Evaluation of the bitter components of bamboo shoots using a metabolomics approach†
Quan
Gao‡
 a,
Hao
Jiang
b,
Feng
Tang
c,
Hai-qun
Cao
a,
Xiang-wei
Wu
d,
Fei-fei
Qi
e,
Jia
Sun‡
*cf and
Jun
Yang‡
*f
a,
Hao
Jiang
b,
Feng
Tang
c,
Hai-qun
Cao
a,
Xiang-wei
Wu
d,
Fei-fei
Qi
e,
Jia
Sun‡
*cf and
Jun
Yang‡
*f
aAnhui Key Laboratory of Agricultural Products, School of Plant Protection, Anhui Agricultural University, Hefei 230036, China
bState Key Laboratory of Tea Plant Biology and Utilization, Anhui Agricultural University, Hefei 230036, China
cState Forestry Administration Key Open Laboratory, International Centre for Bamboo and Rattan, Beijing 100102, China
dAnhui Key Laboratory of Agricultural Products, School of Resource and Environment, Anhui Agricultural University, Hefei 230036, China
eQingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences, Qingdao 266101, China
fDepartment of Entomology and Nematology, UC Davis Comprehensive Cancer Center, University of California, Davis, CA 95616, USA. E-mail: sunjia@icbr.ac.cn; junyang@ucdavis.edu; Fax: +1 530 752 1443; Tel: +1 530 752 5109
First published on 26th November 2018
Abstract
Bamboo shoots, most of which are bitter in taste, are a traditional vegetable eaten in Asia. However, our understanding of the components responsible for this bitterness is limited. In this study, we used metabolomic and phytochemical analyses to scientifically study bamboo shoot bitterness. Based on oral evaluation, 16 common bamboo shoots were categorized into four bitterness groups. The most bitter species was Pleioblastus amarus. Thus, we isolated and performed a detailed spectroscopic analysis of 14 main compounds from this species. The correlation between the concentration of these components and bitterness in four representative bamboo shoot species was then analyzed by UPLC-MRM-MS and PLS-DA. It appears that bamboo shoot bitterness is largely due to L-phenylalanine, uridine, L-omithine, L-tryptophan, and adenine, with L-phenylalanine being the greatest contributor. In addition to identifying the primary component involved in bamboo shoot bitterness, this study also outlines a novel method for evaluating the bitterness of natural foods.
Introduction
Bamboo shoots have been consumed as a traditional delicacy in Asia for more than 2500 years.1 In China, there are more than 100 different species of edible bamboo shoots with different tastes.2,3 However, while most fresh bamboo shoots taste bitter, the level of bitterness varies. For example, bamboo shoots from Pleioblastus amarus have a very bitter taste, while those from Dendrocalamus brandisii (Munro) Kurz. have a barely detectable level of bitterness.4 Although the level of bitterness for some bamboo shoot species is commonly known, many have not been scientifically evaluated.Notably, according to traditional Chinese literature,5,6 bitter foods, such as bamboo shoots, are beneficial for human health.7,8 This largely depends on the level of specific bioactive compounds in these foods. In a previous study, we identified the main nutritional components of Phyllostachys pubescens bamboo shoots, which included specific carbohydrates, amino acids, and nucleotides.9 However, the bioactive compounds in other species are largely unknown. Furthermore, no relevant studies have been conducted to investigate the association between the bioactive chemical constituents of bamboo shoots and their level of bitterness.
In the current study, we evaluated the main molecules that contribute to the level of bitterness in four representative species of bamboo shoots. These species were chosen based on their level of bitterness perceived during oral examination. Furthermore, the major compounds were isolated from the most bitter bamboo shoots of this subset (P. amarus), and their content was then determined in the three other representative species. A multivariate data analysis was conducted to evaluate the correlation between the concentrations of each component and the level of bitterness perceived in the four representative species. The bitterness of these individual compounds was also confirmed via oral taste experiments. To our knowledge, this study is the first to evaluate the bitterness of a natural food using both phytochemical and metabolomic techniques, and it also provides a new method for evaluating the bitter taste and other tastes or nutritional components of natural foods.
Materials and methods
Plant material
Fresh bamboo shoots for 16 species were cultivated and collected from five provinces in China (Table 1). These voucher specimens (designated no. 00-S2016-001 to no. 00-S2016-016) were deposited in the State Forestry Administration Key Open Laboratory, International Centre for Bamboo and Rattan, Beijing 100102, China.| No. | Species | Origin | No. | Species | Origin |
|---|---|---|---|---|---|
| 1 | Phyllostachys sp. | Sichuan, China | 9 | Leleba oldhami | Zhejiang, China |
| 2 | Dendrocalamus latiflorus | Guangdong, China | 10 | Phyllostachys makinoi | Zhejiang, China |
| 3 | Fargesia Franch | Sichuan, China | 11 | Phyllostachys dulcis | Zhejiang, China |
| 4 | Phyllostachys praecox | Zhejiang, China | 12 | Phyllostachys viridis | Shanxi, China |
| 5 | Phyllostachys incarnata | Zhejiang, China | 13 | Pleioblastus amarus | Sichuan, China |
| 6 | Bambusa rutila | Shanxi, China | 14 | Chimonobambusa quadrangularis | Chongqing, China |
| 7 | Bambusa emeiensis | Sichuan, China | 15 | Phyllostachys nigra | Zhejiang, China |
| 8 | Phyllostachys nuda | Zhejiang, China | 16 | Dendrocalamus brandisii | Zhejiang, China |
Bamboo shoot bitterness grading
Sixty healthy volunteers (30 men and 30 women) were asked to taste the sixteen different bamboo shoots. The average age of the study participants was 35, and all of the volunteers had a normal sense of taste (no rhinitis or pharyngitis). The evaluation was based on the International Bitterness Unit (IBU) system, and bitterness scores were assigned as follows: very bitter (+++): 30 to 40, bitter (++): 20 to 30, slightly bitter (+): 10 to 20, and not bitter (−): 0 to 10. Based on the bitterness grade scores, the four most representative bamboo shoots were determined by statistical analysis.General procedures, chemicals, and reagents
Preparative high-performance liquid chromatography (PHPLC) was performed using a Shimadzu LC-6AD instrument with an SPD-20A detector (Shimadzu, Kyoto, Japan) and a YMC-Pack ODS-A column (250 × 20 mm, 5 μm, YMC, Kyoto, Japan). HPLC-PDA analysis was performed using a Waters 2695 system and a PDA detector 2996 (Waters, Milford, USA) with a YMC-Pack ODS-AQ C18 column (250 × 4.6 mm, 5 μm, YMC, Kyoto, Japan), and the PDA detector was set to monitor in the 210 to 400 nm range, and used in conjunction with the following parameters: flow rate: 1 mL min−1, column temperature: 25 °C, and injection volume: 10 μL. Ultraviolet (UV) spectra were obtained using a Waters 2695 HPLC with a PDA detector (Waters, Milford, USA). One- and two-dimensional nuclear magnetic resonance (NMR) spectra were acquired with Bruker 500 spectrometers (Bruker, Zurich, Switzerland) using dimethyl sulfoxide (DMSO)-d6 or deuterium oxide (D2O) as solvents and tetramethylsilane (TMS) as an internal standard to normalize the shift of NMR peaks to 0 ppm for TMS. The system was operated at 500 MHz for 1H and 125 MHz for 13C. Chemical shifts are expressed in δ (ppm) and the coupling constants are reported in hertz. High-resolution electrospray ionization mass spectroscopy (HRESIMS) spectra were obtained using an Agilent 6540 high-resolution time-of-flight (Q-TOF) mass spectrometer (Agilent, Santa Clara, USA).Column chromatography was performed with macroporous resin (Diaion HP-20, Mitsubishi Chemical Corp., Tokyo, Japan), RP-18 (50 μm, YMC, Kyoto, Japan), and Sephadex LH-20 (Pharmacia Fine Chemicals, Uppsala, Sweden) columns. All reagents were purchased from Beijing Chemical Works (Beijing, China) unless otherwise specified. HPLC-grade methanol (MeOH) and ethanol (EtOH) were purchased from Fisher Scientific (Pittsburgh, USA).
Extraction, isolation, purification, and identification of compounds from P. amarus
Fresh P. amarus bamboo shoots (10 kg) were minced into pulp and filtered through a 300-mesh strainer. The filtrate was squeezed out, and the residue was extracted three times with 60% aqueous EtOH at room temperature and under dark conditions (performed over 3 days using 10 L for each extraction). The filtrates were combined and concentrated under reduced pressure to remove the organic solvent. The concentrated aqueous fraction (1.0 kg) was then separated on a macroporous resin column (100 × 10 cm) using a gradient of water–EtOH at five ratios (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, 85
0, 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15, 70
15, 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, 50
30, 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, and 5
50, and 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 95). The 15% EtOH fraction (15.1 g) was further purified on an ODS-A column followed by preparative HPLC using MeOH–water (30
95). The 15% EtOH fraction (15.1 g) was further purified on an ODS-A column followed by preparative HPLC using MeOH–water (30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70) as the elution buffer. This purification yielded compounds 3 (78.5 mg), 4 (2.3 mg), 10 (253.7 mg), 11 (518.3 mg), 12 (6.8 mg), and 13 (5.0 mg). The 30% EtOH fraction (8.5 g) was also separated on an ODS-A preparative HPLC column using MeOH–water (36
70) as the elution buffer. This purification yielded compounds 3 (78.5 mg), 4 (2.3 mg), 10 (253.7 mg), 11 (518.3 mg), 12 (6.8 mg), and 13 (5.0 mg). The 30% EtOH fraction (8.5 g) was also separated on an ODS-A preparative HPLC column using MeOH–water (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 64) as the elution buffer and produced purified compounds 1 (626.5 mg), 2 (7.5 mg), 5 (153.2 mg), 6 (216.2 mg), 7 (5.9 mg), 8 (19.0 mg), and 9 (4.3 mg). Finally, the 50% EtOH fraction (3.4 g) was similarly separated but used a 45
64) as the elution buffer and produced purified compounds 1 (626.5 mg), 2 (7.5 mg), 5 (153.2 mg), 6 (216.2 mg), 7 (5.9 mg), 8 (19.0 mg), and 9 (4.3 mg). Finally, the 50% EtOH fraction (3.4 g) was similarly separated but used a 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 55 ratio of MeOH–water as the elution buffer to yield compound 14 (11.2 mg). The structures of compounds 1–14 were determined by detailed analysis (HRESIMS and NMR) as described below.
55 ratio of MeOH–water as the elution buffer to yield compound 14 (11.2 mg). The structures of compounds 1–14 were determined by detailed analysis (HRESIMS and NMR) as described below.
Compound 1: White powder; HRESIMS (C9H12NO2) m/z 166.0879 [M + H]+ (calculated 166.0868); 1H NMR (D2O, 500 MHz) δ: 7.55 (H-3, 5), 7.52 (H-4), 6.88 (H-2, 6), 3.26 and 3.15 (H-7), and 4.20 (H-8); 13C NMR (D2O, 125 MHz) δ: 174.0 (C-9), 137.19 (C-1), 129.7 (C-3, 5), 129.5 (C-2, 6), 128.0 (C-4), 56.2 (C-8), and 36.7 (C-7).
Compound 2: White powder; HRESIMS (C10H13N2O5) m/z 241.0834 [M − H]− (calculated 241.0824); 1H NMR (D2O, 500 MHz) δ: 7.71 (H-6), 6.18 (H-1′), 4.26 (H-3′), 3.77 (H-4′), 3.59 (H-5′), 2.08 (H-2′), and 1.78 (CH3); 13C NMR (D2O, 125 MHz) δ: 167.2 (C-4), 152.4 (C-2), 138.3 (C-5), 112.3 (C-6), 87.3 (C-4′), 85.8 (C-1′), 71.3 (C-3′), 61.9 (C-5′), 39.4 (C-2′), and 12.3 (CH3).
Compound 3: White powder; HRESIMS (C5H6N5) m/z 136.0613 [M + H]+ (calculated 136.0623); 1H NMR (DMS0-d6, 500 MHz) δ: 12.75 (NH), 8.28 (H-8), 8.15 (H-2), and 7.12 (NH2); 13C NMR (DMS0-d6, 125 MHz) δ: 156.4 (C-6), 153.4 (C-2), 151.6 (C-4), 140.3 (C-8), and 119.1 (C-5).
Compound 4: White powder; HRESIMS (C5H5N2O2) m/z 125.0357 [M − H]− (calculated 125.0351); 1H NMR (D2O, 500 MHz) δ: 7.28 (H-6) and 1.73 (CH3); 13C NMR (D2O, 125 MHz) δ: 168.3 (C-4), 153.7 (C-2), 139.7 (C-5), 110.9 (C-6), and 11.9 (CH3).
Compound 5: White powder; HRESIMS (C10H14N5O5) m/z 284.0986 [M + H]+ (calculated 284.0995); 1H NMR (DMS0-d6, 500 MHz) δ: 10.72 (NH), 7.95 (H-8), 6.53 (NH2), 5.72 (H-1′), 4.44 (H-2′), 4.10 (H-3′), 3.91 (H-4′), and 3.62 and 3.55 (H-5′); 13C NMR (DMS0-d6, 125 MHz) δ: 157.1 (C-6), 153.8 (C-2), 151.5 (C-4), 136.1 (C-8), 116.7 (C-5), 86.7 (C-1′), 85.5 (C-4′), 73.8 (C-2′), 70.5 (C-3′), and 61.7 (C-5′).
Compound 6: White powder; HRESIMS (C11H13N2O2) m/z 205.0969 [M + H]+ (calculated 205.0977); 1H NMR (DMS0-d6, 500 MHz) δ: 11.12 (NH), 7.58 (H-4), 7.36 (H-7), 7.26 (H-2), 7.06 (H-5), 6.97 (H-6), 3.53 (H-11), and 3.33 and 3.05 (H-10); 13C NMR (DMS0-d6, 125 MHz) δ: 174.6 (C-12), 137.3 (C-9), 127.7 (C-8), 125.8 (C-2), 122.9 (C-4), 120.4 (C-5), 119.5 (C-6), 112.7 (C-7), 108.7 (C-3), 56.1 (C-11), and 27.2 (C-10).
Compound 7: White powder; HRESIMS (C10H14N5O3) m/z 252.1090 [M + H]+ (calculated 252.1097); 1H NMR (DMS0-d6, 500 MHz) δ: 8.33(H-2), 8.14 (H-8), 7.30 (NH2), 6.34 (H-1′), 4.42 (H-3′), 3.87 (H-4′), 3.68 (H-5′), and 2.70 (H-2′); 13C NMR (DMS0-d6, 125 MHz) δ: 156.1 (C-6), 153.0 (C-2), 149.1 (C-4), 141.0 (C-8), 119.7 (C-5), 88.2 (C-4′), 85.6 (C-1′), 72.0 (C-3′), 62.4 (C-5′), and 39.9 (C-2′).
Compound 8: White powder; HRESIMS (C9H14N3O5) m/z 244.0942 [M + H]+ (calculated 244.0933); 1H NMR (D2O, 500 MHz) δ: 7.85 (H-6), 6.04 (H-5), 5.90 (H-1′), 4.32 (H-2′), 4.22 (H-3′), 4.15 (H-4′), and 3.95 and 3.84 (H-5′); 13C NMR (D2O, 125 MHz) δ: 165.6 (C-4), 155.5 (C-2), 141.4 (C-6), 93.8 (C-5), 89.2 (C-1′), 84.0 (C-4′), 73.9 (C-2′), 69.4 (C-3′), and 60.8 (C-5′).
Compound 9: White powder; HRESIMS (C9H14N3O4) m/z 228.0990 [M + H]+ (calculated 228.0984); 1H NMR (D2O, 500 MHz) δ: 7.79 (H-6), 6.16 (H-1′), 5.99 (H-5), 4.25 (H-3′), 3.73 (H-4′), 3.58 (H-5′), and 2.10 (H-2′); 13C NMR (D2O, 125 MHz) δ: 165.0 (C-4), 154.8 (C-2), 140.6 (C-6), 93.9 (C-5), 87.4 (C-4′), 83.8 (C-1′), 70.1 (C-3′), 62.1 (C-5′), and 39.8 (C-2′).
Compound 10: White powder; HRESIMS (C9H13N2O6) m/z 245.0768 [M + H]+ (calculated 245.0774); 1H NMR (D2O, 500 MHz) δ: 7.89 (H-6), 5.92 (H-1′), 5.90 (H-6), 4.34 (H-2′), 4.24 (H-3′), 4.14 (H-4′), and 3.93 and 3.82 (H-5′); 13C NMR (D2O, 125 MHz) δ: 163.1 (C-4), 150.6 (C-2), 140.4 (C-6), 101.8 (C-5), 87.6 (C-1′), 84.3 (C-4′), 73.6 (C-2′), 69.6 (C-3′), and 60.7 (C-5′).
Compound 11: White powder; HRESIMS (C5H13N2O2) m/z 133.0985 [M + H]+ (calculated 133.0977); 1H NMR (D2O, 500 MHz) δ: 3.80 (H-2), 3.07 (H-5), 1.95 (H-4), and 1.84 and 1.75 (H-3); 13C NMR (D2O, 125 MHz) δ: 174.7 (C-1), 55.1 (C-2), 39.9 (C-5), 28.2 (C-3), and 23.6 (C-4).
Compound 12: White powder; HRESIMS (C10H14N5O4) m/z 268.1057 [M + H]+ (calculated 268.1046); 1H NMR (D2O, 500 MHz) δ: 7.87 (H-8), 6.07 (H-1′), 4.30 (H-3′), 3.78 (H-4′), 3.55 (H-5′), and 2.48 and 2.17 (H-2′); 13C NMR (D2O, 125 MHz) δ: 159.6 (C-6), 154.6 (C-2), 152.0 (C-4), 138.5 (C-8), 117.4 (C-5), 88.1 (C-4′), 84.8 (C-1′), 72.0 (C-3′), 62.5 (C-5′), and 39.6 (C-2′).
Compound 13: White powder; HRESIMS (C10H14N5O4) m/z 268.1051 [M + H]+ (calculated 268.1046); 1H NMR (DMS0-d6, 500 MHz) δ: 8.38 (H-2), 8.17 (H-8), 7.40 (NH2), 5.91 (H-1′), 4.64 (H-2′), 4.17 (H-3′), 3.98 (H-4′), and 3.70 and 3.58 (H-2′); 13C NMR (DMS0-d6, 125 MHz) δ: 156.3 (C-6), 152.5 (C-2), 149.3 (C-4), 141.2 (C-8), 119.6 (C-5), 88.2 (C-1′), 86.2 (C-4′), 73.8 (C-2′), 70.9 (C-3′), and 62.0 (C-5′).
Compound 14: White powder; HRESIMS (C7H7O3) m/z 137.0247 [M − H]− (calculated 137.0239); 1H NMR (D2O, 500 MHz) δ: 7.84 (H-2, 6) and 6.86 (H-3, 5); 13C NMR (D2O, 125 MHz) δ: 171.0 (C-7), 161.5 (C-4), 132.9 (C-2, 6), 122.1 (C-1), and 116.2 (C-3, 5).
Extraction of the four representative bamboo shoots
An aliquot of each individual sample (200 mg) was precisely weighed and transferred to a 2 mL Eppendorf (EP) tube. After the addition of 1 mL of extraction solution containing 100 nmol L−1L-2-chlorophenylalanine (precooled to −20 °C, in acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH
MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water = 2
water = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (ratio expressed in volume), the samples were vortexed for 30 s, homogenized at 45 Hz for 4 min, and sonicated for 5 min in an ice-water bath. The homogenization and sonication cycle was repeated 3 times after which the samples were incubated at −20 °C for 1 h and centrifuged at 12
1) (ratio expressed in volume), the samples were vortexed for 30 s, homogenized at 45 Hz for 4 min, and sonicated for 5 min in an ice-water bath. The homogenization and sonication cycle was repeated 3 times after which the samples were incubated at −20 °C for 1 h and centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm and 4 °C for 15 min. A 100 μL aliquot of the clear supernatant was then transferred to a new EP tube, and the solvent was evaporated under a gentle nitrogen flow. The residue was reconstituted with 100 μL of 10% MeOH/water and then centrifuged at 12
000 rpm and 4 °C for 15 min. A 100 μL aliquot of the clear supernatant was then transferred to a new EP tube, and the solvent was evaporated under a gentle nitrogen flow. The residue was reconstituted with 100 μL of 10% MeOH/water and then centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm and 4 °C for 15 min. An 80 μL aliquot of the clear supernatant was transferred to an auto-sampler vial for analysis.
000 rpm and 4 °C for 15 min. An 80 μL aliquot of the clear supernatant was transferred to an auto-sampler vial for analysis.
Reference solution preparation
A mixed reference solution containing the 14 compounds previously isolated and identified from P. amarus bamboo shoots was used. Stock solutions were individually prepared by dissolving or diluting each reference solution to give a final concentration of 10 mmol L−1. A 100 μL aliquot of each of the stock solutions was transferred to a 10 mL flask to form a mixed working reference solution. A series of reference solutions for calibration were then prepared by serial dilution of this mixed standard solution. L-2-Chlorophenylalanine (100 nmol L−1) was used as an internal standard.UPLC-MRM-MS/MS analysis
Ultra-performance liquid chromatography (UPLC) separations were carried out using an Agilent 1290 Infinity II series UPLC System (Agilent Technologies, Santa Clara, CA) equipped with a Waters ACQUITY UPLC HSS T3 column (2.1 × 100 mm, 1.8 μm). Mobile phase A was 0.1% acetic acid in water, and mobile phase B was MeOH. The elution gradient was shown in the ESI† and was used in conjunction with the following parameters: flow rate: 300 μL min−1, column temperature: 35 °C, auto-sampler temperature: 10 °C, and injection volume: 1 μL. An Agilent 6460 triple quadrupole mass spectrometer (Agilent Technologies, Santa Clara, CA) equipped with an automatic jet stream electrospray ionization (AJS-ESI) interface was used for assay development. The typical ion source parameters were as follows: capillary voltage: +4000/−3500 V, nozzle voltage: +500/−500 V, gas temperature: 300 °C, gas flow: 5 L min−1, sheath gas temperature: 250 °C, sheath gas flow rate: 11 L min−1, and nebulizer pressure: 45 psi.The multiple reaction monitoring (MRM) parameters for each of the target analytes were optimized by injecting the standard solutions of the individual analytes directly into the API source of the mass spectrometer. Of the two MRM transitions per analyte, the Q1/Q3 pairs that showed the highest sensitivity and selectivity were used as the MRM transitions for quantitative monitoring.
Agilent MassHunter Work Station Software (B.08.00, Agilent Technologies) was utilized for MRM data acquisition and processing.
Calibration curves
Calibration solutions were subjected to UPLC-MRM-MS/MS analysis using the methods described above. A weighting of 1/X was applied for curve fitting as it provided the highest accuracy and correlation coefficients (R2). A minimum of 9 of the 15 concentration levels were included in the final calibration curves. Concentrations were excluded from the calibration plot if their signal-to-noise ratio (S/N) was close to or below 10 or if the accuracy of the calibration was not within 75–120%.10The calibration standard solutions were prepared via serial dilutions with a dilution factor of 2. The signal-to-noise ratios were used to determine the lower limits of detection (LLODs) and lower limits of quantitation (LLOQs). The LLODs and LLOQs were defined as the analyte concentrations that led to peaks with S/N values of 3 and 10, respectively, which is consistent with the US FDA guidelines for bioanalytical method validation.
Statistical analysis
A univariate data analysis was performed using Prism 7 software (GraphPad). Differences with a p < 0.05 were considered statistically significant. Each experiment consisted of eight replicates per set of condition. Hierarchical clustering analysis (HCA) and partial least squares discriminant analysis (PLS-DA) were conducted using MetaboAnalyst 3.11–15Sample preparation for oral examination
For oral evaluation, bitterness was divided into five levels, each of which was assigned a certain range of bitterness according to the literature;16,17 level 1: no bitterness or almost no bitterness (0.5 to 1.5), level 2: slightly bitter (1.5 to 2.5), level 3: bitter but acceptable (2.5 to 3.5), level 4: very bitter but still tolerable (3.5 to 4.5), and level 5: unbearable bitterness (4.5 to 5.5). After several preliminary tests, five different concentrations of berberine hydrochloride (0, 0.01, 0.05, 0.1, and 0.5 mg ml−1 in ultra-pure water) were selected to function as reference solutions 1 to 5, respectively.18 The individual compounds isolated from P. amarus were then formulated to concentrations of 0.05, 0.1, 0.5, 1, and 2 mg ml−1 in ultra-pure water and used for oral evaluation by the volunteer subjects.Evaluation of target compound bitterness
Sixty healthy volunteers (30 men and 30 women) were recruited for this analysis. The average age of the study participants was 35, and all volunteers had a normal sense of taste (no rhinitis or pharyngitis). To establish a reference point, each volunteer took 20 mL of each concentration of the reference solutions into their mouth and held it there for 15 s. During this time, the volunteers were asked to ensure that the solution covered the whole surface of the oral cavity so that the tongue root and tongue were completely exposed to the solution. The volunteers were informed of the bitterness classification of the reference solution and specific bitterness value. After a 15 s tasting period, the volunteers were asked to spit out the solution and rinse their mouths 5 times, until no bitter taste remained. After 15 min, the volunteers tasted another reference solution.The same method outlined above was used for the taste evaluation of the target compound samples. The volunteers then determined the specific bitterness values of each test sample based on the values they experienced with the reference solutions. They recorded these values using a pre-designed bitterness rating evaluation table. After recording the values, the volunteers were asked to spit out the solution and rinse their mouths 5 times, until no bitter taste remained. After 15 min, the volunteers tasted another test solution.
Results and discussion
Based on the experimental bitterness grades, the bamboo shoot species used to represent the very bitter (+++), bitter (++), slightly bitter (+), and not bitter (−) levels are Pleioblastus amarus, Bambusa rutila, Dendrocalamus latiflorus, and Dendrocalamus brandisii, respectively.Component isolation and identification for P. amarus bamboo shoots
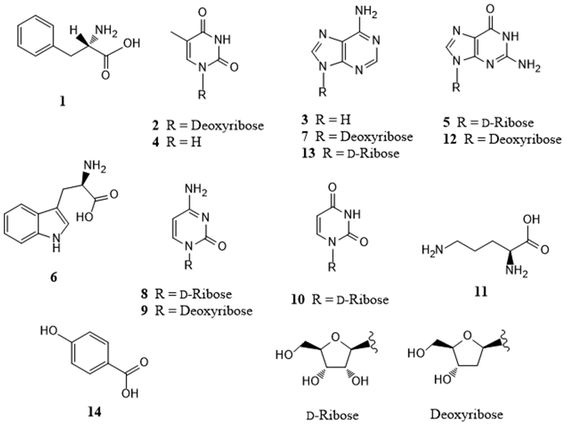
Repeated column chromatography of the extract from fresh P. amarus bamboo shoots resulted in the isolation of 14 compounds. The compounds were identified as L-phenylalanine (1),19 thymidine (2),20 adenine (3),19 thymine (4),20 guanosine (5),19L-tryptophan (6),21 2′-deoxyadenosine (7),19 cytidine (8),19 2′-deoxycytidine (9),22 uridine (10),19L-ornithine (11),19 2′-deoxyguanosine (12),23 adenosine (13),24 and 4-hydroxybenzoic acid (14)19 (Fig. 1) by comparing their spectroscopic and physical data with those previously reported in the literature. Notably, this is the first time these 14 compounds have been isolated from P. amarus bamboo shoots.Separation of the target analytes
Using the optimal gradient elution conditions, the extracted ion chromatographs from a reference solution (2A) and a spiked sample (2B) of all showed excellent peak shapes and separation. Moreover, the retention time and peak shapes for all of the analytes showed good correlation between that of the reference solution and that observed for the real samples. The results of our UPLC-MS analysis are included in the ESI.†Validation of the analysis
The optimized MRM parameters for the target compounds are listed in the ESI,† while the detailed calibration curves for the individual analytes are shown in the ESI.† The ESI† lists the LLODs and LLOQs, which range from 0.1–12.2 nmol L−1 and 0.5–97.7 nmol L−1, respectively, for the 14 target analytes. The linear regression correlation coefficients (R2) were greater than 0.994 for all of the analytes, indicating a good quantitative relationship between the MS responses and the analyte concentrations. Notably, this level is sufficient for targeted metabolomics analysis. The analytical recoveries and relative standard deviations of the quality control samples using five technical replicates are shown in Table 2. The recoveries ranged from 94.9% to 114.8% for all of the analytes, and the relative standard deviation for each analyte was less than 6.4% (n = 5).| Analyte | Concentration | Recovery | RSD |
|---|---|---|---|
| QC = quality control; RSD = relative standard deviation. | |||
| L-Ornithine | 312.5 nmol L−1 | 108.8% | 2.9% |
| Cytidine | 312.5 nmol L−1 | 104.4% | 2.2% |
| Deoxycytidine | 312.5 nmol L−1 | 114.8% | 2.6% |
| Adenine | 312.5 nmol L−1 | 107.0% | 4.1% |
| Uridine | 1562.5 nmol L−1 | 111.5% | 3.1% |
| Thymine | 1562.5 nmol L−1 | 111.1% | 3.2% |
| L-Phenylalanine | 62.5 nmol L−1 | 94.9% | 6.4% |
| Guanosine | 312.5 nmol L−1 | 103.9% | 1.8% |
| Deoxyguanosine | 312.5 nmol L−1 | 104.3% | 1.8% |
| Adenosine | 312.5 nmol L−1 | 104.3% | 1.7% |
| Deoxyadenosine | 62.5 nmol L−1 | 102.3% | 1.8% |
| Thymidine | 1562.5 nmol L−1 | 104.2% | 2.8% |
| L-Tryptophan | 312.5 nmol L−1 | 105.2% | 1.9% |
| 4-Hydroxybenzoic acid | 1562.5 nmol L−1 | 100.2% | 2.1% |
Analyte quantification
The quantification results are shown in the ESI.† For a holistic view of the main components of the extracts, a heatmap based on the 14 metabolites from the bamboo shoot extract was prepared. From this, we observed a group of metabolites, which included L-phenylalanine, L-ornithine, adenine, and guanosine, that tended to increase in concentration as the bitterness level increased (Fig. 2). Additionally, the distances between these metabolites in the Y direction indicates that this group of metabolites contributes in a similar degree to the classification of these bamboo shoot extracts.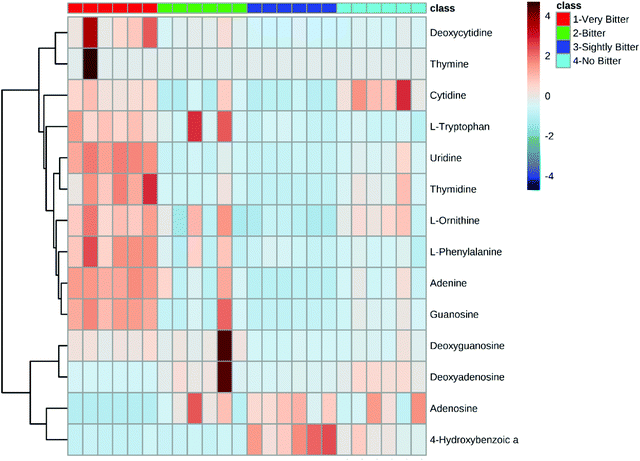 | ||
| Fig. 2 Heatmap highlighting the bitterness scoring of the 14 components from the bamboo shoot extracts. Bitterness scoring was based on four different levels of bitterness. | ||
Although our HCA (Fig. 2) indicates that bitter taste is likely based on the chemical constituents of the bamboo shoot, this analysis did not provide clear evidence concerning which metabolites were the most important contributors to this trait. Thus, we used PLS-DA to identify which metabolites are the major factors controlling bitterness (Fig. 3). PLS-DA is a promising, supervised, machine learning algorithm for identifying classification markers that has been widely used in various metabolomics and chemometrics studies.25,26 As shown in Fig. 3, the extracts from the four representative bamboo shoot species could be easily differentiated on the 3D score plot. The VIP scores calculated in this analysis indicate that bamboo shoot bitterness largely comes from L-phenylalanine, uridine, L-omithine, L-tryptophan, and adenine, with the greatest contributor being L-phenylalanine.
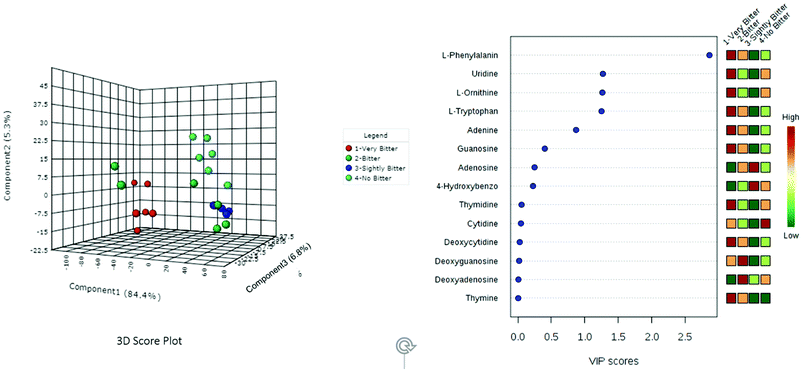 | ||
| Fig. 3 Partial least squares discriminant analysis (PLS-DA) of the metabolites for the four representative bamboo shoot species for each level of bitterness. | ||
It is important to note that the greatest advantage of this type of metabolomics assessment is its use of multivariate analysis, which scientifically characterizes and interprets the unique characteristics of an organism. It is also suitable for studying food–human interactions and assessing the effects of food at the molecular level. When metabolomics are combined with traditional methods for determining phytochemicals, the compounds can be separated, allowing only the main components to be selected and further quantitatively analyzed.
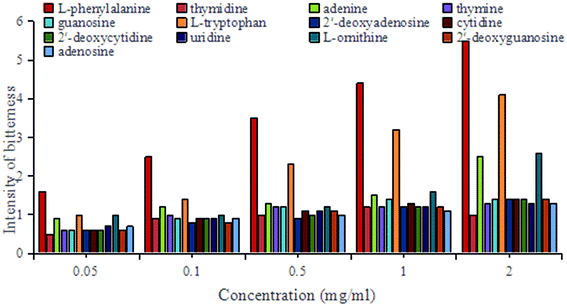
Oral taste evaluation of the individual analyte solutions
Because 4-hydroxybenzoic acid is prohibited for oral ingestion,27 only 13 of the 14 target compounds could be tested orally. The oral taste evaluation experiments show that L-phenylalanine at a concentration of 2 mg ml−1 was the most bitter solution. Its bitterness reached an equivalent level as berberine hydrochloride at a concentration of 0.5 mg ml−1. Almost all of the volunteers were unable to endure this bitter taste. The next compound L-tryptophan at a concentration of 2 mg ml−1 was perceived by the volunteers as exhibiting the same bitterness as berberine hydrochloride at a concentration of 0.2 mg ml−1. The bitterness of this compound was second only to L-phenylalanine. Notably, these findings are consistent with the literature,28,29 in which the bitterness of many amino acids has been established.30,31 In the present study, both adenine and L-ornithine also exhibited some bitterness, but at much lower levels compared with L-phenylalanine and L-tryptophan (Fig. 4).To date, few studies have analyzed the bitterness of bamboo shoots. Some of these investigations suggest that bamboo shoot bitterness is caused by raw cyanogenic glycosides formed when the bamboo shoots are unearthed.32 Other studies suggest that these cyanogenic glycosides are actually the main substances causing the spicy taste of bamboo shoots.33,34 However, as cyanogenic glycosides are highly toxic even in small doses,35 these suggestions are unlikely to be the underlying cause of bitterness. A more recent study has indicated that amino acids may affect the bitter and astringent taste of bamboo shoots depending on their content but did not provide any qualitative or quantitative sensory analysis to justify this conclusion.36 Amino acids, as the main contributors to taste, are often found at relatively high levels in bitter foods,37 especially L-phenylalanine38,39 and L-tryptophan.40,41 Thus, our findings are largely consistent with previous literature concerning bitterness in other foods.
Conclusions
In the present study, we used metabolomic and phytochemical analyses to comprehensively and scientifically study the bitterness of bamboo shoots. Notably, L-phenylalanine was perceived as the most bitter component, and the oral taste evaluation results corresponded with the quantitative LC-MS results. Thus, the content of L-phenylalanine is likely the most important factor involved in the bitterness of bamboo shoots, at least for the four species investigated in the present study. These findings contradict the traditional belief that the main component in the bitter taste of bamboo shoots is hydrocyanide. To our knowledge, this study is the first to evaluate the bitterness of bamboo shoots using these particular analysis techniques. Altogether, this study not only highlights the component involved in bamboo shoot bitterness, but it also provides a new method for evaluating bitterness, and possibly other tastes or nutritional components, of natural foods.Funding
This work was supported by the Basic Science Research Fund Program of the International Centre for Bamboo and Rattan (ICBR) (1632016002) and was sponsored by the China Scholarship Council.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
We are grateful to the staff of the analytical group at the State Forestry Administration Key Open Laboratory in the International Centre for Bamboo and Rattan and the UC Davis Comprehensive Cancer Center.References
- N. Chongtham, M. S. Bisht and S. Haorongbam, Nutritional properties of bamboo shoots: Potential and prospects for utilization as a health food, Compr. Rev. Food Sci. F, 2011, 10(3), 153–169 CrossRef CAS.
- Z. H. Jiang, Conservation of bamboo resources and genetic improvement, in Bamboo and Rattan in the World Beijing, China Forestry Publishing House, China, 2007 Search PubMed.
- C. Jin, Y. Y. Wang, W. J. Zheng, Y. H. Zhang, G. L. Chen and M. S. Lin, Literature analysis and development strategy for bamboo shoot research in China, J. Zhejiang For. Coll., 2000, 17(1), 75–79 Search PubMed.
- D. Ohrnberger, The Bamboos of the World: Annotated Nomenclature and Literature of the Species and the Higher and Lower Taxa, Sci. Hortic., 2001, 87(1), 153–153 Search PubMed.
- M. H. Liu, X. D. Zhou, Q. Li, F. Liu, J. Wang and Y. Q. Zhong, Containment effects of bitter taste naïve ingredient from traditional Chinese medicine on airway inflammation process via bitter taste receptor pathway, Chin. J. Immunol., 2017, 33(9), 1331–1335 Search PubMed.
- Y. X. Hu, W. Cai, H. H. Zhang, C. H. Yu, W. Y. Yu, X. Y. Jin and H. Z. Ying, Establishment and application of screening model for anti-asthmatic natural active ingredients based on bitter taste receptors in lung tissue, Chin. Tradit. Herb. Drugs, 2016, 47(5), 775–780 CAS.
- D. A. Deshpande, W. C. Wang, E. L. Mcllmoyle, K. S. Robinett, R. M. Schillinger, S. S. An, J. S. Sham and S. B. Liggett, Bitter taste receptors on airway smooth muscle bronchodilate by localized calcium signaling and reverse obstruction, Nat. Med., 2010, 16(11), 1299–1304 CrossRef CAS PubMed.
- D. P. Jiang, V. P. Kolosov, J. M. Perelman and X. D. Zhou, IL-13 regulates mucus secretion via FOXA2 in vitro, Chin. J. Immunol., 2011, 27(2), 99–102 Search PubMed.
- J. Sun, Z. Q. Ding, Q. Gao, H. Xun, F. Tang and E. D. Xia, Major chemical constituents of bamboo shoots (Phyllostachys pubescens): Qualitative and quantitative research, J. Agric. Food Chem., 2016, 64(12), 2498–2505 CrossRef CAS PubMed.
- J. Yang, K. Schmelzer, K. Georgi and D. B. Hammock, Quantitative profiling method for oxylipin metabolome by liquid chromatography electrospray ionization tandem mass spectrometry, Anal. Chem., 2015, 81(19), 8085–8093 CrossRef PubMed.
- J. Trygg and S. Wold, Orthogonal projections to latent structures (O-PLS), J. Chemom., 2010, 16(3), 119–128 CrossRef.
- J. Yang, X. Zhao, X. Liu, C. Wang, P. Gao, J. Wang, L. Li, J. Gu, S. Yang and G. Xu, High-performance liquid chromatography-mass spectrometry for metabonomics: Potential biomarkers for acute deterioration of liver function in chronic hepatitis B, J. Proteome Res., 2006, 5(3), 554–561 CrossRef CAS PubMed.
- J. Yang, X. Zhao, X. Lu, X. Lin and G. Xu, A data preprocessing strategy for metabolomics to reduce the mask effect in data analysis, Front. Mol. Biosci., 2015, 2(2), 4–12 Search PubMed.
- J. Xia and D. S. Wishart, Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis, Curr. Protoc. Bioinf., 2016, 55, 14 Search PubMed.
- J. Xia, I. V. Sinelnikov, B. Han and D. S. Wishart, MetaboAnalyst 3.0-making metabolomics more meaningful, Nucleic Acids Res., 2015, 43(W1), W251–W257 CrossRef CAS PubMed.
- L. Li, V. Naini and S. U. Ahmed, Utilization of a modified special-cubic design and an electronic tongue for bitterness masking formulation optimization, J. Pharm. Sci., 2007, 96(10), 2723–2734 CrossRef CAS PubMed.
- C. Apetrei, I. M. Apetrei, S. Villanueva, J. A. De Saja, F. Gutierrez-Rosales and M. L. Rodriguez-Mendez, Combination of an e-nose, an e-tongue and an e-eye for the characterisation of olive oils with different degree of bitterness, Anal. Chim. Acta, 2010, 663(1), 91–97 CrossRef CAS PubMed.
- X. L. Li, X. F. Zhang, R. X. Liu, H. L. Li, J. S. Qiu and Z. D. Wu, Study on quantitation of bitterness intensity and relationship between bitterness intensity and concentration of bitter drug, Chin. Med. Mater. Med., 2013, 15(4), 667–671 Search PubMed.
- E. Pretsch, P. Bühlmann and C. Affolter, Structure Determination of Organic Compounds: Tables of Spectral Data, Springer-Verlag, Berlin Heidelberg, Germany, 2000, pp. 102–103, 150–151, 154–155, 180–181, 234–235, and 237–239 Search PubMed.
- X. F. Shi, X. L. Tang, G. Q. Li, C. Y. Wang and H. S. Guan, Study on chemical constitutes of gorgonian Muriceides collaris from the South China Sea, Chin. Tradit. Herb. Drugs, 2009, 28(2), 18–21 CAS.
- H. M. Shi, J. Wen and P. F. Tu, Chemical constitutes of Abrus cantoniensis, Chin. Tradit. Herb. Drugs, 2006, 37(11), 1610–1613 CAS.
- J. Brzezinska, Z. Gdaniec, L. Popenda and W. T. Markiewicz, Polyaminooligonucleotide: NMR structure of duplex DNA containing a nucleoside with spermine residue, N-[4,9,13- triazatridecan-1-yl]-2′-deoxycytidine, Biochim. Biophys. Acta, 2014, 1840(3), 1163–1170 CrossRef CAS PubMed.
- R. P. Zhuo, H. Z. Fu, L. H. Zhang and W. H. Lin, Nucleosides from Anthopleura stell, Chin. Tradit. Herb. Drugs, 2011, 32(4), 289–291 Search PubMed.
- J. Kinjo, K. Masumoto, M. Inoue, T. Takeshita and T. Nohara, A new sapogenol and other constituents in Abri Semen, the seeds of Abrus precaturius L. I, Chem. Pharm. Bull., 1991, 39(1), 116–119 CrossRef CAS.
- K. L. Yuan, H. W. Kong, Y. F. Guan, J. Yang and G. W. Xu, A GC-based metabonomics investigation of type 2 diabetes by organic acids metabolic profile, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2007, 850(1), 236–240 CrossRef CAS PubMed.
- C. Wang, H. W. Kong, Y. F. Guan, J. Yang, J. R. Gu, S. L. Yang and G. W. Xu, Plasma phospholipid metabolic profiling and biomarkers of type 2 diabetes mellitus based on high-performance liquid chromatography/electrospray mass spectrometry and multivariate statistical analysis, Anal. Chem., 2005, 77(13), 4108–4116 CrossRef CAS PubMed.
- X. L. Jia, J. H. Ye, Q. Zhang, L. Li, Y. L. Hu, M. Z. Zheng, Y. C. Hong, F. Q. Wang and C. Z. Wu, Soil toxicity and microbial community structure of Wuyi rock tea plantation, Allelopathy J., 2017, 41(1), 113–126 CrossRef.
- H. D. Belitz and H. Wieser, Bitter compounds: Occurrence and structure-activity relationships, Food Rev. Int., 1985, 1(2), 271–354 CrossRef CAS.
- S. Kohl, M. Behrens, A. Dunkel, T. Hofmann and W. Meyerhof, Amino acids and peptides activate at least five members of the human bitter taste receptor family, J. Agric. Food Chem., 2013, 61(1), 53–60 CrossRef CAS PubMed.
- H. Wieser and H. D. Belitz, Relations between structure and bitter taste of amino acids and peptides. I. Amino acids and related compounds, Z. Lebensm.-Unters. Forsch., 1975, 159(2), 65–72 CrossRef CAS.
- S. Toelstede and T. Hofmann, Sensomics mapping and identification of the key bitter metabolites in Gouda cheese, J. Agric. Food Chem., 2008, 56(8), 2795–2804 CrossRef CAS PubMed.
- D. Choudhury, J. K. Sahu and G. D. Sharma, Biochemistry of bitterness in bamboo shoots, Assam Univ. J. Sci. Technol., 2010, 6(2), 105–111 Search PubMed.
- S. G. Fu, Y. Yoon and R. Bazemore, Aroma-active components in fermented bamboo shoots, J. Agric. Food Chem., 2002, 50(3), 549–554 CrossRef CAS PubMed.
- K. Sarangthem and T. N. Singh, Microbial bioconversion of metabolites from fermented succulent bamboo shoots into phytosterols, Curr. Sci., 2003, 84(12), 1544–1547 CAS.
- J. Vetter, HCN-containing plant materials: The cyanogen glycosides, Gyógyszerészet, 2000, 44(9), 533–539 CAS.
- X. L. Li, X. C. Ding, S. S. Zhang, Z. Y. Zhang, H. J. Cai and Y. M. Zheng, The distributions of bitter and astringent taste compounds in the bamboo shoot of Denrocalamus latiflorus under different light intensities, J. Nanjing For. Univ., 2015, 39(3), 161–166 Search PubMed.
- G. Chen, J. Li, Z. Sun, S. Zhang, G. Li, C. Song, Y. Suo and J. You, Rapid and sensitive ultrasonic-assisted derivatisation microextraction (UDME) technique for bitter taste-free amino acids (FAA) study by HPLC-FLD, Food Chem., 2014, 143(1), 97–105 CrossRef CAS PubMed.
- S. S. Schiffman, K. Sennewald and J. Gagnon, Comparison of taste qualities and thresholds of D- and L-amino acids, Physiol. Behav., 1981, 27(1), 51–59 CrossRef CAS PubMed.
- M. Kawai, Y. Sekine-Hayakawa, A. Okiyama and Y. Ninomiya, Gustatory sensation of (L)- and (D)-amino acids in humans, Amino Acids, 2012, 43(6), 2349–2358 CrossRef CAS PubMed.
- Y. Miyanaga, A. Tanigake, T. Nakamura, Y. Kobayashi, H. Ikezaki, A. Taniguchi, K. Matsuyama and T. Uchida, Prediction of the bitterness of single, binary- and multiple-component amino acid solutions using a taste sensor, Int. J. Pharm., 2002, 248(1), 207–218 CrossRef CAS PubMed.
- N. Ishibashi, I. Ono, K. Kato, T. Shigenaga, I. Shinoda, H. Okai and S. Fukui, Studies on flavored peptides. Part III. Role of the hydrophobic amino acid residue in the bitterness of peptides, Agric. Biol. Chem., 1988, 52(1), 91–94 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8fo01820k |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2019 |