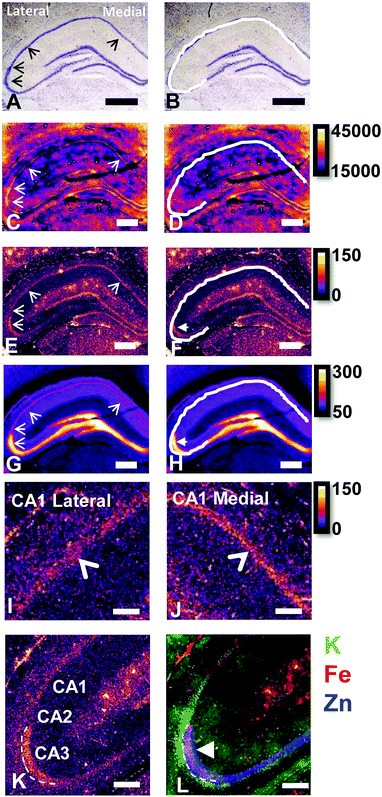
Conventional histochemical Perls Fe staining does not routinely detect Fe in hippocampal pyramidal neurons or in the “Mossy Fibre” sub-region
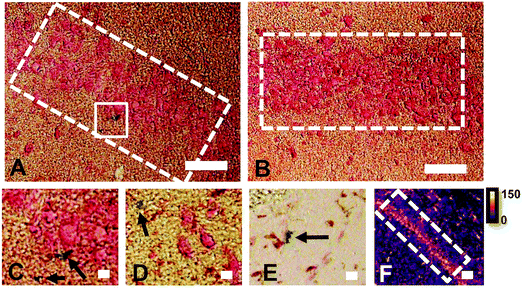
The reaction of Fe3+ with Perls reagent (potassium ferrocyanide) to yield the insoluble Prussian blue pigment (i.e., the Perls stain), is a widely used and accepted method to detect Fe in biological samples. Unfortunately, the results from the method are highly variable, and intrinsically linked to sample preparation.46,49,87,88 It is widely recognised that heme-bound Fe is not detected by Perls, however, less recognition is given to the fact that mobile and labile Fe pools will also not be reproducibly detected, by virtue of redistribution or removal from tissue sections during staining.49 Early studies of the Perls method revealed that perfusion of living animals with the Perls stain produced mild staining of neuronal cytoplasm in addition to dense staining of macrophages, astrocytes and oligodendrocytes.46,49,87,88 Application of the stain ex vivo results in intense staining of macrophages, astrocytes and oligodendrocytes but only mild or no staining of neuronal cytoplasm.46,49,87,88In vivo perfusion of Perls reagent into a living animal is not trivial, and carries substantial ethical considerations. Further, under pathological conditions, there is substantial Fe increase due to increased Ferritin stores within oligodendrocytes, astrocytes and macrophages. Thus, it is not surprising that ex vivo Perls staining has been adopted as standard practice to detect Fe during pathological conditions, but unfortunately, this does not enable robust analysis of neuronal Fe stores. The use of dilute acid in the Perls protocol is likely to liberate loosely coordinated Fe from proteins, enzymes and RNA. Specifically, the pool of Fe that exists as co-factors for enzymes or that is in a labile form within a cell cytoplasm is unlikely to be accurately detected by the Perls stain. This is highlighted by the results of this study (Fig. 1), in which Perls staining fails to stain Fe within the cell cytoplasm of neurons in the hippocampal CA1 layer of a representative rat, whereas XFM elemental mapping revealed this layer was enriched in Fe (Fig. 1). While neurons did not stain positive for Fe, Perls staining shown in Fig. 1 did indicates the presence of Fe within smaller cell bodies. These are likely to be glial cells, such as astrocytes or oligodendrocytes, which are known to contain abundant pools of Ferritin that are easily detected with Perls methods.46,49,87,88XFI elemental maps of K and Zn demarcate the anatomical locations of the CA1, CA2, or CA3 sub-regions of the hippocampus
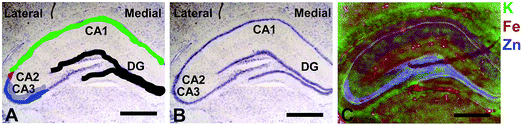
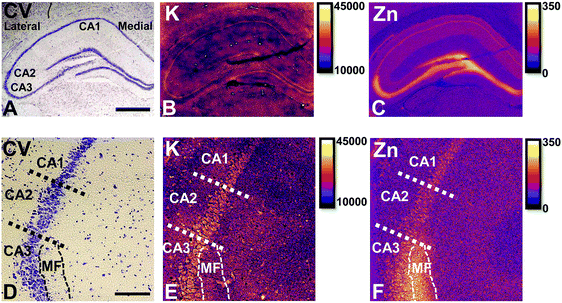
Elemental mapping at the cellular level (3 point moving average applied to data collected at 1 μm pixel resolution) provides rich chemical and anatomic insight. As shown in Fig. 2, a tri-colour overlay of K, Fe and Zn distribution reveals substantial anatomical detail within the rat hippocampus, which is similar to that previously reported for the hippocampus of an accelerated ageing mouse strain.18 In this study, the XFI elemental map of K was used to define the anatomical boundaries of individual cells, similar to previous reports.53 As can be seen in Fig. 3, the ability to visually identify the location of the cell layer and cell bodies with K elemental maps, is similar to that which can be observed with cresyl violet histology (Fig. 3A, B, D and E) i.e., a map of K distribution can be used to identify the neuronal layer of the hippocampus. Routine histology, microscopy and Zn histochemistry is well established to identify several well-characterized anatomical differences between neurons of the CA1, CA2 and CA3 sub-regions.21,27 Specifically, these characteristics are: (i) neurons of the CA1 sub-region have a small soma and their dendrites contain minimal “spiny processes”, which are not rich in histochemically detectable labile Zn. Even though the dendrites are not visible at this spatial resolution with XFI, their localisation to specific sub-regions allows identification of tissue sub-regions in which the dendrites reside;21,27,89 (ii) the CA1 pyramidal neuron cell layer is thinner than CA2 or CA3 layers; (iii) neurons of the CA3 sub-region have a relatively large soma (compared to CA1 neurons), and their dendrites which are confined to the mossy fibre layer have numerous “spiny processes”, which are rich in histochemically detectable labile Zn. Although the individual fibers can not be identified, the mossy fiber sub-region can be identified through the Zn enrichment in this location;89 and (iv) neurons of the CA2 sub-region seem to have properties intermediate between those of CA1 and CA3 neurons, and are characterized by relatively large soma that resemble CA3 neurons, but the dendrites of CA2 neurons contain few spiny processes that are not rich in histochemically detectable labile Zn, and resemble more closely the dendrites of the CA1 neuron.21,27,89 In this study, cells of the pyramidal CA3 sub-region (Fig. 3D and E) appear larger than cells of the CA1 sub-region (Fig. 3D and E) in both the XFI elemental K maps and in cresyl violet histology, consistent with the known morphology of these cells. The thickness of the CA3 layer is also greater than that of the CA1 layer, which is evident in both XFI K elemental maps and cresyl violet histology. Further, the tissue region immediately adjacent to the CA3 pyramidal neuron layer, which contains neuron dendrites, is enriched in Zn, confirming the location of the mossy fibres, and thus, the CA3 layer (Fig. 3C and F). At the spatial resolution used in this study, we cannot conclusively confirm localisation of the Zn to individual dendrites, although, the distribution closely resembles the well established Zn distribution known to occur in the dendrite rich “mossy fibre” sub-region.43 A distinct region exists between the CA3 and CA1 layer that contains larger neuron soma and a thicker cell layer (observed in CV histology and XFI K elemental maps), however no elevation of Zn is observed in the nearby dendrite rich tissue sub-layer. These observations are consistent with the location of the CA2 sub-region. Therefore, XFI elemental maps of K and Zn, in combination with cresyl violet histology allows approximate identification of the CA1, CA2 or CA3 sub-regions of the rat hippocampus (Fig. 3).In this study no attempt has been made to demarcate any differences between the most medial extent of the CA1 region that extends into the subiculum. To avoid reporting data for medial CA1 neurons that are actually neurons of the subiculum, a conservative location was chosen for measurements of medial CA1 neurons (lateral side of the crack in the tissue section shown in Fig. 2B and C). Likewise, quantitative analysis of neurons from CA1, CA2 and CA3 sub-regions purposefully avoided selection of neurons at the interface between sub-regions.
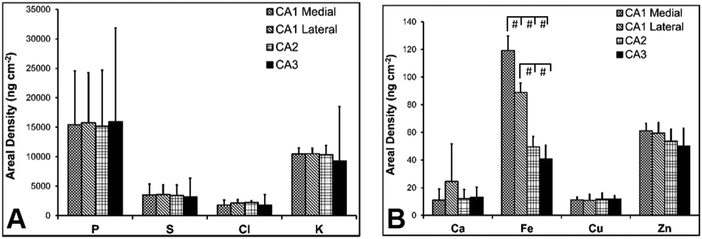
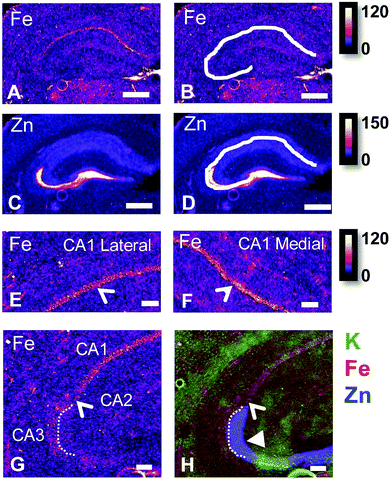
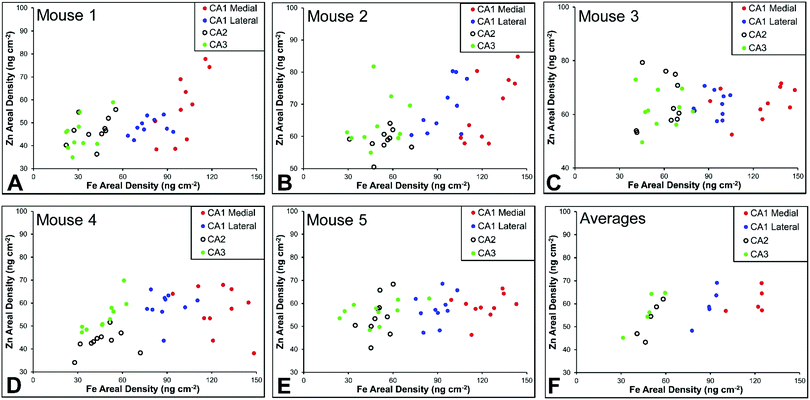
Hippocampal sub-regions display differential Fe and Zn content in the rat, and differential Fe content in the mouse
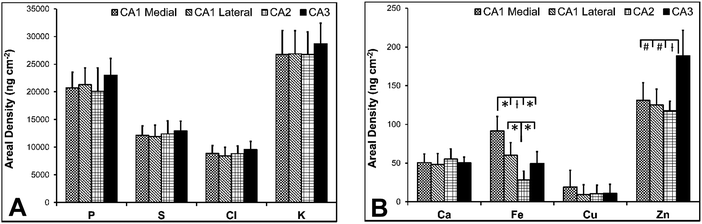
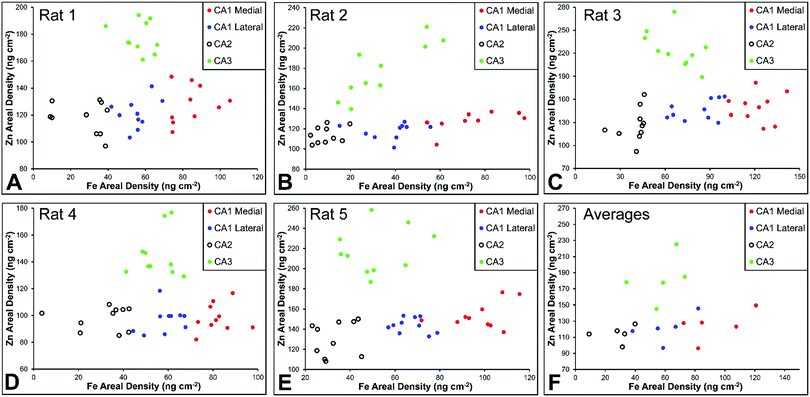
There are several well-characterised anatomic and functional differences between the CA1, CA2 and CA3 hippocampal sub-regions, as already described above. Although there is little argument that a functionally distinct sub-region exists between the CA1 and CA3 sub-regions (i.e., the CA2 sub-region), it has been long debated if neurons within this sub-region comprise a distinct population of unique cells, or if the sub-region is simply a transition tissue zone, containing a mixed population of CA1 and CA3 neurons.27–29 The morphology of neurons within this region appear to be a hybrid between CA1 and CA3 neurons, as the soma resemble CA3 neurons, and the dendrites resemble CA1 neurons.27–29 However, recent in situ-hybridization studies have shown that CA2 neurons express a unique RNA and protein profile, and display several proteins that are not expressed in CA1 and CA3 neurons.23–26 Therefore, the in situ hybridization evidence appears to have resolved this long-standing question, and strongly suggests the CA2 neurons are indeed a unique subset of pyramidal neurons. Our study further validates that in the rat hippocapmus CA2 neurons are chemically different from CA1 and CA3 neurons. XFI elemental mapping was used to compare the elemental composition of cells within each of the CA1, CA2, and CA3 sub-regions of the rat and murine hippocampus. As can be seen in Fig. 4 and 5, Fe and Zn content of neurons (but not P, S, Cl, K, Ca or Cu) was found to be significantly different between sub-regions of the rat hippocampus (One Way ANOVA details, Fig. 4). In the murine hippocampus (Fig. 6 and 7) a significant difference in the elemental content of cells, between sub-regions was only observed for Fe (One way ANOVA details, Fig. 6). Post hoc testing indicates that a statistically significant difference in Fe content exists between all hippocampal sub-regions in the rat, with the CA1 sub-regions containing the highest Fe content, and the CA2 sub-region containing the lowest (Fig. 4). In mice the CA1 sub-regions were found to have the greatest Fe concentration, but differences between CA2 and CA3 regions was not observed (Fig. 6). These differences are further supported by inspection of Fe versus Zn scatterplots of the elemental content of the individual neurons in each animal (Fig. 8 and 9). The scatterplots highlight that a unique Fe and Zn signature exists for neurons within each sub-region of the rat hippocampus, enabling visual discrimination between the cells of each sub-region (Fig. 8). However, in the murine hippocampus, no differentiation between CA2 and CA3 neurons is observed, but CA1 neurons appear separated from CA2 and CA3 neurons (Fig. 9). Importantly, cells from the CA2 sub-region in the rat hippocampus do not appear in the scatterplot as a transition between cells of the CA1 and CA3 sub-regions, suggesting that CA2 neurons are elementally distinct from CA1 and CA3 neurons in the rat hippocampus. Additional replicate images supporting Fig. 5 and 7 are presented in Fig. S1 (rat) and Fig. S2 (mouse) (ESI†).As already discussed, tissue regions in which the dendrites of CA3 neurons are found (mossy fibre layer), are enriched with Zn, in both mice and rats, which is consistent with previous XFI and histochemical studies of Zn within the hippocampus.67,68,90 In addition, in this study we show a relatively high Zn content within the cell bodies of neurons within the CA3 sub-region, and not just the mossy fibre layer. This finding was only observed in rats, and not mice. Zn content within the soma of CA3 neurons in rats was found to be significantly greater than Zn levels in the soma of neurons in the CA1 or CA2 sub-regions (Fig. 4, 5 and 8). While this is not unexpected given the high content of Zn in the adjacent mossy fibre sub-region, which contains the dendrites of the CA3 neurons, most studies that report the histochemical detection of labile Zn do not normally indicate the presence of Zn within the soma of CA3 neurons.43,91 This suggests that the neuronal Zn may not be labile, or, can not be detected by Zn-sensitive fluorophores which may not penetrate cell membranes in vivo. Immuno-histochemistry has revealed that the soma of neurons within the CA3 sub-region are enriched in Zn transport proteins, particularly ZnT-1 and ZnT-3,59,92–95 which may help explain the high Zn content observed within the neurons in this study. At this stage we have no explanation as to why CA3 neuron with elevated Zn content relative to CA2 and CA1 neurons is observed in the rat, but not mouse hippocampus (Fig. 6, 7 and 9). The findings may suggest a higher Zn demand in the rat hippocampus, consistent with the higher memory functions of rats compared to mice; however, clearly more studies are required.
This study did not identify any other significant differences for elements other than Fe and Zn. Distinct patterns of Cu distribution in tissue regions that surround the hippocampus, specifically the lateral ventricle sub-ventricular zones and corpus callosum, have been reported by others,11–13,16,96 and differences in the distribution of Cu in these regions, between mice and rats, have been shown.11,13 Although not a focus of this study, our elemental maps support the recently published findings, and indicate the presences of numerous Cu enriched cells (reported to be a sub-set of astrocytes) in the corpus callosum of rats, but not mice (Fig. S3, ESI†).
The mossy fibre CA3 sub-region is enriched in Fe, not just Zn, in rats, but not mice
It is well established that the dendrites of CA3 neurons – the “mossy fibres” – contain a substantial pool of labile Zn, which can be detected with histochemical methods.43,44 However, the co-localisation of other metals within this hippocampal sub-region, such as Fe, has not previously been reported, to the best of our knowledge. XFI analysis of Zn and Fe, in combination with routine histology in this investigation has revealed that the “mossy fibre” region of the hippocampus contains both Fe and Zn, as shown by the magenta co-localisation of Zn (blue) and Fe (red) in Fig. 5 and Fig. S1 (ESI†). Interestingly, no such co-localisation was observed in the mouse hippocampus, i.e. mossy fibres zone appears blue, not magenta (Fig. 7 and Fig. S2, ESI†). Line plots of K, Fe and Zn content through the CA3 cell layer and the mossy fibre zone provide further support of the Fe co-localisation with Zn in the mossy fibre region of rats, but not mice (Fig. 10). Specifically, the highest K content (CA3 cell layer) does not co-localise with the highest Fe or Zn content, but the highest Fe and Zn content does co-localise in line plots of elemental content in the rat (Fig. 10A, and Fig. S4, ESI†). In contrast, in the mouse, K and Fe co-localise in the CA3 cell layer, but with less Fe observed in the Zn rich mossy fibre sub-region (Fig. 10B and Fig. S4, ESI†). The chemical form of the Fe within the mossy fibre sub-region of the rat hippocampus is as yet unknown. Previous studies that have used methods to precipitate or chelate labile metals within this region have failed to demonstrate the presence of Fe, despite the existence of substantial levels of Zn.44,97 This would suggest that the Fe within this sub-region is not in a labile form. It is possible that the Fe within this zone is in fact not contained within the dendrites themselves, but it could also be contained within the processes of glial cells that occur in this zone. Future sub-cellular XFI analysis at sub-micron resolution will be required to investigate this result further.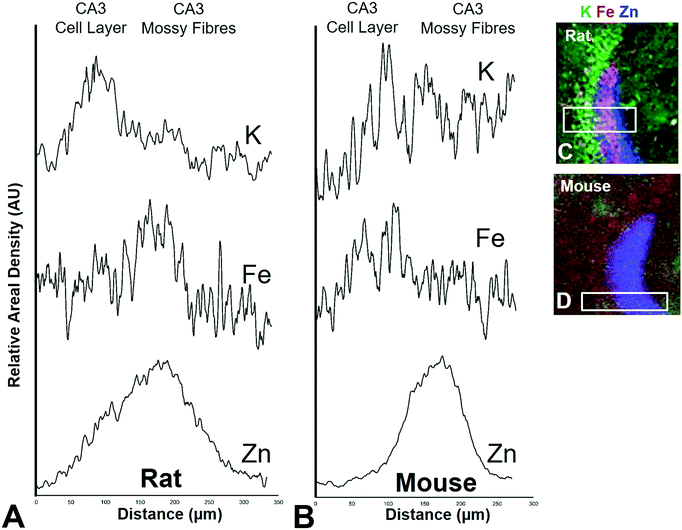 | ||
| Fig. 10 Rat (A) and mouse (B) K, Fe, and Zn profiles (line-plots) of a cross section through the CA3 pyramidal neuron cell layer and mossy fibre region. Correspond XFI tri-colour overlay (green = K, red = Fe, blue Zn) for rat (C) and mouse (D) is shown. Additional profiles are shown in Fig. S4 (ESI†). | ||
A lateral-to-medial trend in Fe content exists across the CA1 sub-region of rats and mice
Numerous studies of global brain ischemia have demonstrated that a lateral-to-medial trend in cell death occurs within the CA1 sub-region of the hippocampus, with medial cells being the most susceptible.37,41 Therefore, in this investigation of healthy control rats (i.e., non-degenerating hippocampus), we sought to determine if CA1 neurons contained an elemental signature distinct from neurons of the CA2 and CA3 sub-regions, and if there was a lateral to medial trend in the elemental content of CA1 neurons, which may underlie the enhanced vulnerability of this region. As discussed above, the Fe versus Zn scatterplot shows CA1 neurons are distinct from both CA2 and CA3 neurons, in the rat and mouse hippocampus. Specifically, neurons within the CA1 sub-region contain increased Fe content relative to CA2 and CA3 neurons. In addition, it can be seen that medial CA1 neurons contain significantly more Fe than lateral CA1 neurons, both the rat (Fig. 4, 5 and 8) and mouse (Fig. 6, 7 and 9) hippocampus. The lateral to medial Fe gradient across the CA1 sub-region, is intriguing, as it reflects the same pattern of cell death that is established to occur in this region during neurodegenerative disease. Therefore, the findings of this study open the door for future investigations of the role that the endogenous Fe within the CA1 sector may play in neurodegeneration. At this stage we can only speculate if such a role exists, however, due to the known involvement of Fe in the production of free radicals through Fenton chemistry, increased Fe levels in the medial CA1 neurons may generate greater oxidative stress under ischemic conditions, which could cause increased cell death within this region. Future studies are required to determine the bioavailability, chemical form and oxidation state of Fe in this hippocampal sub-region.Conclusions
This study further contributes to a growing body of work that demonstrates the immense potential for application of XFI to the field of neuroscience, as an analytical tool for studying the elemental composition of individual cells, in situ, within tissue sections. Our data has revealed key characteristics of metal distribution within the hippocampus that appears conserved across both mice and rats, in addition, more subtle differences between species was found to exist within the CA3 sub-regions. Specifically, our data has revealed for the first time that cells within the CA2 sub-region of the rat hippocampus have a distinct elemental signature, which differentiates these cells from those in the CA1 and CA3 sub-regions. Further, in healthy rats and mice, cells in the CA1 sub-region of the hippocampus contain the highest Fe content, compared to CA2 and CA3 sub-regions, and a lateral-to-medial trend in Fe concentration is observed across CA1 sub-region in both mice and rats. The lateral to medial trend matches the unexplained vulnerability to cell death that has been reported for this region, highlighting future studies should investigate the chemical form and oxidation state of Fe within the CA1 sector, to help determine if it could be a contributing factor to selective degeneration. Lastly, this study identified that the mossy fibre sub-region of the rat hippocampus, a region known to be rich in labile Zn, also contains substantial levels of Fe.Author contributions
Ashley Hollings (PhD student): assisted with sample preparation of mouse samples and performed detailed data analysis of mouse data set. Sally Caine (post-doctoral fellow, co-investigator): generated rat tissue samples, assisted with sample preparation and collection of XFI elemental maps. Performed histology of rat tissue sections. Brian E. Bewer (Research Associate, co-investigator): assisted in collection of XFI data, analysis of XFI data (data fitting) and design of statistical methods. Michael Jones (Beamline Scientist, co-investigator): assisted with XFI experiments at Australian Synchrotron, including optimisation of beamline, essential for analyses of dilute elemental concentrations in thin biological samples. Martin de Jonge (Beamline Scientist, co-investigator): assisted in XFI data collection and data analysis at the Australian Synchrotron. Ryu Takechi (Research Fellow, co-investigator). MJH/RT/JCLM designed mouse experiment. RT assisted with collection of mouse XFI data. John C. L. Mamo (co-investigator), co-supervisor of AH. MJH/RT/JCLM designed mouse experiment. JCLM edited manuscript, pertaining to mouse data. Mariam A. Alaverdashvilli (co-investigator). Provided expertise in neuroanatomy of the hippocampus. Phyllis G. Paterson (co-investigator), Supervisor of MJH when MJH was a postdoctoral fellow. Provided key expertise in fundamental function of the hippocampus and use of rat animal models. Ingrid J. Pickering (co-investigator), supervisor of MJH when MJH was a postdoctoral fellow. Provided key expertise in XFI experimental design. Graham N. George (co-investigator), supervisor of MJH when MJH was a postdoctoral fellow. Responsible for design of rat experiments (experiments designed by MJH, GNG, IJP and PGP).Conflicts of interest
There are no conflicts to declare.Acknowledgements
MJH is supported by the Dementia Australia Research Foundation, Mamutil New Investigator Project Grant (11646). This work was funded in part by the Canadian Institutes of Health Research (CIHR)/Heart and Stroke Foundation of Canada Synchrotron Medical Imaging Team Grant No. 99472 (IJP, GNG, PGP and others). IJP and GNG acknowledge Natural Science and Engineering Research Council of Canada (GNG, IJP) and the Government of Saskatchewan (IJP). SC is a CIHR-THRUST Fellow and MA is a CIHR-THRUST Associate. GNG and IJP are Canada Research Chairs. MJH was supported by postdoctoral Fellowships from the CIHR, from the Saskatchewan Health Research Foundation, and from the CIHR Training grant in Health Research Using Synchrotron Techniques (CIHR-THRUST) for components of this research. JM and RT are supported by the Australian National Health and Medical Research Council, Department of Health, Australian Government (APP1064567). We gratefully acknowledge travel funding provided by ANSTO, funded by the Australian Government. Components of this research were undertaken at the X-ray fluorescence microscopy (XFM) beamline at the Australian Synchrotron, ANSTO, Victoria, Australia. This work was performed in part at the Melbourne Centre for Nanofabrication (MCN) in the Victorian Node of the Australian National Fabrication Facility (ANFF). We would like to thank other members of the Australian Synchrotron XFM beamline, David Paterson, Daryl Howland, and Simon James for their assistance in experiments performed at the Australian Synchrotron. The authors gratefully acknowledge the surgical expertise of Dr Shari Smith and Angela Cooper who performed the animal surgeries for this research.References
- K. Chung and K. Deisseroth, CLARITY for mapping the nervous system, Nat. Methods, 2013, 10, 508–513 CrossRef CAS PubMed.
- E. Gray, Electron microscopy of synaptic contacts on dendrite spines of the cerebral cortex, Nature, 1959, 183, 1592–1593 CrossRef CAS PubMed.
- M. Colonnier, Synaptic patterns on different cell types in the different laminae of the cat visual cortex. An electron microscope study, Brain Res., 1968, 9, 268275–273287 CrossRef.
- M. Marin-Padilla, Structural abnormalities of the cerebral cortex in human chromosomal aberrations: a Golgi study, Brain Res., 1972, 44, 625–629 CrossRef CAS PubMed.
- M. J. Hackett, J. A. McQuillan, F. El-Assaad, J. B. Aitken, A. Levina and D. D. Cohen, et al., Chemical alterations to murine brain tissue induced by formalin fixation: implications for biospectroscopic imaging and mapping studies of disease pathogenesis, Analyst, 2011, 136, 2941–2952 RSC.
- V. Zohdi, D. R. Whelan, B. R. Wood, J. T. Pearson, K. R. Bambery and M. J. Black, Importance of tissue preparation methods in FTIR micro-spectroscopical analysis of biological tissues: ‘Traps for new users’, PLoS One, 2015, 10, e0116491 CrossRef PubMed.
- Q. Jin, T. Paunesku, B. Lai, S. C. Gleber, S. Chen and L. Finney, et al., Preserving elemental content in adherent mammalian cells for analysis by synchrotron-based X-ray fluorescence microscopy, J. Microsc., 2016, 1–13 CAS.
- M. J. Hackett, S. E. Smith, P. G. Paterson, H. Nichol, I. J. Pickering and G. N. George, X-ray Absorption Spectroscopy at the Sulfur K-Edge: A New Tool to Investigate the Biochemical Mechanisms of Neurodegeneration, ACS Chem. Neurosci., 2012, 3, 178–185 CrossRef CAS PubMed.
- L. Perrin, A. Carmona, S. Roudeau and R. Ortega, Evaluation of sample preparation methods for single cell quantitative elemental imaging using proton or synchrotron radiation focused beams, J. Anal. At. Spectrom., 2015, 30, 2525–2532 RSC.
- M. J. Hackett, C. J. Britz, H. Nichol, P. G. Paterson, I. J. Pickering and G. N. George, In situ Bio-Spectroscopic Investigation of Rapid Ischemic and Post-mortem Induced Biochemical Alterations in the Rat Brain, ACS Chem. Neurosci., 2015, 6, 226–238 CrossRef CAS PubMed.
- B. Sullivan, G. Robison, J. Osborn, M. Kay, P. Thompson and K. Davis, et al., On the nature of the Cu-rich aggregates in brain astrocytes, Redox Biol., 2017, 11, 231–239 CrossRef CAS PubMed.
- Y. Pushkar, G. Robison, B. Sullivan, S. X. Fu, M. Kohne and W. Jiang, et al., Aging results in copper accumulations in glial fibrillary acidic protein-positive cells in the subventricular zone, Aging Cell, 2013, 12, 823–832 CrossRef CAS PubMed.
- B. Sullivan, G. Robison, Y. Pushkar, J. K. Young and K. F. Manaye, Copper accumulation in rodent brain astrocytes: A species difference, J. Trace Elem. Med. Biol., 2017, 39, 6–13 CrossRef CAS PubMed.
- D. J. Hare, E. J. New, M. D. de Jonge and G. McColl, Imaging metals in biology: balancing sensitivity, selectivity and spatial resolution, Chem. Soc. Rev., 2015, 44, 5941–5958 RSC.
- N. Braidy, A. Poljak, C. Marjo, H. Rutlidge, A. Rich and B.-E. Jugder, et al., Identification of Cerebral Metal Ion Imbalance in the Brain of Aging Octodon degus, Front. Aging Neurosci., 2017, 9, 66 Search PubMed.
- B. R. Lins, J. M. Pushie, M. Jones, D. L. Howard, J. G. Howland and M. J. Hackett, Mapping Alterations to the Endogenous Elemental Distribution within the Lateral Ventricles and Choroid Plexus in Brain Disorders Using X-Ray Fluorescence Imaging, PLoS One, 2016, 11, e0158152 CrossRef PubMed.
- S. Caine, M. J. Hackett, H. Hou, S. Kumar, J. Maley and Z. Ivanishvili, et al., A novel multi-modal platform to image molecular and elemental alterations in ischemic stroke, Neurobiol. Dis., 2016, 91, 132–142 CrossRef CAS PubMed.
- N. Fimognari, A. Hollings, V. Lam, R. J. Tidy, C. M. Kewish and M. A. Albrecht, et al., Biospectroscopic Imaging Provides Evidence of Hippocampal Zn Deficiency and Decreased Lipid Unsaturation in an Accelerated Aging Mouse Model, ACS Chem. Neurosci., 2018 DOI:10.1021/acschemneuro.8b00193.
- S. Zola-Morgan, L. R. Squire and D. G. Amaral, Human amnesia and the medial temporal region: enduring memory impairment following a bilateral lesion limited to field CA1 of the hippocampus, J. Neurosci., 1986, 6, 2950–2967 CrossRef CAS PubMed.
- L. E. Jarrard, On the role of the hippocampus in learning and memory in the rat, Behav. Neural Biol., 1993, 60, 9–26 CrossRef CAS PubMed.
- R. Lorente de Nó, Studies on the structure of the cerebral cortex. II. Continuation of the study of the ammonic system, J. Psychol. Neurol., 1934, 46, 113–177 Search PubMed.
- J. O'keefe and L. Nadel, The hippocampus as a cognitive map, Oxford University Press, USA, 1978 Search PubMed.
- D. E. Pellegrini-Giampietro, R. S. Zukin, M. Bennett, S. Cho and W. A. Pulsinelli, Switch in glutamate receptor subunit gene expression in CA1 subfield of hippocampus following global ischemia in rats, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 10499–10503 CrossRef CAS.
- X. Zhao, E. S. Lein, A. He, S. C. Smith, C. Aston and F. H. Gage, Transcriptional profiling reveals strict boundaries between hippocampal subregions, J. Comp. Neurol., 2001, 441, 187–196 CrossRef CAS PubMed.
- E. S. Lein, X. Zhao and F. H. Gage, Defining a molecular atlas of the hippocampus using DNA microarrays and high-throughput in situ hybridization, J. Neurosci., 2004, 24, 3879–3889 CrossRef CAS PubMed.
- C. Burger, Region-specific genetic alterations in the aging hippocampus: implications for cognitive aging, Front. Aging Neurosci., 2010, 2, 140 Search PubMed.
- B. J. Claiborne, D. G. Amaral and W. M. Cowan, A light and electron microscopic analysis of the mossy fibers of the rat dentate gyrus, J. Comp. Neurol., 1986, 246, 435–458 CrossRef CAS PubMed.
- S. Tole, C. Christian and E. A. Grove, Early specification and autonomous development of cortical fields in the mouse hippocampus, Development, 1997, 124, 4959–4970 CAS.
- P. L. Woodhams, M. R. Celio, N. Ulfig and M. P. Witter, Morphological and functional correlates of borders in the entorhinal cortex and hippocampus, Hippocampus, 1993, 3, 303–311 Search PubMed.
- M. Celio, Calbindin D-28k and parvalbumin in the rat nervous system, Neuroscience, 1990, 35, 375–475 CrossRef CAS PubMed.
- C. Leranth and C. Ribak, Calcium-binding proteins are concentrated in the CA2 field of the monkey hippocampus: a possible key to this region's resistance to epileptic damage, Exp. Brain Res., 1991, 85, 129–136 CrossRef CAS PubMed.
- L. Wodicka, H. Dong, M. Mittmann, M.-H. Ho and D. J. Lockhart, Genome-wide expression monitoring in Saccharomyces cerevisiae, Nat. Biotechnol., 1997, 15, 1359–1367 CrossRef CAS PubMed.
- D. J. Lockhart and E. A. Winzeler, Genomics, gene expression and DNA arrays, Nature, 2000, 405, 827–836 CrossRef CAS PubMed.
- D. J. Lockhart and C. Barlow, Expressing what's on your mind: DNA arrays and the brain, Nat. Rev. Neurosci., 2001, 2, 63–68 CrossRef CAS PubMed.
- D. E. Pellegrini-Giampietro, R. S. Zukin, M. V. Bennett, S. Cho and W. A. Pulsinelli, Switch in glutamate receptor subunit gene expression in CA1 subfield of hippocampus following global ischemia in rats, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 10499–10503 CrossRef CAS.
- T. L. Babb, W. J. Brown, J. Pretorius, C. Davenport, J. P. Lieb and P. H. Crandall, Temporal lobe volumetric cell densities in temporal lobe epilepsy, Epilepsia, 1984, 25, 729–740 CrossRef CAS PubMed.
- M. Nakamura, M. Araki, K. Oguro and T. Masuzawa, Differential distribution of 68 Kd and 200 Kd neurofilament proteins in the gerbil hippocampus and their early distributional changes following transient forebrain ischemia, Exp. Brain Res., 1992, 89, 31–39 CrossRef CAS PubMed.
- P. Lipton, Ischemic cell death in brain neurons, Physiol. Rev., 1999, 79, 1431–1568 CrossRef CAS PubMed.
- D. Corbett and S. Nurse, The problem of assessing effective neuroprotection in experimental cerebral ischemia, Prog. Neurobiol., 1998, 54, 531–548 CrossRef CAS PubMed.
- T. Kirino, A. Tamura and K. Sano, Delayed neuronal death in the rat hippocampus following transient forebrain ischemia, Acta Neuropathol., 1984, 64, 139–147 CrossRef CAS PubMed.
- B. J. Grain, W. D. Westerkam, A. H. Harrison and J. V. Nadler, Selective neuronal death after transient forebrain ischemia in the mongolian gerbil: A silver impregnation study, Neuroscience, 1988, 27, 387–402 CrossRef.
- D. L. McCullers, P. G. Sullivan, S. W. Scheff and J. P. Herman, Mifepristone protects CA1 hippocampal neurons following traumatic brain injury in rat, Neuroscience, 2002, 109, 219–230 CrossRef CAS PubMed.
- C. Frederickson, B. Rampy, S. Reamy-Rampy and G. Howell, Distribution of histochemically reactive zinc in the forebrain of the rat, J. Chem. Neuroanat., 1992, 5, 521–530 CrossRef CAS PubMed.
- G. Danscher and J. Zimmer, An improved Timm sulphide silver method for light and electron microscopic localization of heavy metals in biological tissues, Histochem. Cell Biol., 1978, 55, 27–40 CAS.
- R. Meguro, Y. Asano, S. Odagiri, C. Li, H. Iwatsuki and K. Shoumura, The presence of ferric and ferrous iron in the nonheme iron store of resident macrophages in different tissues and organs: histochemical demonstrations by the perfusion-Perls and -Turnbull methods in the rat, Arch. Histol. Cytol., 2005, 68, 171–183 CrossRef CAS PubMed.
- R. Meguro, Y. Asano, S. Odagiri, C. Li and K. Shoumura, Cellular and subcellular localizations of nonheme ferric and ferrous iron in the rat brain: a light and electron microscopic study by the perfusion-Perls and -Turnbull methods, Arch. Histol. Cytol., 2008, 71, 205–222 CrossRef CAS PubMed.
- L. Pilloni, S. Lecca, P. Van Eyken, C. Flore, L. Demelia and G. Pilleri, et al., Value of histochemical stains for copper in the diagnosis of Wilson's disease, Histopathology, 1998, 33, 28–33 CrossRef CAS PubMed.
- J. W. Meeusen, H. Tomasiewicz, A. Nowakowski and D. H. Petering, TSQ (6-Methoxy-8-p-Toluenesulfonamido-Quinoline), a Common Fluorescent Sensor for Cellular Zinc, Images Zinc Proteins, Inorg. Chem., 2011, 50, 7563–7573 CrossRef CAS PubMed.
- S. A. Sands, R. Leung-Toung, Y. Wang, J. Connelly and S. M. LeVine, Enhanced Histochemical Detection of Iron in Paraffin Sections of Mouse Central Nervous System Tissue: Application in the APP/PS1 Mouse Model of Alzheimer's Disease, ASN Neuro, 2016, 8 DOI:10.1177/1759091416670978.
- L. M. Hyman and K. J. Franz, Probing oxidative stress: Small molecule fluorescent sensors of metal ions, reactive oxygen species, and thiols, Coord. Chem. Rev., 2012, 256, 2333–2356 CrossRef CAS PubMed.
- E. J. New, Tools to study distinct metal pools in biology, Dalton Trans., 2013, 42, 3210–3219 RSC.
- E. Malucelli, M. Fratini, A. Notargiacomo, A. Gianoncelli, L. Merolle and A. Sargenti, et al., Where is it and how much? Mapping and quantifying elements in single cells, Analyst, 2016, 141, 5221–5235 RSC.
- S. Bohic, D. Hare, A. Daoust, P. Cloetens and E. L. Barbier. Spatially resolved imaging methods to probe metals in the brain: from subcellular to organ level, Metal Ions in Neurological Systems, Springer, 2012, pp. 211–222 Search PubMed.
- D. J. Hare, M. W. M. Jones, V. C. Wimmer, N. L. Jenkins, M. D. de Jonge and A. I. Bush, et al., High-resolution complementary chemical imaging of bio-elements in Caenorhabditis elegans, Metallomics, 2016, 8, 156–160 RSC.
- M. D. de Jonge, C. G. Ryan and C. J. Jacobsen, X-ray nanoprobes and diffraction-limited storage rings: opportunities and challenges of fluorescence tomography of biological specimens, J. Synchrotron Radiat., 2014, 21, 1031–1047 CrossRef CAS PubMed.
- S. A. James, M. D. de Jonge, D. L. Howard, A. I. Bush, D. Paterson and G. McColl, Direct in vivo imaging of essential bioinorganics in Caenorhabditis elegans, Metallomics, 2013, 5, 627–635 RSC.
- M. J. Pushie, I. J. Pickering, M. Korbas, M. J. Hackett and G. N. George, Elemental and Chemically Specific X-ray Fluorescence Imaging of Biological Systems, Chem. Rev., 2014, 114, 8499–8541 CrossRef CAS PubMed.
- M. Alaverdashvili, M. J. Hackett, I. J. Pickering and P. G. Paterson, Laminar-specific distribution of zinc: Evidence for presence of layer IV in forelimb motor cortex in the rat, NeuroImage, 2014, 103, 502–510 CrossRef CAS PubMed.
- D. H. Linkous, J. M. Flinn, J. Y. Koh, A. Lanzirotti, P. M. Bertsch and B. F. Jones, et al., Evidence that the ZNT3 protein controls the total amount of elemental zinc in synaptic vesicles, J. Histochem. Cytochem., 2008, 56, 3–6 CrossRef CAS PubMed.
- M. W. Bourassa and L. M. Miller, Metal imaging in neurodegenerative diseases, Metallomics, 2012, 4, 721–738 RSC.
- A. C. Leskovjan, A. Kretlow, A. Lanzirotti, R. Barrea, S. Vogt and L. M. Miller, Increased brain iron coincides with early plaque formation in a mouse model of Alzheimer's disease, NeuroImage, 2011, 55, 32–38 CrossRef CAS PubMed.
- A. C. Leskovjan, A. Lanzirotti and L. M. Miller, Amyloid plaques in PSAPP mice bind less metal than plaques in human Alzheimer's disease, NeuroImage, 2009, 47, 1215–1220 CrossRef PubMed.
- L. M. Miller, Q. Wang, T. P. Telivala, R. J. Smith, A. Lanzirotti and J. Miklossy, Synchrotron-based infrared and X-ray imaging shows focalized accumulation of Cu and Zn co-localized with beta-amyloid deposits in Alzheimer's disease, J. Struct. Biol., 2006, 155, 30–37 CrossRef CAS PubMed.
- G. D. Ciccotosto, S. A. James, M. Altissimo, D. Paterson, S. Vogt and B. Lai, et al., Quantitation and localization of intracellular redox active metals by X-ray fluorescence microscopy in cortical neurons derived from APP and APLP2 knockout tissue, Metallomics, 2014, 6, 1894–1904 RSC.
- J. Chwiej, D. Adamek, M. Szczerbowska-Boruchowska, A. Krygowska-Wajs, S. Wojcik and G. Falkenberg, et al., Investigations of differences in iron oxidation state inside single neurons from substantia nigra of Parkinson's disease and control patients using the micro-XANES technique, JBIC, J. Biol. Inorg. Chem., 2007, 12, 204–211 CrossRef CAS PubMed.
- B. F. G. Popescu, M. J. George, U. Bergmann, A. V. Garachtchenko, M. E. Kelly and R. P. E. McCrea, et al., Mapping metals in Parkinson's and normal brain using rapid-scanning X-ray fluorescence, Phys. Med. Biol., 2009, 54, 651 CrossRef PubMed.
- J. Chwiej, J. Dulinska, K. Janeczko, K. Appel and Z. Setkowicz, Variations in elemental compositions of rat hippocampal formation between acute and latent phases of pilocarpine-induced epilepsy: an X-ray fluorescence microscopy study, JBIC, J. Biol. Inorg. Chem., 2012, 17, 731–739 CrossRef CAS PubMed.
- J. Chwiej, J. Kutorasinska, K. Janeczko, K. Gzielo-Jurek, L. Uram and K. Appel, et al., Progress of elemental anomalies of hippocampal formation in the pilocarpine model of temporal lobe epilepsy: an X-ray fluorescence microscopy study, Anal. Bioanal. Chem., 2012, 404, 3071–3080 CrossRef CAS PubMed.
- J. Chwiej, W. Winiarski, M. Ciarach, K. Janeczko, M. Lankosz and K. Rickers, et al., The role of trace elements in the pathogenesis and progress of pilocarpine-induced epileptic seizures, JBIC, J. Biol. Inorg. Chem., 2008, 13, 1267–1274 CrossRef CAS.
- J. Chwiej, A. Sarapata, K. Janeczko, Z. Stegowski, K. Appel and Z. Setkowicz, X-ray fluorescence analysis of long-term changes in the levels and distributions of trace elements in the rat brain following mechanical injury, JBIC, J. Biol. Inorg. Chem., 2011, 16, 275–283 CrossRef CAS.
- M. J. Hackett, M. DeSouza, S. Caine, B. Bewer, H. Nichol and P. G. Paterson, et al., A New Method To Image Heme-Fe, Total Fe, and Aggregated Protein Levels after Intracerebral Hemorrhage, ACS Chem. Neurosci., 2015, 6, 761–770 CrossRef CAS.
- G. Silasi, A. C. Klahr, M. J. Hackett, A. M. Auriat, H. Nichol and F. Colbourne, Prolonged therapeutic hypothermia does not adversely impact neuroplasticity after global ischemia in rats, J. Cereb. Blood Flow Metab., 2012, 32, 1525–1534 CrossRef CAS.
- R. A. Colvin, B. Lai, W. R. Holmes and D. Lee, Understanding metal homeostasis in primary cultured neurons. Studies using single neuron subcellular and quantitative metallomics, Metallomics, 2015, 7, 1111–1123 RSC.
- S. Roudeau, A. Carmona, L. Perrin and R. Ortega, Correlative organelle fluorescence microscopy and synchrotron X-ray chemical element imaging in single cells, Anal. Bioanal. Chem., 2014, 406, 6979–6991 CrossRef CAS.
- A. Grubman, S. James, J. James, C. Duncan, I. Volitakis and J. Hickey, et al., X-ray fluorescence imaging reveals subcellular biometal disturbances in a childhood neurodegenerative disorder, Chem. Sci., 2014, 5, 2503–2516 RSC.
- R. A. Colvin, Q. Jin, B. Lai and L. Kiedrowski, Visualizing Metal Content and Intracellular Distribution in Primary Hippocampal Neurons with Synchrotron X-Ray Fluorescence, PLoS One, 2016, 11, e0159582 CrossRef.
- G. Robison, T. Zakharova, S. Fu, W. Jiang, R. Fulper and R. Barrea, et al., X-ray fluorescence imaging of the hippocampal formation after manganese exposure, Metallomics, 2013, 5, 1554–1565 RSC.
- M. J. Hackett, S. E. Smith, S. Caine, H. Nichol, G. N. George and I. J. Pickering, et al., Novel bio-spectroscopic imaging reveals disturbed protein homeostasis and thiol redox with protein aggregation prior to hippocampal CA1 pyramidal neuron death induced by global brain ischemia in the rat, Free Radicals Biol. Med., 2015, 89, 806–818 CrossRef CAS.
- F. Colbourne and D. Corbett, Delayed postischemic hypothermia: a six month survival study using behavioral and histological assessments of neuroprotection, J. Neurosci., 1995, 15, 7250–7260 CrossRef CAS.
- D. Paterson, M. De Jonge, D. Howard, W. Lewis, J. McKinlay and A. Starritt, et al., The X-ray Fluorescence Microscopy Beamline at the Australian Synchrotron, AIP Conf. Proc., 2011, 1365, 219–222 CrossRef.
- C. Ryan, D. Siddons, R. Kirkham, Z. Li, M. de Jonge and D. Paterson, et al., MAIA X-ray fluorescence imaging: capturing detail in complex natural samples, J. Phys.: Conf. Ser., 2014, 499, 012002 CrossRef.
- C. Ryan, Quantitative trace element imaging using PIXE and the nuclear microprobe, Int. J. Imag. Syst. Tech., 2000, 11, 219–230 CrossRef.
- B. Bewer, Quantification estimate methods for synchrotron radiation X-ray fluorescence spectroscopy, Nucl. Instrum. Methods Phys. Res., Sect. B, 2015, 347, 1–6 CrossRef CAS.
- M. Szczerbowska-Boruchowska, Sample thickness considerations for quantitative X-ray fluorescence analysis of the soft and skeletal tissues of the human body – theoretical evaluation and experimental validation, X-Ray Spectrom., 2012, 41, 328–337 CrossRef CAS.
- A. Grubman, P. Guennel, K. A. Vessey, M. W. M. Jones, S. A. James and M. D. de Jonge, et al., X-ray fluorescence microscopic measurement of elemental distribution in the mouse retina with age, Metallomics, 2016, 8, 1110–1121 RSC.
- M. W. M. Jones, D. J. Hare, S. A. James, M. D. de Jonge and G. McColl, Radiation Dose Limits for Bioanalytical X-ray Fluorescence Microscopy, Anal. Chem., 2017, 89, 12168–12175 CrossRef CAS PubMed.
- S. van Duijn, R. J. A. Nabuurs, S. G. van Duinen and R. Natté, Comparison of Histological Techniques to Visualize Iron in Paraffin-embedded Brain Tissue of Patients with Alzheimer's Disease, J. Histochem. Cytochem., 2013, 61, 785–792 CrossRef PubMed.
- R. Meguro, Y. Asano, S. Odagiri, C. Li, H. Iwatsuki and K. Shoumura, Nonheme-iron histochemistry for light and electron microscopy: a historical, theoretical and technical review, Arch. Histol. Cytol., 2007, 70, 1–19 CrossRef CAS.
- M. Cassell and M. Brown, The distribution of Timm's stain in the nonsulphide-perfused human hippocampal formation, J. Comp. Neurol., 1984, 222, 461–471 CrossRef CAS.
- S. A. James, Q. I. Churches, M. D. de Jonge, I. E. Birchall, V. Streltsov and G. McColl, et al., Iron, Copper, and Zinc Concentration in Aβ Plaques in the APP/PS1 Mouse Model of Alzheimer's Disease Correlates with Metal Levels in the Surrounding Neuropil, ACS Chem. Neurosci., 2017, 8, 629–637 CrossRef CAS.
- C. J. Frederickson, S. W. Suh, D. Silva, C. J. Frederickson and R. B. Thompson, Importance of Zn in the central nervous system: the Zinc-containing neuron, J. Nutr., 2000, 130, 1471–1483 CrossRef.
- T. B. Cole, H. J. r. Wenzel, K. E. Kafer, P. A. Schwartzkroin and R. D. Palmiter, Elimination of zinc from synaptic vesicles in the intact mouse brain by disruption of the ZnT3 gene, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 1716–1721 CrossRef CAS.
- I. Nydegger, S. M. Rumschik, J. Zhao and A. R. Kay, Evidence for an Extracellular Zinc-Veneer in Rodent Brains from Experiments with Zn-Ionophores and ZnT3 Knockouts, ACS Chem. Neurosci., 2012, 3, 761–766 CrossRef CAS.
- H. J. Wenzel, T. B. Cole, D. E. Born, P. A. Schwartzkroin and R. D. Palmiter, Ultrastructural localization of zinc transporter-3 (ZnT-3) to synaptic vesicle membranes within mossy fiber boutons in the hippocampus of mouse and
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) monkey, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 12676–12681 CrossRef CAS.
monkey, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 12676–12681 CrossRef CAS. - I. Sekler, A. Moran, M. Hershfinkel, A. Dori, A. Margulis and N. Birenzweig, et al., Distribution of the zinc transporter ZnT-1 in comparison with chelatable zinc in the mouse brain, J. Comp. Neurol., 2002, 447, 201–209 CrossRef CAS.
- M. J. Pushie, I. J. Pickering, G. R. Martin, S. Tsutsui, F. R. Jirik and G. N. George, Prion protein expression level alters regional copper, iron and zinc content in the mouse brain, Metallomics, 2011, 3, 206–214 RSC.
- G. Danscher, Histochemical demonstration of heavy metals, Histochemistry, 1981, 71, 1–16 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8mt00230d |
| ‡ Current affiliation: Central Analytical Research Facility, Institute of Future Environments, Queensland University of Technology, Brisbane, Queensland 4000, Australia. |
| This journal is © The Royal Society of Chemistry 2019 |