Modification of wheat bran particle size and tissue composition affects colonisation and metabolism by human faecal microbiota†
Kim
De Paepe
 a,
Joran
Verspreet‡
b,
Mohammad Naser
Rezaei
b,
Silvia Hidalgo
Martinez
c,
Filip
Meysman
c,
Davy
Van de Walle
d,
Koen
Dewettinck
d,
Christophe M.
Courtin
a,
Joran
Verspreet‡
b,
Mohammad Naser
Rezaei
b,
Silvia Hidalgo
Martinez
c,
Filip
Meysman
c,
Davy
Van de Walle
d,
Koen
Dewettinck
d,
Christophe M.
Courtin
 b and
Tom
Van de Wiele
*a
b and
Tom
Van de Wiele
*a
aCenter for Microbial Ecology and Technology (CMET), Department of Biotechnology, Faculty of Bioscience Engineering, Ghent University, Ghent, Belgium. E-mail: Tom.VandeWiele@UGent.be; Tel: +32 9 264 59 76
bLaboratory of Food Chemistry and Biochemistry, Leuven Food Science and Nutrition Research Centre (LFoRCe), Faculty of Bioscience Engineering, KU Leuven, Heverlee, Belgium
cEcosystem Management Research Group (ECOBE), Department of Biology, Faculty of Sciences, Universiteit Antwerpen, Antwerp, Belgium
dLaboratory of Food Technology and Engineering (FTE), Department of Food technology, Safety and Health, Faculty of Bioscience Engineering, Ghent University, Ghent, Belgium
First published on 20th December 2018
Abstract
Dietary modulation can alter the gut microbiota composition and activity, in turn affecting health. Particularly, dietary fibre rich foods, such as wheat bran, are an important nutrient source for the gut microbiota. Several processing methods have been developed to modify the functional, textural and breadmaking properties of wheat bran, which can affect the gut microbiota. We therefore studied the effect of enzyme treatment, particle size reduction and wheat kernel pearling on the faecal microbiota of ten healthy individuals. The most commonly studied health marker, associated to the gut microbiota activity is Short Chain Fatty Acid (SCFA) production. This study shows that modifying wheat bran physicochemical properties allows control over the extent and the rate of SCFA production by the faecal microbiota. Wheat bran pericarp fractions, depleted in starch and enriched in cellulose and highly branched arabinoxylans, were poorly fermentable compared to unmodified wheat bran, thus resulting in a reduced SCFA production with up to 20 mM. The nature of the SCFA, however, largely depends on the donor and can be linked to the individual's gut microbiota composition. The latter changed in an individually dependent manner in response to wheat bran modification. Some product dependent significant differences could still be identified across the ten donors. This product effect is more pronounced in the microbial community attached to the wheat bran residue as compared to the luminal microbial community. Generally, we find lower levels of Firmicutes, Bacteroidetes and Bifidobacterium and a higher abundance of Proteobacteria in the pericarp enriched wheat bran fractions, compared to unmodified wheat bran.
Introduction
The human gut is colonised by a dense and diverse microbial population, living in close symbiosis with the human host and playing a vital role in human health.1 The composition and activity of this gut microbial community is shaped by the complex interplay between host and environmental factors, with diet being recognised as a main driver.2–6 Dietary fibre (DF) is considered a key element in sustaining health, with dietary recommendations set at 25–38 g per day in adults.7 The term “dietary fibre” is, however, ill-defined.7–9 DF comprises a chemically and structurally diverse range of plant polysaccharides, resistant to digestion and absorption in the human upper digestive tract and partly available for fermentation by the gut microbiota.10 It was not until the 1970s that the colon was identified as the main site of DF breakdown and gut bacteria as the key players.11 It was soon hypothesized that the chemical composition and structure of DF determine the extent of fermentation and faecal bulking.11,12Wheat based foods are one of the main DF sources in the Western diet, delivering both fermentable and less readily fermentable fibre to the gut microbiota.13,14 DF is particularly concentrated in the outer histological layers of the wheat grain kernel, consisting of the pericarp, testa, nucellar epidermis and aleurone, representing 14–19% of the kernel.15 During the milling process, these tissue layers, collectively termed wheat bran, are separated from the germ and endosperm fraction, used to produce white flour.16,17 Wheat bran is considered a by-product and it is therefore an inexpensive source of chemically diverse fibres, entangled in a porous insoluble network. Arabinoxylans are the main dietary fibres present in wheat bran, representing 17–33% of the total bran fraction.18 Arabinoxylans are very heterogeneous, highly branched, complex molecules consisting of a xylopyranosyl backbone substituted with arabinofuranosyl residues, often cross-linked with other arabinoxylan chains or cell wall components, such as cellulose (10%), β-glucans or proteins through the formation of diferulic acid bridges.19–24 Wheat bran can contain considerable levels of starch ranging from 6–30%, due to the incomplete separation of the starchy endosperm from the bran during grain milling.18 Next to proteins, adding up to 14–26% of the wheat bran composition, some minor constituents include lipids, phenolic compounds, lignin, minerals and vitamins.18
Albeit being more defined than DF, wheat bran is not a standardized, homogeneous product. Variations in the chemical composition arise from wheat cultivar differences and additional processing steps.25,26 The latter include micronization, enzymatic treatment and pearling or debranning.10,27–29 Micronization of wheat bran, through milling, reduces wheat bran particle size and consequently affects surface area and porosity. Pearling sequentially removes the outer wheat kernel layers by friction and abrasion, resulting in different pearling fractions with a specific tissue composition, distinguished by the proportions of pericarp and aleurone.29,30 In this paper, all the tested wheat derived products are referred to as wheat bran, even though strictly speaking, the definition of bran implies milling, which does not apply to the pearlings. The various processing methods are developed out of economic and technological interests and evaluated for their functional, textural and organoleptic bread making properties.25,27 However, chemical and structural modifications should also be evaluated for their physiological effects, amongst which the effect stemming from colonic fermentation by the gut microbiota. Many studies found the beneficial bulking properties of bran to be confined to coarse particles, due to the lower hydration capacity of fine bran.31–33 On the other hand, fermentability is expected to increase upon particle size reduction.11 Next to fermentability, the nature of the produced fermentation products, such as SCFA, has also been shown to vary with DF chemical and structural features.5,7,10,34–40 These DF type dependent differences in fermentation products can partly be accounted for by DF type dependent shifts in the gut microbial community composition.41
We previously showed that insoluble wheat bran particles are colonised by a specific subset of bacteria from the luminal microbial community in suspension.42 Here, we investigated how processes like wheat kernel pearling, wheat bran micronization and enzymatic treatment affect that colonisation process and the resulting fermentation activity from faecal microbiota of ten healthy individuals during in vitro batch incubations.
Materials & methods
Wheat bran modification and characterisation
Commercial wheat bran, referred to as ‘unmodified wheat bran’, was obtained from Dossche Mills (Deinze, Belgium) and was produced from the roller milling of standard EU baking varieties (E, A and B type according to the German classification system) grown in France and Germany, harvest of September 2012. The particle size of this unmodified wheat bran was reduced with a Cyclotec 1093 Sample mill (FOSS, Höganäs, Sweden) in which a grinding ring and a sieve with a 500 μm mesh size was installed, as described by Jacobs et al. (2015).28 The resulting ‘micronized wheat bran’ had an average particle size of 150 μm. The histological and chemical composition of wheat bran was modified by pearling or by an enzyme-based method. Pearlings from sound wheat (cv. Akteur, harvest 2012, Dossche Mills, Deinze, Belgium) were obtained from Satake Europe (Bredbury, UK). Briefly, using the PeriTec debranning technology, wheat kernels were treated to sequentially remove a total of 3 or 6% of the kernel weight, producing the corresponding pearling fractions pearlings 0–3% (P 0–3%) and pearlings 3–6% (P 3–6%).27 The enzyme based method to modify the wheat bran chemical composition consisted of a combined amylase (Termamyl, Novozymes, Bagsvaerd, Denmark) and xylanase (Grindamyl H640, Danisco, Copenhagen, Denmark) treatment, as described by Swennen et al. (2006).43 This method, developed for large scale arabinoxylan oligosaccharides (AXOS) production, yields an insoluble residue as a side-product, termed Destarched Pericarp Enriched (DSPE) wheat bran. DSPE bran was obtained from Fugeia N.V. (Heverlee, Belgium). As for the unmodified bran, the DSPE bran particle size (367 μm) was reduced using the Cyclotec method to produce micronized DSPE bran (171 μm).All wheat bran samples were characterised. The average particle size of bran was measured by sieving 20 g of dry bran on a set of sieves with different mesh sizes and weighing the amount of bran remaining on each sieve.28 The cumulative particle size distribution is displayed in Fig. S1.† The water retention capacity was determined in triplicate according to AACC (American Association of Cereal Chemists) international method 56-11-02.44 The strongly bound water was estimated using a drainage centrifugation technique.28 The arabinoxylan, starch, β-glucan, fructan, cellulose, lipid, protein and ash levels were measured in triplicate and the DF levels were measured in duplicate as described by De Paepe et al. (2017).42 Briefly, the dietary fibre level of wheat bran was determined according to AOAC Method 991.43 using the Megazyme dietary fiber assay (K-TDFR, Megazyme, Bray, Ireland).45 β-Glucan was determined with the Megazyme mixed linkage β-glucan kit (K-BGLU, Megazyme, Bray, Ireland), as described in AACC method 32-23.44 Fructan was quantified by high performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD) on a Dionex ICS3000 chromatography system (Sunnyvale, CA, USA) after mild acid hydrolysis.46 The remaining sugars were measured after acid hydrolysis using sulfuric acid (cellulose) and trifluoroacetic acid (arabinoxylans and starch), followed by sugar derivatization and gas chromatographic analysis. Protein concentrations were measured by the Dumas combustion method, an adapted version of the AOAC Official Method 990.03.45 An automated Dumas protein analysis system (EAS VarioMax N/CN, Elt, Gouda, The Netherlands) was used with 6.25 as the nitrogen to protein conversion factor. Lipid content was gravimetrically determined after extraction of lipids from bran with water saturated butanol using an ASE 200 device (Dionex, Amsterdam, The Netherlands) and a subsequent purification step.47 Ash content was measured with AACC method 08-01.01.44
Experimental set-up
The six different wheat bran products were used to study the impact of wheat bran modification on the fermentation activity and microbial community composition of the human faecal microbiota during in vitro batch incubations.Prior to in vitro fermentation, all wheat bran products underwent in vitro batch digestion, according to an adaptation to the method from Minekus et al. (2014),48 described by De Paepe et al. (2017),42 to mimic stomach and small intestine passage. Briefly, wheat bran samples were incubated at 37 °C with simulated saliva fluid containing salivary α-amylase at pH 7 for 15 min (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 w/v), simulated gastric fluid containing porcine pepsin at pH 3 for 2 h (1
2 w/v), simulated gastric fluid containing porcine pepsin at pH 3 for 2 h (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) and simulated pancreatic juice containing porcine pancreatin and bile salts at pH 7 for 2 h (1
1 v/v) and simulated pancreatic juice containing porcine pancreatin and bile salts at pH 7 for 2 h (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 w/v). Insoluble, undigested material was collected by centrifugation at 500g for 5 min and lyophilised for preservation until chemical characterisation or batch incubation with faecal slurries from the ten donors. This course of action may lead to the loss of soluble, indigestible β-glucan and fructan, which is not entirely representative for the in vivo situation where these compounds would reach the colon and be available to gut microorganisms for colonic fermentation.
1 w/v). Insoluble, undigested material was collected by centrifugation at 500g for 5 min and lyophilised for preservation until chemical characterisation or batch incubation with faecal slurries from the ten donors. This course of action may lead to the loss of soluble, indigestible β-glucan and fructan, which is not entirely representative for the in vivo situation where these compounds would reach the colon and be available to gut microorganisms for colonic fermentation.
Faecal samples from ten healthy individuals, without a history of antibiotic use, six months prior to the study, were used to account for inter-individual variability. All donors consumed a mixed Western diet, apart from subject 8, who is vegetarian. The five male (2, 3, 5, 6, 9) and five female (1, 4, 7, 8, 10) subjects were of the age 23–37. Incubation research with faecal microbiota from human origin was approved by the ethical committee of the Ghent University hospital under registration number B670201214538. Written informed consent was obtained from all participants.
Faecal samples were collected under anoxic conditions in sterile airtight containers, comprising an AnaeroGen™ sachet (Oxoid Ltd, Basingstoke, Hampshire, UK). Within an hour, a 20% (w/v) faecal slurry was prepared from each donor stool separately by homogenising the faecal sample for 10 min (LabBlender 400, Seward Ltd, Worthing, West Sussex, UK) in 0.1 M phosphate buffer at pH 7, containing 1 g L−1 sodium thioglycolate (C2H3O2SNa) as reducing agent (Sigma Aldrich, St Louis, MO, US). Particulate material was removed by centrifugation for 2 min at 500g.49 Faecal slurries were inoculated for each donor separately at a fivefold dilution in Hungate tubes (Glasgeratebau Ochs Gmbh, Bovenden-Lenglern, Finland), containing a carbohydrate-low medium supplemented with 1% wheat bran. Six different wheat bran products were evaluated along with a control without wheat bran product. All Hungate tubes from one donor were inoculated using the same faecal slurry. Anaerobic conditions were obtained by flushing the tubes with N2 during 30 cycles. The tubes were set at atmospheric pressure before incubation at 37 °C and 150 rpm on an orbital shaker (KS 4000 i control, IKA, Staufen, Germany). The carbohydrate-low medium consisted of 3 g L−1 yeast extract (Oxoid Ltd, Basingstoke, Hampshire, UK), 1 g L−1 peptone (Oxoid Ltd, Basingstoke, Hampshire, UK) and 1 g L−1 mucin from porcine stomach Type II (Sigma Aldrich, St Louis, MO, US), dissolved in 0.1 M phosphate buffer at pH 6.8.42
To follow the course of fermentation, the tubes were sampled at 0, 6, 24 and 48 h. For each time point, product and donor a separate tube was incubated. The gas pressure was measured (Infield7, UMS, Munich, Germany) and 8 mL of the headspace was sampled in vacuette tubes (Greiner Bio One, Kremsmünster, Austria) to analyse the gas phase composition using a compact GC (Global Analyser Solutions, Breda, The Netherlands), equipped with a Molsieve 5A pre-column, a Porabond column (CH4, O2, H2 and N2) and a thermal conductivity detector. The unfermented wheat bran residue was separated from the supernatant, termed luminal suspension hereinafter, by centrifugation for 5 min at 700g. Aliquots of 0.1 g wheat bran residue were stored at −20 °C for DNA extraction, after three washing steps with phosphate buffer to remove non-attached and loosely attached micro-organisms, as suggested by Leitch et al. (2007).50 This washing procedure was slightly adapted, as described by De Paepe et al. (2017).42 Briefly, thirty mL of 0.1 M phosphate buffer at pH 6.8 was added to the bran pellet, vigorously vortex-mixed and removed by centrifugation for 2 min at 500g. The luminal suspension was aliquoted for DNA extraction (250 μL), SCFA and Total Ammonium Nitrogen (TAN) analysis and the pH was measured. Samples were stored at −20 °C until further analysis. For DNA extraction, the pellet obtained after centrifuging the sample for 10 min at 5000g was stored.
In order to allow for chemical characterisation of the wheat bran residue after fermentation, the batch fermentation was repeated at large scale for one donor, increasing the volume from 10 mL in Hungate tubes to 1 L in a 2 L Schott bottle. The same carbohydrate-low medium and wheat bran concentration (1%) were used. The 20% (w/v) faecal slurry was tenfold diluted to obtain a final concentration of 2% (w/v). One bottle for each wheat bran product was incubated and used to measure the gas pressure and sample the gas and liquid phase over time. The wheat bran residue was collected only after 48 h and chemically characterised, as described above.
SCFA and TAN measurement
SCFA levels were measured using capillary gas chromatography coupled to a flame ionization detector (Shimadzu, Hertogenbosch, the Netherlands) after diethyl ether extraction, as described by De Paepe et al. (2017).42 Samples were twofold diluted with Milli-Q water. TAN was determined as an indicator for protein fermentation through steam distillation according to Standard methods (4500-NH3 B; APHA, 1992) using a Vapodest 30 s (C. Gerhardt GmbH & Co. KG, Königswinter, Germany).51SEM microscopy
Scanning electron microscopy (SEM) was performed to examine the presence and spatial distribution of gut microorganisms on the residual wheat bran particles after fermentation. To fix and preserve the shape of bacterial cells, the bran samples were chemically dried with hexamethyldisilazane (HMDS) as described by Araujo et al. (2003).52 After complete evaporation of the HMDS, samples were mounted on an aluminium pin (diameter: 12 mm) using double sided carbon tape and subsequently gold sputtered for 45 s at 30 mA (Agar Sputter Coater B7340, Agar Scientific, UK). Images were collected using a Phenom Pro X SEM microscope (Phenom-World B. V., the Netherlands) with a beam intensity of 10 keV.Cryo-SEM microscopy
As an alternative to SEM microscopy, samples were also visualised using a Jeol JSM 7100F scanning electron microscope (JEOL Ltd, Tokyo, Japan). A small amount of wheat bran was placed on a sticky carbon surface mounted on an aluminium stub, vitrified in a nitrogen slush and transferred under vacuum conditions into the cryo-preparation chamber (PP3010T Cryo-SEM Preparation System, Quorum Technologies, UK) conditioned at −140 °C. Subsequently, the sample was sublimated for 20 min at −70 °C to remove frost artefacts, sputter-coated with platinum using argon gas, transferred to the SEM stage at −140 °C and electron beam targeted at 3 keV.Microbial community analysis
The microbial community after 48 h of incubation with unmodified, micronized, DSPE micronized bran and P 0–3% was assessed using Illumina next generation 16S rRNA gene amplicon sequencing. Both the luminal suspension and the washed bran residue were analysed.A phenol–chloroform DNA extraction was performed, following a mechanical lysis by multidirectional beating, according to De Paepe et al. (2017).42 As starting material, the pellet obtained after centrifuging 250 μL of luminal sample at 5000g for 10 min or 0.1 g bran aliquots were used. The DNA quality was verified by electrophoresis on a 1.5% (w/v) agarose gel. DNA was quantified by a fluorescence assay using the QuantiFluor® dsDNA kit (Promega, Madison, WI, US) and Glomax®-Multi+ system (Promega, Madison, WI, US). Samples were sent out to LGC Genomics (Teddington, Middlesex, UK) for library preparation and sequencing on an Illumina Miseq platform. To assess sequencing quality, four mock community samples were included. The mock community consisted of the genomic DNA of 12 species, belonging to 4 different phyla (Table S1†), pooled to reach an equimolar concentration of 16S rRNA gene copies based on qPCR with the primers listed below.
The V3–V4 region of the 16S rRNA gene was amplified by PCR using primers derived from Klindworth et al. (2013), with a slight modification to the reverse primer by introducing another degenerated position (K) to make it more universal.53 The library preparation was performed by means of adaptor ligation using the Ovation Rapid DR Multiplex System 1-96 (NuGEN Technologies, Inc., San Carlos, CA, US), as described by De Paepe et al. (2017).42 The sequence data has been submitted to the NCBI (National Center for Biotechnology Information) database under accession number SRP091975.
Bioinformatics analysis of Illumina next generation 16S rRNA gene amplicon sequencing data
The mothur software package (v.1.39.5) and guidelines were used to process the amplicon data generated by LGC Genomics.54In a first step, forward and reverse reads were assembled into contigs by means of a heuristic approach based on the Phred quality scores. After removing contigs with any ambiguous base calls or unsatisfying overlap, 52% of the data was retained. The remaining contigs, with a length between 402 and 427 bases, were aligned to the mothur formatted silva_seed release 123 alignment database trimmed between positions 6428 and 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 440, to be compatible with the 341F–785R primers.55 Any sequences not aligning within this region or containing homopolymer stretches of more than 12 bases were removed, resulting in an additional 0.7% of the data being culled out. In a next step, data was preclustered allowing up to 4 differences between sequences to be merged. A chimera check was performed using UCHIME, after which only 46% of the original reads were retained.56 Subsequently, sequences were classified by means of a naive Bayesian classifier against the RDP 16S rRNA gene training set, version 16 with an 80% cut-off for the pseudobootstrap confidence score. Sequences annotated as Chloroplast, Mitochondria, unknown, Archaea or Eukaryota at the kingdom level were excluded. Additionally, the sequences from the mock community samples were omitted and separately analysed. A total of 4
440, to be compatible with the 341F–785R primers.55 Any sequences not aligning within this region or containing homopolymer stretches of more than 12 bases were removed, resulting in an additional 0.7% of the data being culled out. In a next step, data was preclustered allowing up to 4 differences between sequences to be merged. A chimera check was performed using UCHIME, after which only 46% of the original reads were retained.56 Subsequently, sequences were classified by means of a naive Bayesian classifier against the RDP 16S rRNA gene training set, version 16 with an 80% cut-off for the pseudobootstrap confidence score. Sequences annotated as Chloroplast, Mitochondria, unknown, Archaea or Eukaryota at the kingdom level were excluded. Additionally, the sequences from the mock community samples were omitted and separately analysed. A total of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700
700![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 213 sequences were withheld (44% of the data). Sequences were binned into Operational Taxonomic Units (OTUs) within each order identified by the preceding classification step. The OptiClust clustering algorithm was used with a cut-off set at 0.15.57 Eventually, a 3% dissimilarity level was used to bin sequences into OTUs to generate a shared file at species level. Accordingly, an OTU is defined in this manuscript as a collection of sequences with a length between 402 and 427 nucleotides that are found to be more than 97% similar to one another in the V3–V4 region of their 16S rRNA gene after applying OptiClust clustering.58–61 44
213 sequences were withheld (44% of the data). Sequences were binned into Operational Taxonomic Units (OTUs) within each order identified by the preceding classification step. The OptiClust clustering algorithm was used with a cut-off set at 0.15.57 Eventually, a 3% dissimilarity level was used to bin sequences into OTUs to generate a shared file at species level. Accordingly, an OTU is defined in this manuscript as a collection of sequences with a length between 402 and 427 nucleotides that are found to be more than 97% similar to one another in the V3–V4 region of their 16S rRNA gene after applying OptiClust clustering.58–61 44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 OTUs were identified. Finally, taxonomy assignment was obtained according to the RDP version 16 and silva.nr_v123 database.55,62,63 The shared file, containing the number of reads observed for each OTU in each sample and the taxonomic annotation were loaded into R, version 3.4.0 (2017-04-21).64 The read count table was summarized at genus and phylum level. ‘Unclassified’ taxa at these levels were replaced by the deepest taxon classification. Four samples did not obtain sufficient reads and were removed from the dataset. Reads occurring only once in all samples (singletons) were removed.65 5488 OTUs were retained. For the most abundant OTUs the sequences retrieved from the 3% dissimilarity level fasta file, obtained in mothur, were classified through the RDP web interface using the RDP SeqMatch tool restricting the database search to type strains with only near-full-length, good quality sequences and blasted in NCBI against the 16S rRNA gene sequences, selecting only type material, with optimisation of the BLAST algorithm for highly similar sequences (accession date: April 2017).62,63,66 Although identification to the species level based on short 300 bp reads may involve some ambiguity, the most likely species classification of some interesting OTUs is reported in the results section. In the event of inconsistencies in the results of the RDP SeqMatch tool and NCBI BLAST, no species level classification is provided. The results for the 30 most abundant OTUs (best hit as well as the next best hit) are shown in Table S2.†
280 OTUs were identified. Finally, taxonomy assignment was obtained according to the RDP version 16 and silva.nr_v123 database.55,62,63 The shared file, containing the number of reads observed for each OTU in each sample and the taxonomic annotation were loaded into R, version 3.4.0 (2017-04-21).64 The read count table was summarized at genus and phylum level. ‘Unclassified’ taxa at these levels were replaced by the deepest taxon classification. Four samples did not obtain sufficient reads and were removed from the dataset. Reads occurring only once in all samples (singletons) were removed.65 5488 OTUs were retained. For the most abundant OTUs the sequences retrieved from the 3% dissimilarity level fasta file, obtained in mothur, were classified through the RDP web interface using the RDP SeqMatch tool restricting the database search to type strains with only near-full-length, good quality sequences and blasted in NCBI against the 16S rRNA gene sequences, selecting only type material, with optimisation of the BLAST algorithm for highly similar sequences (accession date: April 2017).62,63,66 Although identification to the species level based on short 300 bp reads may involve some ambiguity, the most likely species classification of some interesting OTUs is reported in the results section. In the event of inconsistencies in the results of the RDP SeqMatch tool and NCBI BLAST, no species level classification is provided. The results for the 30 most abundant OTUs (best hit as well as the next best hit) are shown in Table S2.†
Statistical analysis
All statistical analysis were performed in R, version 3.4.0.64 All formal hypothesis tests were conducted on the 5% significance level (α = 0.05).Statistical hypothesis testing to assess the effect of the products on the SCFA and TAN production was performed by using the Kruskal–Wallis rank sum test, followed by Pairwise Wilcoxon Rank Sum Tests with Holm correction for multiple testing.
Differences in microbial community composition between the donors, products and niches (luminal versus bran-attached) were explored using ordination and clustering techniques. For these purposes, the shared file was filtered based on the arbitrary cut-off's described by McMurdie and Holmes (2014),65 whereby OTUs observed in less than 5% of the samples and with read counts below 0.5 times the number of samples were removed. The data was normalised by rescaling to proportions.65 Principle Coordinate Analysis (PCoA; package stats3.4.0)67–71 was performed on the abundance based jaccard dissimilarity matrix (package vegan 2.4-3)72–74 and visualised with ggplot 2.2.1.75 On the genus level, weighed averages of genera abundances were a posteriori added to the ordination plot, using the wascores function in vegan.74 Clustering was performed by means of Unweighted Pair-Grouped Method, using arithmetic Averages (UPGMA) clustering (cluster 2.0.6).76 Ordination and clustering analysis were conducted on the complete dataset, as well as for the luminal and bran-attached communities separately.
DESeq2 was applied on the filtered, unnormalised community composition data for either the luminal or the bran-attached communities to detect statistically significant differences in OTU abundances at species, genus or phylum level between the different wheat bran products.65,77 The factors donor and wheat bran product were included in the design formula of the full model. The effect of the wheat bran product was determined by a likelihood ratio test on the difference in deviance between a full and reduced model formula. Pairwise significant differences were obtained using Wald tests, specifying all pairwise combinations of products as the contrast argument.
Partial Least Squares (PLS) regression, available in the R-package mixOmics, was performed to establish the relation between the DESeq2 normalised and variance stabilised microbial community composition data (X) and the molar acetate, propionate, butyrate, valerate, branched SCFA and ammonium concentrations (Y). A model with two components was selected based on a Leave One Out (LOO) and Mfold validation.78,79 In a next step, smart feature selection was applied to look for specific species or genus level associations with the propionate or butyrate production by using sparse Partial Least Squares (sPLS) regression, implemented in the R-package mixOmics.78 The initial model was built in regression mode with the molar propionate and butyrate ratio as response (Y) variables in function of the DESeq2 transformed variance stabilised read counts (X). 2 Dimensions, 2 Y variables and 25 or 15 X variables (OTUs) at species, respectively, genus level were selected on each of the components after tuning based on a LOO validation.78,79 In an attempt to refine the clustering of samples, the wheat bran product was supplied as a factor variable (Y) in a supervised PLS Discriminant Analysis (PLS-DA) with 2 components in mixOmics.78
Results
The effect of wheat bran particle size and histological and chemical composition on the fermentation activity and microbial community composition of the human faecal microbiota of ten healthy individuals was assessed in an in vitro batch experiment.Characterisation of the wheat bran fermentation substrate
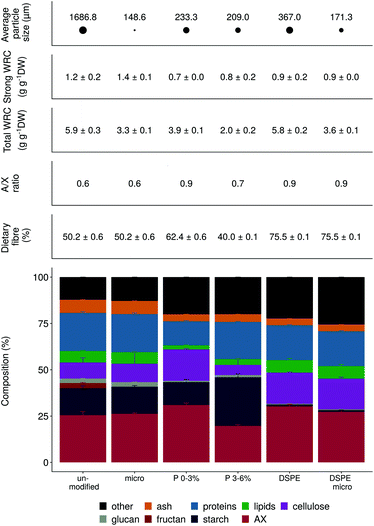
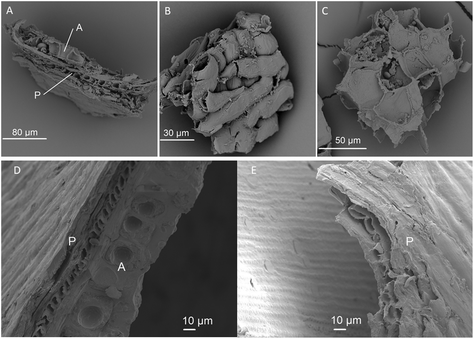
The six wheat bran products varied in particle size, ranging from 149 to 1687 μm (Fig. 1). Micronization results in some cases in tissue disintegration, as illustrated by the SEM microscopy (Fig. 2). Wheat kernel pearling and wheat bran enzymatic treatment with amylase and xylanase were applied to modify amongst others the arabinoxylan, starch, cellulose and protein content (Fig. 1). DSPE bran was depleted in starch, as a result of the amylase treatment, and enriched in cellulose and arabinoxylans with a higher degree of substitution (A/X ratio), which corresponded to the removal of the aleurone layer with a low A/X ratio (Fig. 2). The first pearling fraction (P 0–3%), mainly consisting of the outer pericarp layer of the wheat kernel, was also enriched in highly substituted arabinoxylans and cellulose, but had a higher starch and lower protein content compared to DSPE bran. The increased starch content emanates from endosperm contamination due to the crude, imprecise pearling process. Further pearling of the wheat bran kernel resulted in an aleurone-rich pearling fraction (P 3–6%), with an even higher starch concentration. All processing methods affected the total water retention capacity (WRC) (Fig. 1). Unmodified and DSPE bran displayed the highest total WRC (5.8 g g−1 DW), consistent with the microporous structure present in the pericarp. Pearling and micronization decreased the total WRC due to a decrease in micrometer size pores. The strongly bound water was less affected by micronization and chemical modification.Prior to fermentation, all wheat bran products underwent in vitro batch digestion to mimic human digestion in the stomach and small intestine. The activity of the digestive enzymes (amylase, pepsin and pancreatin) reduced the protein content and increased the dietary fibre percentage with up to 15%. The proportion of starch and cellulose remained unaffected (Fig. S2†). In contrast to the in vivo situation, soluble indigestible β-glucan and fructan (present at <2.5%) were also removed. Hence, the ‘pre-digested wheat bran’ used hereinafter, refers to the insoluble wheat bran fraction remaining after pre-digestion.
Fermentation of the wheat bran substrate
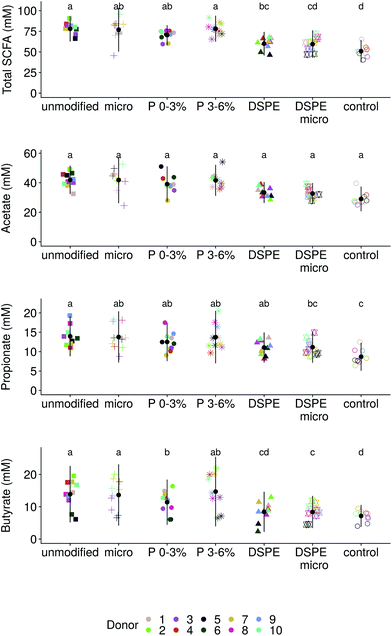
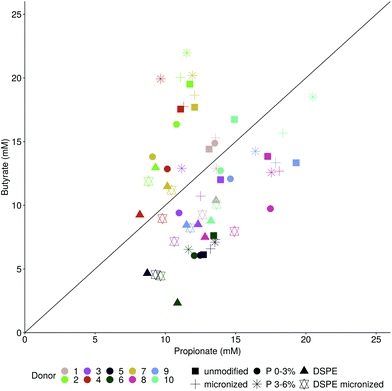
Separately incubating these pre-digested wheat bran products for 6, 24 and 48 h with the faecal material of ten donors in a carbohydrate-low pH buffered medium resulted in a stable course of fermentation with gas pressure built-up and increasing SCFA and ammonium production over time (Fig. S3†). Some acidification, related to the formation of SCFA, occurred but the pH never dropped below 6.15, starting from pH 6.81 (data not shown). The control medium supported an average SCFA production of 51 mM after 48 h, corresponding to a minor pH decrease to minimally 6.47 (Fig. 3). On average, 78 ± 8 mM of SCFA were produced by the ten donors on the carbohydrate-low medium supplemented with unmodified wheat bran after 48 h incubation. Micronizing wheat bran did not alter the SCFA production (79 ± 8 mM) (Fig. 3). Enzymatic treatment of wheat bran significantly reduced average SCFA concentrations for ten donors by 18 mM (p = 0.03), to 60 ± 7 mM (DSPE) and 60 ± 8 mM (micronized DSPE) (Fig. 3). Addition of P 0–3% yields an intermediary production of 70 ± 6 mM. Finally, P 3–6% supplementation supports a production of 78 ± 8 mM of SCFAs, comparable to unmodified and micronized wheat bran (Fig. 3). The observed reduction in total SCFA production with DSPE bran and P 0–3% is mainly attributed to acetate and butyrate, with an observed drop of 8 mM in average acetate and 6 mM in average butyrate production. This resulted in a shift in average butyrate to propionate ratio from 1 (unmodified and micronized bran) to 0.95 for P 0–3%, 0.78, for DSPE and 0.76 for micronized DSPE bran. Incubation with P 3–6% increases the ratio to 1.13, indicating a stimulation of butyrate. Yet, Fig. 3, also indicates large inter-individual differences in propionate and butyrate production. Donors 2, 4 and 7 tended to produce more butyrate than propionate, particularly for unmodified bran, micronized bran and P 3–6%, while P 0–3% and DSPE bran resulted in butyrate/propionate ratios closer to 1 (Fig. 4). In contrast, donors 3, 5, 6, 8, 9 and 10 consistently displayed higher propionate than butyrate production. For donor 1, the ratio is close to 1. Ammonium and branched SCFA levels, as markers for protein fermentation, are donor-dependent and not significantly different between wheat bran products (Fig. S4†). Bran micronization of unmodified or DSPE bran did not affect chemical composition or protein and carbohydrate fermentation after 48 h (Fig. 1, 3 and S4†). However, when averaging carbohydrate fermentation activity over time for the ten donors, it became clear that micronized wheat bran particles were more rapidly fermented (Fig. S5†). This particle size effect was only significant for butyrate and was absent in donors 7 and 9 (Fig. S5†). Furthermore, smaller differences in particle size, as in the case of DSPE (367 μm) and micronized DSPE bran (171 μm), did not affect the rate of carbohydrate fermentation (Fig. S5†).Impact of wheat bran modification on the luminal microbial community composition
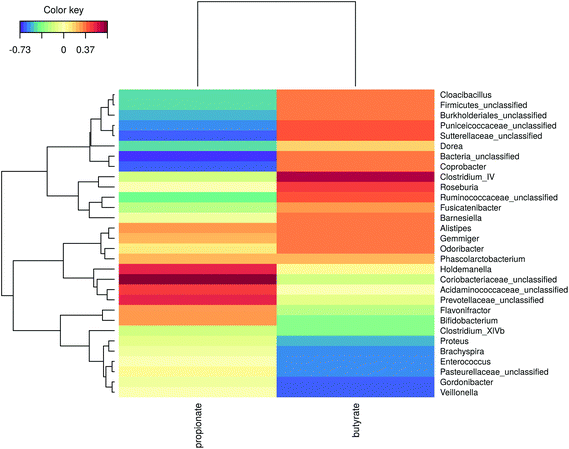
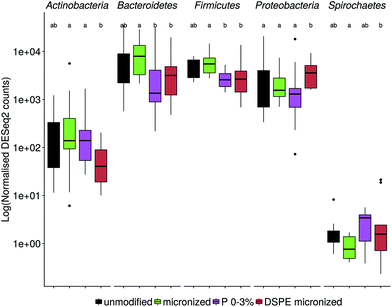
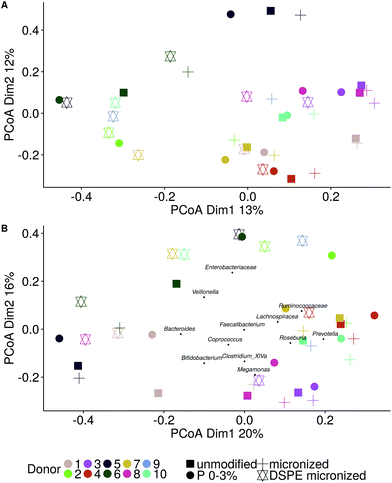
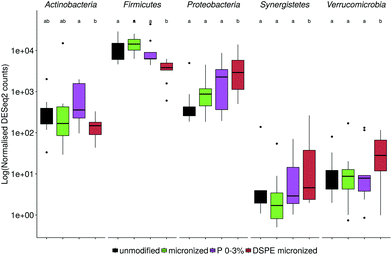
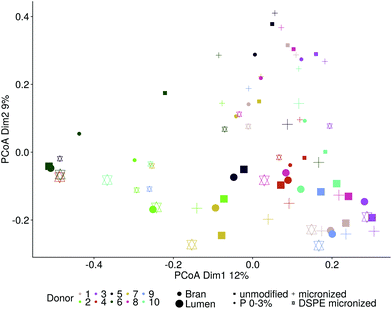
The microbial community composition after 48 h of incubation was analysed using Illumina 16S rRNA gene amplicon sequencing. A Principle Coordinate Analysis (PCoA) of the luminal microbial community at species and genus level indicated that donor origin is the main factor determining the grouping of samples (Fig. S6†). The close proximity of donors 2, 4 and 7 in these explorations coincides with the observed similarity in metabolic output (Fig. 3 and 4). Combining functional data and microbial community composition data at species or genus level, applying Partial Least Squares (PLS) regression, confirmed the donor based clustering of samples (data not shown). A sparse Partial Least Squares (sPLS) model, retaining 2 components, was used to select the 25 OTUs at species or the 15 OTUs at genus level most predictive of the propionate and butyrate ratio on each of the components (Fig. 5 and S7†). Clostridium cluster IV, Roseburia, Barnesiella, Gemmiger, Alistipes Odoribacter and unclassified genera belonging to the Ruminococcaceae, Puniceicoccaceae and Sutterellaceae families were associated with high butyrate production whereas Holdemanella and unclassified genera belonging to the Coriobacteriaceae, Acidaminococcaceae and Prevotellaceae families were all positively correlated with high propionate production (Fig. 5).While ordination methods showed donor effects predominating over product effects, DESeq2 analysis revealed significant differences between wheat bran products. Considering all ten donors, particle size reduction significantly stimulated Anaerostipes (p = 0.046), Bifidobacterium (p = 0.033), Blautia (p = 0.011), Clostridium cluster XVIII (p = 0.001) and Roseburia (p = 0.046) compared to unmodified bran (Table S3†). Chemical wheat bran modification (enzymatic treatment and wheat kernel pearling) induces more pronounced changes in the luminal microbial community composition. To exclude the particle size effect, pairwise significant differences due to modification of the chemical composition were evaluated compared to micronized bran, as these products are more similar in size. DSPE bran was characterised by a significant decrease in Actinobacteria (p = 9 e−04), Bacteroidetes (p = 3 e−03) and Firmicutes (p = 4 e−03) and an increase in Proteobacteria (p = 6 e−03), compared to the micronized bran (Fig. 6). At genus level, this corresponds to a significant increase in Enterobacteriaceae (p = 0.022), and significant decreases in Alistipes, Bacteroides, Blautia, Clostridium cluster XIVa, Clostridium cluster XVI, Faecalibacterium, Lachnospiraceae, Roseburia and Ruminococcus (Fig. S8†). A significant reduction in the aforementioned genera was also observed for P 0–3% relative to the micronized bran, causing a phylum level reduction of Bacteroidetes (p = 4.38 e−06) and Firmicutes (p = 1.06 e−03) (Fig. 6 and S8†). The corresponding species level significant differences are displayed in Table S4.†
Impact of wheat bran modification on the bran-attached microbial community composition
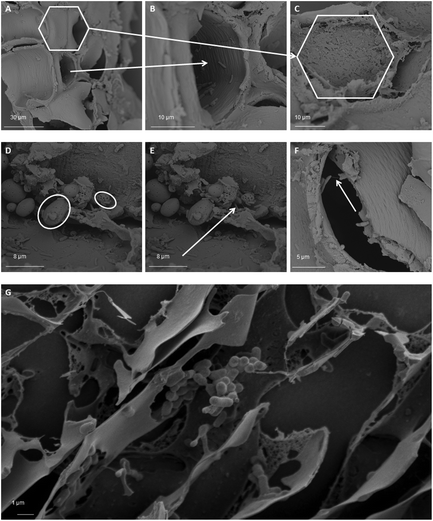
Cryo-SEM and SEM microscopy (Fig. 7) confirmed wheat bran colonisation. We therefore analysed the microbial community attached to the wheat bran residue after washing off loosely associated bacteria. The impact of the wheat bran product on the bran-attached communities is more distinct than on the luminal community, as can be seen from the PCoA at species and genus level, which is less dominated by inter-individual variability (Fig. 8). Micronization significantly increased the colonisation of Blautia (p = 0.002), Clostridium cluster XVIII (p = 1.8 e−06) and Fusicatenibacter (p = 0.004) on the bran compared to unmodified bran (Table S5†). More differences were to be found in the community colonising the DSPE bran, with significant increases in Proteobacteria and Verrucomicrobia and decreases in Firmicutes (Fig. 9). The corresponding significant genus level shifts compared to micronized bran comprised Blautia, Butyricicoccus, Clostridium cluster XVIII and XIVa, Faecalibacterium, Ruminococcus, Coprococcus, Fusicatenibacter, Lachnospiraceae, Roseburia and Bifidobacterium and Citrobacter (Fig. S9†). With P 0–3%, no significant phylum and less significant genus level differences were observed, encompassing Blautia, Butyriciccocus, Clostridium cluster XVIII, Dorea, Fusicatenibacter, Lachnospiraceae and Roseburia (Fig. 9 and S9†). The significant differences at species level are displayed in Table S6.†Comparing the luminal and bran-attached microbial community
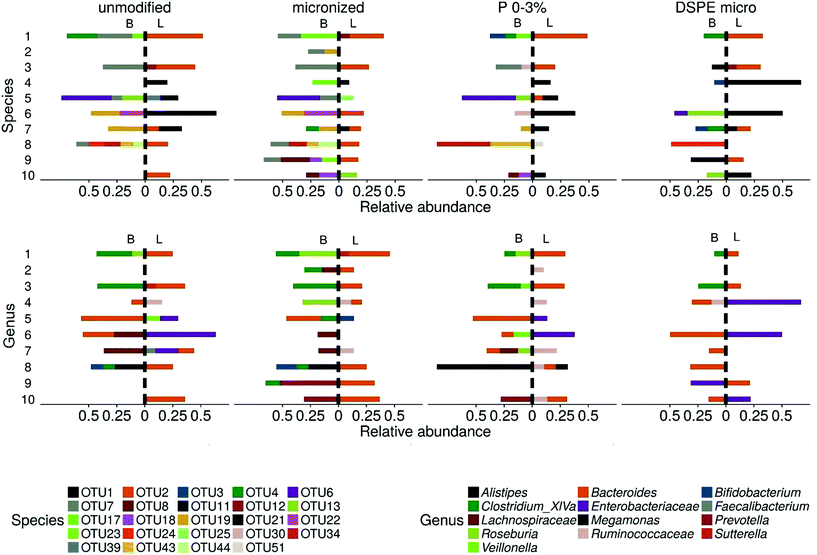
The luminal and bran-attached communities clearly differed, as indicated by a PCoA on species level (Fig. 10). The largest proportional shifts, i.e. more than 10% difference in relative abundance between the luminal and bran community, at genus and species level are shown in Fig. 11. The relative abundance of Clostridium cluster XIVa, Lachnospiraceae, Prevotella, Roseburia and Megamonas was increased on the bran at the expense of Enterobacteriaceae, Alistipes and Sutterella. However, there is a large inter-individual variability, as indicated by shifts in Prevotella that were characteristic for donor 9 and 10, while Megamonas enrichment on the bran was only observed for donor 8. At genus level, Bacteroides was predominantly retrieved in the luminal community, except for donors 5 and 6. This can be explained by species level differences in bran colonisation potential. B. dorei (OTU2), present at high abundances in the luminal community of donors 1, 3, 6, 7, 8, 9 (up to 55%), was not successfully colonising any of the bran products (not exceeding 6.6%), as opposed to B. ovatus (OTU6), B. cellulosilyticus (OTU22) and B. xylanisolvens (OTU24), that preferentially colonised the bran in donors 5, 6 and 8 respectively. Within the Firmicutes phylum, the dominant bran colonising species were Clostridium xylanolyticum (OTU7) in donors 1, 2, 3, 5, 8, 9, R. faecis (OTU17) in donors 1 and 4 and Hungatella hathewayi (OTU19) in donors 6, 7, 8.Next to donor dependent shifts, wheat bran modifications affected wheat bran colonisation preference. In donor 1, the luminal microbial community composition is very similar between different bran types, whereas the magnitude of bran colonisation by B. uniformis (OTU3), B. stercoris (OTU4), B. xylanolyticus (OTU7) and Roseburia faecis (OTU17) depended on the bran type (Fig. 11). Similarly, in donor 3, depending on the bran type Clostridium xylanolyticum (OTU7) or Ruminococcus lactaris (OTU21) enrichment prevailed. In donor 8, the Megamonas on the bran residue, enriched with 27% (unmodified and micronized bran) up to 85% (P 0–3%), was substituted by B. xylanisolvens (OTU24) in the DSPE bran-attached community.
Characterisation of the wheat bran residue after fermentation
To determine the fermentability of the various wheat bran components, for donor 10 an additional upscaled fermentation was performed in 2 L bottles, containing 10 g of the different bran products. On a dry weight basis, 70% of the unmodified and micronized wheat bran and P 3–6% disappeared after 48 h. P 0–3% and DSPE bran were less fermented (60%, respectively, 50%). Starch was efficiently fermented, as indicated by the remaining starch content of around 1% in all bran products, compared with initial proportions of up to 26% (P 3–6%) (Fig. 1 and S10†). Proteins represented about 10% of the fermented wheat bran residue, corresponding to a reduction of approximately 10% compared to the original product. The increased AX and cellulose fraction after fermentation reflect their lower fermentability and contributed to the higher DF levels of about 80%. The remaining AX fraction was characterised by an increased A/X ratio.Discussion
To assess the impact of structural and chemical wheat bran modifications on human gut microbiome metabolism and community composition, we incubated six wheat bran products with separate faecal microbial slurries from ten healthy individuals. The microbial community composition was probed by means of 16S rRNA gene amplicon sequencing, thereby including microbiota members regardless of their activity and viability. We found that modification of wheat bran physicochemical properties largely affects the amount, but not the ratio of produced SCFA and that inter-individual variability dictates the functional and composition response from the luminal microbiota to wheat bran supplementation. Yet, the wheat bran-attached microbiome composition was more affected by wheat bran structure.Micronization of unmodified bran from 1687 μm to 149 μm resulted in a higher SCFA production after 24 h for all donors, except donor 7 and 9. This difference between micronized and unmodified bran disappeared again after 48 h and was not observed at 6 h. This suggests that particle size only affects the rate of fermentation, confirming the finding from Stewart and Slavin (2009)80 that a reduction in average wheat bran particle size from 1239 μm to 551 μm increased SCFA levels starting from 8 h up till 24 h. The authors attributed the increased SCFA production to an increased surface area, providing a larger contact area for bacterial enzymes to access the substrate. Others claim however that bran porosity, more than surface area, determines substrate accessibility to enzymes.37,81 Extracellularly secreted enzymes are able to penetrate in nanometer size pores, whereas membrane-bound enzyme complexes, which are suggested to play a major role in the rate limiting primary degradation of wheat bran, are restricted to micrometer size pores.10,42,81 Changes in porosity may partly offset the effect of an increased surface area on enzyme accessibility, limiting the effect of micronization on fermentability. Indeed, SEM pictures from micronized bran in this study illustrated the loss of micrometer size pores, located in the empty pericarp cells and the voids in between stacked tissue layers, due to tissue segregation. The higher porosity of unmodified wheat bran resulted in an increased total water retention capacity, which determines the faecal bulking capacity of wheat bran by increasing faecal and stool moisture content.31,33,82 Gaining control over stool consistency by wheat bran particle size modification, could offer the possibility to alter the gut microbiota composition, as Van de Putte et al. (2016)83 recently showed that stool consistency correlates with the gut enterotype landscape.
Wheat bran enzymatic treatment reduced the wheat bran fermentability and significantly decreased for all ten donors the 48 h average SCFA production with 18 mM compared to unmodified wheat bran. The poor fermentability of the DSPE bran is likely related to starch depletion, the high cellulose content and the highly branched arabinoxylans (high A/X ratio). A high cellulose content and highly substituted arabinoxylans were also found to be characteristic of the pearlings 0–3% (P 0–3%) fraction. In contrast to DSPE bran, however, P 0–3% still contained 10% starch, which was reflected in an intermediary fermentability and only an 8 mM drop in average SCFA production after 48 h, compared to unmodified bran. Further pearling of the wheat kernel produced a P 3–6% fraction. Despite the increased A/X ratio, this structure resulted in similar fermentation activity as the unmodified and micronized wheat bran. This can be attributed to a higher starch content residing from starchy endosperm contamination during pearling.30 These results are in line with the expectations: starch being more readily fermentable as opposed to cellulose and the arabinoxylan fermentability being inversely related to the degree of arabinoxylan substitution, resulting in a low pericarp and high endosperm and aleurone fermentability.22,37,84–88
Besides the total SCFA production, we investigated whether wheat bran modification also affects the molar ratio of the individual SCFA. Acetate, propionate and butyrate have distinct physiological effects on glucose, cholesterol, lipid and colonocyte energy metabolism, satiety, proliferation and differentiation of human colonocytes, carcinogenesis, inflammation and gut barrier function.38,89–93 We only found minor effects from wheat bran modification on the SCFA profiles, which were predominantly donor-dependent. The production of SCFA by the gut microbiota in response to different substrates is a poorly understood complex process, governed by many factors.88 Substrate structure and chemical composition, substrate flux and environmental conditions such as pH, transit time and gas phase composition have all been shown to act as a switch between alternative metabolic pathways at the individual bacterial species level.89,94–98 Moreover, in a complex gut microbial community, the favourable growth of specific species with a specific metabolic capacity determines the SCFA proportions.5,40,89 Inter-individual variability in microbiome composition is therefore an often overlooked, though important, confounding factor in studies characterising the fermentation of various dietary fibre types and can even give rise to contradictory results. Moreover, a majority of studies concluding that dietary fibre type determines the SCFA profile typically used pooled faecal inocula, masking inter-individual differences.39,99–105 Only a limited number of studies pinpoint the importance of inter-individual variability in dietary fibre fermentation.106–108 This is supported by our observation that, regardless of the wheat bran product, donors 3, 5, 6, 8, 9 and 10 responded with high propionate production, whereas butyrate prevailed for donors 2, 4 and 7. This was further underscored by the observed differences in microbial community composition with donors 2, 4 and 7 clustering together. Relating specific members of the community with specific metabolites is challenging, given the metabolic versatility within a single species, cross-feeding or competitive interactions between species and inter-individual differences in microbial community composition.5,41,109,110 At the genus level, sPLS regression revealed the strongest association of butyrate with Clostridium cluster IV, which encompasses several butyrate producers.38 Propionate production was correlated mostly with Holdemanella, Coriobacteriaceae and Prevotellaceae, genera containing species producing propionate or lactate and succinate, both acting as intermediates in propionate production pathways.98,111–114
It is well documented that the initial composition of an individual's gut microbiota affects the response to dietary fibre supplementation.6,41,115 Despite the large inter-individual differences, the substrate modifications affected the luminal and to a bigger extent the wheat bran-attached microbial community, which is in more intimate contact with the substrate. In general, micronization of unmodified bran caused a significant increase in Bifidobacterium, Clostridium cluster XVIII, Roseburia, Anaerostipes and Blautia. The largest differences, however, were found between micronized bran and P 0–3% or micronized DSPE bran. Enzymatic treatment of bran stimulated Proteobacteria at the expense of Actinobacteria, Bacteroidetes and Firmicutes. The elevated Proteobacteria levels may be the result of the decreased wheat bran fermentability, leading to a decrease in other taxa and a concurrent increase of proteolytic Enterobacteriaceae species in the protein rich, carbohydrate-low medium.116–118 Similar shifts were observed with the P 0–3% fraction, except that the shift towards Enterobacteriaceae was not significant. Our results are in agreement with the selective Proteobacteria enrichment observed on poorly fermentable wheat bran cellulose fractions.119 This finding is relevant from a health perspective as Proteobacterial blooms are a marker for a dysbiotic gut microbiota and have been associated with intestinal inflammation.120–123 Besides, the proteolytic activity displayed by Enterobacteriaceae members produces potentially harmful metabolites, that have been implicated in the development of colorectal cancer, particularly in the distal colon region.118,124–126 Our results suggest that readily fermentable wheat bran products (pearlings 3–6%, micronized and unmodified bran) are most useful to delay this proteolytic fermentation.
Finally, we found that luminal and bran-attached microbial communities clearly differed, confirming our previous results.42 Regardless of the structural modifications, wheat bran was primarily colonised by Clostridium cluster XIVa, and depending on the donor Prevotella, Roseburia, Megamonas, Bifidobacterium and Bacteroides species. The Clostridium cluster XIVa, Roseburia and Lachnospiraceae enrichment was most marked for micronized and unmodified bran. The Megamonas enrichment was particularly pronounced for P 0–3%, while DSPE bran was characterised by a dominant Bacteroides colonisation. As stated in our previous paper, little is known regarding the driving forces and the mechanisms of attachment.42 It can be expected that bacteria with wheat bran degrading enzymes are located in close proximity of the insoluble wheat bran substrate. Besides the attraction of bacteria that can use wheat bran as a nutrient source, flagella might play a role in enabling bacteria to reach the wheat bran substrate. We proposed bacterial cell surface structures (EPS, pili, enzyme systems and adhesins) as possible mechanisms of the attachment to insoluble wheat bran. In this study, using SEM microscopy, substances resembling pilus like structures were identified.
Conclusions
Fermentation of an insoluble substrate, such as wheat bran, by gut microbes is the result of substrate–enzyme interactions, in the first place governed by the substrate accessibility to enzymes. Judged by the modest impact of wheat bran micronization on fermentation activity and microbial community composition, we showed that substrate accessibility is not the limiting factor, which can be explained by the trade-off between surface area and micro-porosity. In addition, we found that SCFA profiles are primarily affected by inter-individual differences in microbial community composition, rather than bran structure. Finally, the micro-environment represents the largest effect on the microbial community, offsetting the influence of both donor and product type, confirming our previous finding of a clearly distinct wheat bran-attached microbial community. The composition of the luminal microbial community is largely donor-dependent, whereas wheat product related shifts become more apparent in the bran-attached community. The latter observation is important as it suggests that wheat bran modifications can steer the microbial community.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors would like to acknowledge Jolien De Paepe for reviewing this manuscript. This research was funded by the Research Foundation Flanders (FWO, Grant id = IWT130028, title = SBO BRANDING) and the Special Research Fund (BOF) Concerted Research Actions (GOA, BOF12/GOA/008) from the Flemish Government. The funding agency did not take part in designing and conducting the experiment, processing or interpreting the data or the submission for publication. The Phenom SEM instrument was supported by funding from Research Foundation Flanders (FWO grant G031416N to FJRM). The Hercules foundation is acknowledged for its financial support in the acquisition of the scanning electron microscope JEOL JSM-7100F equipped with the cryo-transfer system Quorum PP3010T (grant no. AUGE-09-029).References
- F. Bäckhed, R. E. Ley, J. L. Sonnenburg, D. A. Peterson and J. I. Gordon, Host-bacterial mutualism in the human intestine, Science, 2005, 307, 1915–1920 CrossRef PubMed.
- M. A. Conlon and A. R. Bird, The impact of diet and lifestyle on gut microbiota and human health, Nutrients, 2014, 7, 17–44 CrossRef PubMed.
- L. A. David, C. F. Maurice, R. N. Carmody, D. B. Gootenberg, J. E. Button, B. E. Wolfe, A. V. Ling, A. S. Devlin, Y. Varma, M. A. Fischbach, S. B. Biddinger, R. J. Dutton and P. J. Turnbaugh, Diet rapidly and reproducibly alters the human gut microbiome, Nature, 2014, 505, 559–563 CrossRef CAS PubMed.
- K. P. Scott, S. W. Gratz, P. O. Sheridan, H. J. Flint and S. H. Duncan, The influence of diet on the gut microbiota, Pharmacol. Res., 2013, 69, 52–60 CrossRef CAS PubMed.
- H. J. Flint, S. H. Duncan, K. P. Scott and P. Louis, Interactions and competition within the microbial community of the human colon: links between diet and health, Environ. Microbiol., 2007, 9, 1101–1111 CrossRef CAS PubMed.
- G. D. Wu, J. Chen, C. Hoffmann, K. Bittinger, Y. Y. Chen, S. A. Keilbaugh, M. Bewtra, D. Knights, W. A. Walters, R. Knight, R. Sinha, E. Gilroy, K. Gupta, R. Baldassano, L. Nessel, H. Li, F. D. Bushman and J. D. Lewis, Linking long-term dietary patterns with gut microbial enterotypes, Science, 2011, 334, 105–108 CrossRef CAS PubMed.
- N. Efsa, Panel on Dietetic Products and Allergies, Scientific Opinion on Dietary Reference Values for carbohydrates and dietary fibre, EFSA J., 2010, 8, 1462–1539 Search PubMed.
- J. M. Jones, Dietary Fiber Future Directions: Integrating New Definitions and Findings to Inform Nutrition Research and Communication, Adv. Nutr., 2013, 4, 8–15 CrossRef CAS PubMed.
- J. Slavin and D. R. Jacobs, in Nutrition Guide for Physicians, ed. T. Wilson, G. A. Bray, N. J. Temple and M. B. Struble, Humana Press, Totowa, NJ, 2010, pp. 13–24, DOI:10.1007/978-1-60327-431-9_2.
- F. Guillon and M. Champ, Structural and physical properties of dietary fibres, and consequences of processing on human physiology, Food Res. Int., 2000, 33, 233–245 CrossRef.
- J. H. Cummings, Microbial Digestion of Complex Carbohydrates in Man, Proc. Nutr. Soc., 1984, 43, 35–44 CrossRef CAS PubMed.
- E. N. Bergman, Energy Contributions of Volatile Fatty-Acids from the Gastrointestinal-Tract in Various Species, Physiol. Rev., 1990, 70, 567–590 CrossRef CAS PubMed.
- K. O'Sulllivan, The superior benefits of wheat bran fibre in digestive health., Eur. Gastroenterol. Hepatol. Rev., 2012, 8, 90–93 Search PubMed.
- A. Fardet, New hypotheses for the health-protective mechanisms of whole-grain cereals: what is beyond fibre?, Nutr. Res. Rev., 2010, 23, 65–134 CrossRef CAS PubMed.
- Z. Merali, S. R. A. Collins, A. Elliston, D. R. Wilson, A. Kasper and K. W. Waldron, Characterization of cell wall components of wheat bran following hydrothermal pretreatment and fractionation, Biotechnol. Biofuels, 2015, 8, 1–13 CrossRef PubMed.
- C. Antoine, S. Peyron, F. Mabille, C. Lapierre, B. Bouchet, J. Abecassis and X. Rouau, Individual contribution of grain outer layers and their cell wall structure to the mechanical properties of wheat bran, J. Agric. Food Chem., 2003, 51, 2026–2033 CrossRef CAS PubMed.
- L. Stevenson, F. Phillips, K. O'Sullivan and J. Walton, Wheat bran: its composition and benefits to health, a European perspective, Int. J. Food Sci. Nutr., 2012, 63, 1001–1013 CrossRef CAS PubMed.
- S. Hemdane, P. J. Jacobs, E. Dornez, J. Verspreet, J. A. Delcour and C. M. Courtin, Wheat (Triticum aestivum L.) Bran in Bread Making: A Critical Review, Compr. Rev. Food Sci. Food Saf., 2016, 15, 28–42 CrossRef.
- J. M. Lattimer and M. D. Haub, Effects of Dietary Fiber and Its Components on Metabolic Health, Nutrients, 2010, 2, 1266–1289 CrossRef CAS PubMed.
- M. S. Izydorczyk and C. G. Biliaderis, Cereal arabinoxylans: Advances in structure and physicochemical properties, Carbohydr. Polym., 1995, 28, 33–48 CrossRef CAS.
- M. Bunzel, J. Ralph, J. M. Marita, R. D. Hatfield and H. Steinhart, Diferulates as structural components in soluble and insoluble cereal dietary fibre, J. Sci. Food Agric., 2001, 81, 653–660 CrossRef CAS.
- S. Edwards, M. F. Chaplin, A. D. Blackwood and P. W. Dettmar, Primary structure of arabinoxylans of ispaghula husk and wheat bran, Proc. Nutr. Soc., 2003, 62, 217–222 CrossRef CAS PubMed.
- D. Dodd and I. K. O. Cann, Enzymatic deconstruction of xylan for biofuel production, GCB Bioenergy, 2009, 1, 2–17 CrossRef CAS PubMed.
- D. Dodd, R. I. Mackie and I. K. O. Cann, Xylan degradation, a metabolic property shared by rumen and human colonic Bacteroidetes, Mol. Microbiol., 2011, 79, 292–304 CrossRef CAS PubMed.
- D. C. Zhang and W. R. Moore, Effect of wheat bran particle size on dough rheological properties, J. Sci. Food Agric., 1997, 74, 490–496 CrossRef CAS.
- M. E. Seyer and P. Gelinas, Bran characteristics and wheat performance in whole wheat bread, Int. J. Food Sci. Technol., 2009, 44, 688–693 CrossRef CAS.
- S. Hemdane, N. A. Langenaeken, P. J. Jacobs, J. Verspreet, J. A. Delcour and C. M. Courtin, Study of the intrinsic properties of wheat bran and pearlings obtained by sequential debranning and their role in bran-enriched bread making, J. Cereal Sci., 2016, 71, 78–85 CrossRef.
- P. J. Jacobs, S. Hemdane, E. Dornez, J. A. Delcour and C. M. Courtin, Study of hydration properties of wheat bran as a function of particle size, Food Chem., 2015, 179, 296–304 CrossRef CAS PubMed.
- J. E. Dexter and P. J. Wood, Recent applications of debranning of wheat before milling, Trends Food Sci. Technol., 1996, 7, 35–41 CrossRef CAS.
- N. De Brier, S. Hemdane, E. Dornez, S. V. Gomand, J. A. Delcour and C. M. Courtin, Structure, chemical composition and enzymatic activities of pearlings and bran obtained from pearled wheat (Triticum aestivum L.) by roller milling, J. Cereal Sci., 2015, 62, 66–72 CrossRef CAS.
- A. J. M. Brodribb and C. Groves, Effect of Bran Particle-Size on Stool Weight, Gut, 1978, 19, 60–63 CrossRef CAS PubMed.
- W. O. Kirwan, A. N. Smith, A. A. Mcconnell, W. D. Mitchell and M. A. Eastwood, Action of Different Bran Preparations on Colonic Function, BMJ [Br. Med. J.], 1974, 4, 187–189 CrossRef CAS.
- K. L. Wrick, J. B. Robertson, P. J. Van Soest, B. A. Lewis, J. M. Rivers, D. A. Roe and L. R. Hackler, The influence of dietary fiber source on human intestinal transit and stool output, J. Nutr., 1983, 113, 1464–1479 CrossRef CAS PubMed.
- V. Salvador, C. Cherbut, J. L. Barry, D. Bertrand, C. Bonnet and J. Delort-Laval, Sugar composition of dietary fibre and short-chain fatty acid production during in vitro fermentation by human bacteria, Br. J. Nutr., 1993, 70, 189–197 CrossRef CAS PubMed.
- N. M. Anson, R. Havenaar, W. Vaes, L. Coulier, K. Venema, E. Selinheimo, A. Bast and G. R. Haenen, Effect of bioprocessing of wheat bran in wholemeal wheat breads on the colonic SCFA production in vitro and postprandial plasma concentrations in men, Food Chem., 2011, 128, 404–409 CrossRef CAS PubMed.
- A. M. Berggren, I. M. E. Bjorck, G. L. Nyman and B. O. Eggum, Short-Chain Fatty-Acid Content and Ph in Cecum of Rats Given Various Sources of Carbohydrates, J. Sci. Food Agric., 1993, 63, 397–406 CrossRef CAS.
- A. Fardet, F. Guillon, C. Hoebler and J. L. Barry, In vitro fermentation of beet fibre and barley bran, of their insoluble residues after digestion and of ileal effluents, J. Sci. Food Agric., 1997, 75, 315–325 CrossRef CAS.
- S. E. Pryde, S. H. Duncan, G. L. Hold, C. S. Stewart and H. J. Flint, The microbiology of butyrate formation in the human colon, FEMS Microbiol. Lett., 2002, 217, 133–139 CrossRef CAS PubMed.
- V. Lebet, E. Arrigoni and R. Amado, Measurement of fermentation products and substrate disappearance during incubation of dietary fibre sources with human faecal flora, Food Sci. Technol., 1998, 31, 473–479 CAS.
- B. Q. Cheng, R. P. Trimble, R. J. Illman, B. A. Stone and D. L. Topping, Comparative Effects of Dietary Wheat Bran and Its Morphological Components (Aleurone and Pericarp-Seed Coat) on Volatile Fatty-Acid Concentrations in the Rat, Br. J. Nutr., 1987, 57, 69–76 CrossRef CAS PubMed.
- A. W. Walker, J. Ince, S. H. Duncan, L. M. Webster, G. Holtrop, X. L. Ze, D. Brown, M. D. Stares, P. Scott, A. Bergerat, P. Louis, F. McIntosh, A. M. Johnstone, G. E. Lobley, J. Parkhill and H. J. Flint, Dominant and diet-responsive groups of bacteria within the human colonic microbiota, ISME J., 2011, 5, 220–230 CrossRef CAS PubMed.
- K. De Paepe, F.-M. Kerckhof, J. Verspreet, C. M. Courtin and T. Van de Wiele, Inter-individual differences determine the outcome of wheat bran colonization by the human gut microbiome, Environ. Microbiol., 2017, 19, 3251–3267 CrossRef CAS PubMed.
- K. Swennen, C. M. Courtin, G. C. J. E. Lindemans and J. A. Delcour, Large-scale production and characterisation of wheat bran arabinoxylooligosaccharides, J. Sci. Food Agric., 2006, 86, 1722–1731 CrossRef CAS.
- AACC, Approved Methods of the American Association of Cereal Chemists, AACC, Inc., St. Paul, MN, USA, 16th edn, 1999 Search PubMed.
- AOAC, Official Methods of Analysis of the Association of Official Analytical Chemists, The Association of Official Analytical Chemists, Washington, DC, USA, 16th edn, 1995 Search PubMed.
- J. Verspreet, A. Pollet, S. Cuyvers, R. Vergauwen, W. Van den Ende, J. A. Delcour and C. M. Courtin, A Simple and Accurate Method for Determining Wheat Grain Fructan Content and Average Degree of Polymerization, J. Agric. Food Chem., 2012, 60, 2102–2107 CrossRef CAS PubMed.
- L. R. Gerits, B. Pareyt and J. A. Delcour, Single run HPLC separation coupled to evaporative light scattering detection unravels wheat flour endogenous lipid redistribution during bread dough making, LWT–Food Sci. Technol., 2013, 53, 426–433 CrossRef CAS.
- M. Minekus, M. Alminger, P. Alvito, S. Ballance, T. Bohn, C. Bourlieu, F. Carriere, R. Boutrou, M. Corredig, D. Dupont, C. Dufour, L. Egger, M. Golding, S. Karakaya, B. Kirkhus, S. Le Feunteun, U. Lesmes, A. Macierzanka, A. Mackie, S. Marze, D. J. McClements, O. Menard, I. Recio, C. N. Santos, R. P. Singh, G. E. Vegarud, M. S. Wickham, W. Weitschies and A. Brodkorb, A standardised static in vitro digestion method suitable for food - an international consensus, Food Funct., 2014, 5, 1113–1124 RSC.
- P. De Boever, B. Deplancke and W. Verstraete, Fermentation by gut microbiota cultured in a simulator of the human intestinal microbial ecosystem is improved by supplementing a soygerm powder, J. Nutr., 2000, 130, 2599–2606 CrossRef CAS PubMed.
- E. C. M. Leitch, A. W. Walker, S. H. Duncan, G. Holtrop and H. J. Flint, Selective colonization of insoluble substrates by human faecal bacteria, Environ. Microbiol., 2007, 9, 667–679 CrossRef PubMed.
- A. E. Greenberg, L. S. Clesceri and A. D. Eaton, Standard methods for the examination of water and wastewater, 18th edn, 1992 Search PubMed.
- J. C. Araujo, F. C. Teran, R. A. Oliveira, E. A. A. Nour, M. A. P. Montenegro, J. R. Campos and R. F. Vazoller, Comparison of hexamethyidisilazane and critical point drying treatments for SEM analysis of anaerobic biofilms and granular sludge, J. Electron Microsc., 2003, 52, 429–433 CrossRef CAS PubMed.
- A. Klindworth, E. Pruesse, T. Schweer, J. Peplies, C. Quast, M. Horn and F. O. Glockner, Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies, Nucleic Acids Res., 2013, 41, 1–11 CrossRef PubMed.
- J. J. Kozich, S. L. Westcott, N. T. Baxter, S. K. Highlander and P. D. Schloss, Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform, Appl. Environ. Microbiol., 2013, 79, 5112–5120 CrossRef CAS PubMed.
- C. Quast, E. Pruesse, P. Yilmaz, J. Gerken, T. Schweer, P. Yarza, J. Peplies and F. O. Glöckner, The SILVA ribosomal RNA gene database project: improved data processing and web-based tools, Nucleic Acids Res., 2013, 41, D590–D596 CrossRef CAS PubMed.
- R. C. Edgar, B. J. Haas, J. C. Clemente, C. Quince and R. Knight, UCHIME improves sensitivity and speed of chimera detection, Bioinformatics, 2011, 27, 2194–2200 CrossRef CAS PubMed.
- S. L. Westcott and P. D. Schloss, OptiClust, an Improved Method for Assigning Amplicon-Based Sequence Data to Operational Taxonomic Units, mSphere, 2017, 2, e00073–e00017 CrossRef PubMed.
- P. D. Schloss and S. L. Westcott, Assessing and Improving Methods Used in Operational Taxonomic Unit-Based Approaches for 16S rRNA Gene Sequence Analysis, Appl. Environ. Microbiol., 2011, 77, 3219–3226 CrossRef CAS PubMed.
- P. D. Schloss, S. L. Westcott, T. Ryabin, J. R. Hall, M. Hartmann, E. B. Hollister, R. A. Lesniewski, B. B. Oakley, D. H. Parks, C. J. Robinson, J. W. Sahl, B. Stres, G. G. Thallinger, D. J. Van Horn and C. F. Weber, Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities, Appl. Environ. Microbiol., 2009, 75, 7537–7541 CrossRef CAS PubMed.
- W. Chen, C. K. Zhang, Y. Cheng, S. Zhang and H. Zhao, A comparison of methods for clustering 16S rRNA sequences into OTUs, PLoS One, 2013, 8, e70837 CrossRef CAS PubMed.
- X. Y. Wang, Y. P. Cai, Y. J. Sun, R. Knight and V. Mai, Secondary structure information does not improve OTU assignment for partial 16s rRNA sequences, ISME J., 2012, 6, 1277–1280 CrossRef CAS PubMed.
- J. R. Cole, Q. Wang, J. A. Fish, B. L. Chai, D. M. McGarrell, Y. N. Sun, C. T. Brown, A. Porras-Alfaro, C. R. Kuske and J. M. Tiedje, Ribosomal Database Project: data and tools for high throughput rRNA analysis, Nucleic Acids Res., 2014, 42, D633–D642 CrossRef CAS PubMed.
- Q. Wang, G. M. Garrity, J. M. Tiedje and J. R. Cole, Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy, Appl. Environ. Microbiol., 2007, 73, 5261–5267 CrossRef CAS PubMed.
- R Core Team, R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria, https://www.R-project.org/, 2016 Search PubMed.
- P. J. McMurdie and S. Holmes, Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible, PLoS Comput. Biol., 2014, 10, 1–12 CrossRef PubMed.
- S. F. Altschul, W. Gish, W. Miller, E. W. Myers and D. J. Lipman, Basic Local Alignment Search Tool, J. Mol. Biol., 1990, 215, 403–410 CrossRef CAS PubMed.
- F. Cailliez, The Analytical Solution of the Additive Constant Problem, Psychometrika, 1983, 48, 305–308 CrossRef.
- T. F. Cox, Multidimensional scaling used in multivariate statistical process control, J. Appl. Stat., 2001, 28, 365–378 CrossRef.
- J. C. Gower, Some Distance Properties of Latent Root and Vector Methods Used in Multivariate Analysis, Biometrika, 1966, 53, 325–338 CrossRef.
- R. A. Becker, J. M. Chambers and A. R. Wilks, The new S language: a programming environment for data analysis and graphics, Wadsworth and Brooks/Cole Advanced Books & Software, 1988 Search PubMed.
- A. Ramette, Multivariate analyses in microbial ecology, FEMS Microbiol. Ecol., 2007, 62, 142–160 CrossRef CAS PubMed.
- M. J. Anderson, K. E. Ellingsen and B. H. McArdle, Multivariate dispersion as a measure of beta diversity, Ecol. Lett., 2006, 9, 683–693 CrossRef PubMed.
- D. Borcard, F. Gillet and P. Legendre, Numerical Ecology with R, Springer Sciece, 2011 Search PubMed.
- J. Oksanen, G. Blanchet, M. Friendly, R. Kindt, P. Legendre, D. McGlinn, P. R. Minchin, R. B. O'Hara, G. L. Simpson, P. Solymos, H. Stevens, E. Szoecs and H. Wagner, vegan: Community Ecology Package. R package version 2.4-0, https://CRAN.R-project.org/package=vegan, 2016 Search PubMed.
- H. Wickham, ggplot2: Elegant Graphics for Data Analysis, 2009 Search PubMed.
- M. Maechler, P. Rousseeuw, A. Struyf, M. Hubert and K. Hornik, cluster: Cluster Analysis Basics and Extensions. R package version 2.0.4, 2016 Search PubMed.
- M. I. Love, W. Huber and S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol., 2014, 15, 550–571 CrossRef PubMed.
- K. A. Le Cao, F. Rohart, I. Gonzalez, S. Dejean, B. Gautier, F. Bartolo, P. Monget, J. Coquery, F. Yao and B. Liquet, mixOmics: Omics Data Integration Project, R package version 6.0.0, 2016, https://CRAN.R-project.org/package=mixOmics Search PubMed.
- K. A. Le Cao, D. Rossouw, C. Robert-Granie and P. Besse, A Sparse PLS for Variable Selection when Integrating Omics Data, Stat. Appl. Genet. Mol., 2008, 7, 35 Search PubMed.
- M. L. Stewart and J. L. Slavin, Particle size and fraction of wheat bran influence short-chain fatty acid production in vitro, Br. J. Nutr., 2009, 102, 1404–1407 CrossRef CAS PubMed.
- F. Guillon, A. Auffret, J. A. Robertson, J. F. Thibault and J. L. Barry, Relationships between physical characteristics of sugar-beet fibre and its fermentability by human faecal flora, Carbohydr. Polym., 1998, 37, 185–197 CrossRef CAS.
- N. Efsa, Panel on Dietetic Products and Allergies, Scientific Opinion on the substantiation of health claims related to wheat bran fibre and increase in faecal bulk (ID 3066), reduction in intestinal transit time (ID 828, 839, 3067, 4699) and contribution to the maintenance or achievement of a normal body weight (ID 829) pursuant to Article 13(1) of Regulation (EC) No 1924/2006, European Food Safety Authority (EFSA) Panel on Dietetic Products, Nutrition and Allergies (NDA), 2010 Search PubMed.
- D. Vandeputte, G. Falony, S. Vieira-Silva, R. Y. Tito, M. Joossens and J. Raes, Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates, Gut, 2016, 65, 57–62 CrossRef CAS PubMed.
- K. E. B. Knudsen, S. Steenfeldt, C. F. Borsting and B. O. Eggum, The Nutritive-Value of Decorticated Mill Fractions of Wheat .1. Chemical-Composition of Raw and Enzyme-Treated Fractions and Balance Experiments with Rats, Anim. Feed Sci. Technol., 1995, 52, 205–225 CrossRef CAS.
- T. M. Amrein, P. Granicher, E. Arrigoni and R. Amado, In vitro digestibility and colonic fermentability of aleurone isolated from wheat bran, Lebensm.-Wiss. Technol., 2003, 36, 451–460 CrossRef CAS.
- F. Brouns, Y. Hemery, R. Price and N. M. Anson, Wheat Aleurone: Separation, Composition, Health Aspects, and Potential Food Use, Crit. Rev. Food Sci., 2012, 52, 553–568 CrossRef CAS PubMed.
- H. N. Englyst, S. Hay and G. T. Macfarlane, Polysaccharide Breakdown by Mixed Populations of Human Fecal Bacteria, FEMS Microbiol. Ecol., 1987, 45, 163–171 CrossRef CAS.
- J. H. Cummings, G. T. Macfarlane and H. N. Englyst, Prebiotic digestion and fermentation, Am. J. Clin. Nutr., 2001, 73, 415s–420s CrossRef CAS PubMed.
- P. Louis, K. P. Scott, S. H. Duncan and H. J. Flint, Understanding the effects of diet on bacterial metabolism in the large intestine, J. Appl. Microbiol., 2007, 102, 1197–1208 CrossRef CAS PubMed.
- D. Ríos-Covián, P. Ruas-Madiedo, A. Margolles, M. Gueimonde, C. G. de los Reyes-Gavilán and N. Salazar, Intestinal Short Chain Fatty Acids and their Link with Diet and Human Health, Front. Microbiol., 2016, 7, 253–270 Search PubMed.
- G. den Besten, K. Lange, R. Havinga, T. H. van Dijk, A. Gerding, K. van Eunen, M. Muller, A. K. Groen, G. J. Hooiveld, B. M. Bakker and D. J. Reijngoud, Gut-derived short-chain fatty acids are vividly assimilated into host carbohydrates and lipids, Am. J. Physiol.: Gastrointest. Liver Physiol., 2013, 305, G900–G910 CrossRef CAS PubMed.
- E. Puertollano, S. Kolida and P. Yaqoob, Biological significance of short-chain fatty acid metabolism by the intestinal microbiome, Curr. Opin. Clin. Nutr. Metab. Care, 2014, 17, 139–144 CrossRef CAS PubMed.
- H. M. Hamer, D. Jonkers, K. Venema, S. Vanhoutvin, F. J. Troost and R. J. Brummer, Review article: the role of butyrate on colonic function, Aliment. Pharmacol. Ther., 2008, 27, 104–119 CrossRef CAS PubMed.
- S. Macfarlane and G. T. Macfarlane, Regulation of short-chain fatty acid production, Proc. Nutr. Soc., 2003, 62, 67–72 CrossRef CAS PubMed.
- K. P. Scott, J. C. Martin, G. Campbell, C. D. Mayer and H. J. Flint, Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium “Roseburia inulinivorans”, J. Bacteriol., 2006, 188, 4340–4349 CrossRef CAS PubMed.
- J. L. Sonnenburg, J. Xu, D. D. Leip, C. H. Chen, B. P. Westover, J. Weatherford, J. D. Buhler and J. I. Gordon, Glycan foraging in vivo by an intestine-adapted bacterial symbiont, Science, 2005, 307, 1955–1959 CrossRef CAS PubMed.
- C. Engels, H. J. Ruscheweyh, N. Beerenwinkel, C. Lacroix and C. Schwab, The Common Gut Microbe Eubacterium hallii also Contributes to Intestinal Propionate Formation, Front. Microbiol., 2016, 7, 713 Search PubMed.
- H. J. Strobel, Vitamin-B12-Dependent Propionate Production by the Ruminal Bacterium Prevotella-Ruminicola-23, Appl. Environ. Microbiol., 1992, 58, 2331–2333 CAS.
- S. Karppinen, K. Liukkonen, A. M. Aura, P. Forssell and K. Poutanen, In vitro fermentation of polysaccharides of rye, wheat and oat brans and inulin by human faecal bacteria, J. Sci. Food Agric., 2000, 80, 1469–1476 CrossRef CAS.
- M. Aguirre, J. Ramiro-Garcia, M. E. Koenen and K. Venema, To pool or not to pool? Impact of the use of individual and pooled fecal samples for in vitro fermentation studies, J. Microbiol. Methods, 2014, 107, 1–7 CrossRef CAS PubMed.
- E. C. Titgemeyer, L. D. Bourquin, G. C. Fahey and K. A. Garleb, Fermentability of Various Fiber Sources by Human Fecal Bacteria Invitro, Am. J. Clin. Nutr., 1991, 53, 1418–1424 CrossRef CAS PubMed.
- E. Nordlund, A. M. Aura, I. Mattila, T. Kosso, X. Rouau and K. Poutanen, Formation of Phenolic Microbial Metabolites and Short-Chain Fatty Acids from Rye, Wheat, and Oat Bran and Their Fractions in the Metabolical in Vitro Colon Model, J. Agric. Food Chem., 2012, 60, 8134–8145 CrossRef CAS PubMed.
- D. C. Hernot, T. W. Boileau, L. L. Bauer, I. S. Middelbos, M. R. Murphy, K. S. Swanson and G. C. Fahey, In Vitro Fermentation Profiles, Gas Production Rates, and Microbiota Modulation as Affected by Certain Fructans, Galactooligosaccharides, and Polydextrose, J. Agric. Food Chem., 2009, 57, 1354–1361 CrossRef CAS PubMed.
- J. L. Casterline, C. J. Oles and Y. Ku, In vitro fermentation of various food fiber fractions, J. Agric. Food Chem., 1997, 45, 2463–2467 CrossRef CAS.
- A. M. Aura, S. Karppinen, H. Virtanen, P. Forssell, S. M. Heinonen, T. Nurmi, H. Adlercreutz and K. Poutanen, Processing of rye bran influences both the fermentation of dietary fibre and the bioconversion of lignans by human faecal flora in vitro, J. Sci. Food Agric., 2005, 85, 2085–2093 CrossRef CAS.
- L. D. Bourquin, E. C. Titgemeyer, K. A. Garleb and G. C. Fahey, Short-Chain Fatty-Acid Production and Fiber Degradation by Human Colonic Bacteria - Effects of Substrate and Cell-Wall Fractionation Procedures, J. Nutr., 1992, 122, 1508–1520 CrossRef CAS PubMed.
- M. I. Mcburney and L. U. Thompson, Effect of Human Fecal Donor on Invitro Fermentation Variables, Scand. J. Gastroenterol., 1989, 24, 359–367 CrossRef CAS PubMed.
- G. A. Weaver, J. A. Krause, T. L. Miller and M. J. Wolin, Cornstarch Fermentation by the Colonic Microbial Community Yields More Butyrate Than Does Cabbage Fiber Fermentation - Cornstarch Fermentation Rates Correlate Negatively with Methanogenesis, Am. J. Clin. Nutr., 1992, 55, 70–77 CrossRef CAS PubMed.
- R. E. Ley, P. J. Turnbaugh, S. Klein and J. I. Gordon, Microbial ecology - Human gut microbes associated with obesity, Nature, 2006, 444, 1022–1023 CrossRef CAS PubMed.
- A. H. Franks, H. J. M. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut and G. W. Welling, Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-Targeted oligonucleotide probes, Appl. Environ. Microbiol., 1998, 64, 3336–3345 CAS.
- A. Kageyama and Y. Benno, Emendation of genus Collinsella and proposal of Collinsella stercoris sp nov and Collinsella intestinalis sp nov., Int. J. Syst. Evol. Microbiol., 2000, 50, 1767–1774 CrossRef CAS PubMed.
- M. Rajilic-Stojanovic and W. M. de Vos, The first 1000 cultured species of the human gastrointestinal microbiota, FEMS Microbiol. Rev., 2014, 38, 996–1047 CrossRef CAS PubMed.
- C. De Maesschalck, F. Van Immerseel, V. Eeckhaut, S. De Baere, M. Cnockaert, S. Croubels, F. Haesebrouck, R. Ducatelle and P. Vandamme, Faecalicoccus acidiformans gen. nov., sp nov., isolated from the chicken caecum, and reclassification of Streptococcus pleomorphus (Barnes et al. 1977), Eubacterium biforme (Eggerth 1935) and Eubacterium cylindroides (Cato et al. 1974) as Faecalicoccus pleomorphus comb. nov., Holdemanella biformis gen. nov., comb. nov and Faecalitalea cylindroides gen. nov., comb. nov., respectively, within the family Erysipelotrichaceae, Int. J. Syst. Evol. Microbiol., 2014, 64, 3877–3884 CrossRef PubMed.
- P. Kovatcheva-Datchary, A. Nilsson, R. Akrami, Y. S. Lee, F. De Vadder, T. Arora, A. Hallen, E. Martens, I. Bjorck and F. Backhed, Dietary Fiber-Induced Improvement in Glucose Metabolism Is Associated with Increased Abundance of Prevotella, Cell Metab., 2015, 22, 971–982 CrossRef CAS PubMed.
- P. J. Turnbaugh, V. K. Ridaura, J. J. Faith, F. E. Rey, R. Knight and J. I. Gordon, The Effect of Diet on the Human Gut Microbiome: A Metagenomic Analysis in Humanized Gnotobiotic Mice, Sci. Transl. Med., 2009, 1, 1–19 Search PubMed.
- A. J. Richardson, N. McKain and R. J. Wallace, Ammonia production by human faecal bacteria, and the enumeration, isolation and characterization of bacteria capable of growth on peptides and amino acids, BMC Microbiol., 2013, 13, 6 CrossRef CAS PubMed.
- M. Marzorati, R. Vilchez-Vargas, J. V. Bussche, P. Truchado, R. Jauregui, R. A. El Hage, D. H. Pieper, L. Vanhaecke and T. Van de Wiele, High-fiber and high-protein diets shape different gut microbial communities, which ecologically behave similarly under stress conditions, as shown in a gastrointestinal simulator, Mol. Nutr. Food Res., 2017, 61, 1–13 CrossRef PubMed.
- C. K. Yao, J. G. Muir and P. R. Gibson, Review article: insights into colonic protein fermentation, its modulation and potential health implications, Aliment. Pharmacol. Ther., 2016, 43, 181–196 CrossRef CAS PubMed.
- K. D'hoe, L. Conterno, F. Fava, G. Falony, S. Vieira-Silva, J. Vermeiren, K. Tuohy and J. Raes, Prebiotic Wheat Bran Fractions Induce Specific Microbiota Changes, Front. Microbiol., 2018, 9, 31 CrossRef PubMed.
- B. Stecher, L. Maier and W. D. Hardt, ‘Blooming’ in the gut: how dysbiosis might contribute to pathogen evolution, Nat. Rev. Microbiol., 2013, 11, 277–284 CrossRef CAS PubMed.
- N. R. Shin, T. W. Whon and J. W. Bae, Proteobacteria: microbial signature of dysbiosis in gut microbiota, Trends Biotechnol., 2015, 33, 496–503 CrossRef CAS PubMed.
- B. Stecher, R. Denzler, L. Maier, F. Bernet, M. J. Sanders, D. J. Pickard, M. Barthel, A. M. Westendorf, K. A. Krogfelt, A. W. Walker, M. Ackermann, U. Dobrindt, N. R. Thomson and W. D. Hardt, Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 1269–1274 CrossRef CAS PubMed.
- N. Rolhion and B. Chassaing, When pathogenic bacteria meet the intestinal microbiota, Philos. Trans. R. Soc., B, 2016, 371, 1–9 CrossRef PubMed.
- J. H. Cummings and G. T. Macfarlane, The control and consequences of bacterial fermentation in the human colon, J. Appl. Bacteriol., 1991, 70, 443–459 CrossRef CAS PubMed.
- K. Windey, V. De Preter and K. Verbeke, Relevance of protein fermentation to gut health, Mol. Nutr. Food Res., 2012, 56, 184–196 CrossRef CAS PubMed.
- A. M. Davila, F. Blachier, M. Gotteland, M. Andriamihaja, P. H. Benetti, Y. Sanz and D. Tome, Intestinal luminal nitrogen metabolism: role of the gut microbiota and consequences for the host, Pharmacol. Res., 2013, 68, 95–107 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8fo01272e |
| ‡ Current affiliation: Flemish Institute for Technological Research (VITO), Mol, Belgium. |
| This journal is © The Royal Society of Chemistry 2019 |