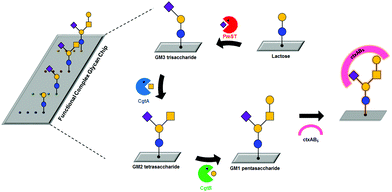
On-chip biosynthesis of GM1 pentasaccharide-related complex glycans†
Chang Sup
Kim‡
 a,
Hye Ryoung
Heo‡
b,
Jeong Hyun
Seo
c and
Hyung Joon
Cha
a,
Hye Ryoung
Heo‡
b,
Jeong Hyun
Seo
c and
Hyung Joon
Cha
 *bd
*bd
aSchool of Chemistry and Biochemistry, Yeungnam University, Gyeongsan 38541, Korea
bSchool of Interdisciplinary Bioscience and Bioengineering, Pohang University of Science and Technology, Pohang 37673, Korea
cSchool of Chemical Engineering, Yeungnam University, Gyeongsan 38541, Korea
dDepartment of Chemical Engineering, Pohang University of Science and Technology, Pohang 37673, Korea. E-mail: hjcha@postech.ac.kr
First published on 27th November 2018
Abstract
A functional glycan chip combined with on-chip enzymatic glycosylation was developed to prepare complex glycan sources and to apply glycan-involved applications simultaneously. GM3 trisaccharide, GM2 tetrasaccharide, and GM1 pentasaccharide were successfully directly biosynthesized on lactose-immobilized surfaces through three consecutive glycosyltransferase reactions along with small amounts of enzymes and donors, without any additional processes. Biosynthesized GM1 pentasaccharide-related complex glycans were demonstrated to provide information on the substrate specificity of whole cholera toxin. Thus, the proposed on-chip glycan biosynthesis system can provide a new direction toward obtaining complex glycan sources and complex glycan-involved applications such as glycan–protein interaction analysis and glycan biomarker-based diagnosis.
Glycan–protein interactions greatly influence a number of biological processes, including cell-to-cell interactions, ligand–receptor recognition, transport of biological molecules, blood group typing, and immune responses.1–3 However, many roles of glycans in biology remain unknown and glycomics is still at an early stage compared to genomics and proteomics.4 The major reason might be the limitation of access to glycan libraries with diverse structures,5,6 which hampers extensive studies on in vivo molecular interactions of glycans. Thus, effective glycan synthesis approaches are required for understanding the biological functions of glycans.
Glycan chips have been used for high-throughput analyses of glycan–protein interactions.7–10 Considering successful applications of on-surface synthesis of oligonucleotides and peptides into chip platforms for genomics and proteomics,11,12 on-chip synthesis of glycans would be an attractive tool for glycomics studies. Previously, 21 disaccharides were constructed on a chip using a chemical synthesis strategy to present a potential on-chip glycan synthesis approach.13 However, this method has difficulty controlling the stereochemistry of glycosidic bond formation, which is a typical drawback of chemical glycan synthesis. Thus, the on-chip chemical synthesis strategy could be feasible for only simple glycans such as disaccharides. This limitation could be circumvented by using glycosyltransferase enzymes that have specific substrate specificity, regio-specificity, and stereo-specificity.14,15
Chip-based platforms could provide obvious advantages for enzymatic glycosylation. First, immobilized carbohydrates with proper space and functional orientation are able to mimic glycans on the membrane surface, and thus, they could interact with glycosyltransferases efficiently. Second, complex glycans can be synthesized by using small amounts of expensive glycosyltransferases and nucleotide donors. Finally, the planar surface commonly used in chip platforms provides high accessibility for the enzyme to immobilized substrates compared to porous polymers used in solid-phase chemistry, including TantaGel™, controlled pore glass, and poly(ethylene glycol) acrylate.16–18 Due to these merits, glycan chips have been widely used for assaying the activities and substrate specificities of glycosyltransferases.9,13,19 These successful applications indicate that the glycan chip platform can be a good tool for the enzymatic synthesis of glycans and biochemical analysis of glycan–protein interactions at the same time. However, glycan chips have rarely been utilized for the enzymatic synthesis of complex glycans possibly due to the difficulty of chip preparation and limited access to commercially available glycosyltransferases; thus, only one case has been reported yet with a limited number and limited structures of glycans.20
Previously, we developed a facile and efficient method for modifying glycans and immobilizing them onto glass or gold.10,21 Here, we developed a functional complex glycan chip combined with enzymatic glycosylations for simultaneous on-chip synthesis and application of complex glycans (Table 1 and Fig. 1). For effective on-chip enzymatic glycosylation, we produced recombinant glycosyltransferases as active forms in the Escherichia coli system, which enables the biosynthesis of diverse complex glycans. Our approach takes advantage of the combination of an on-chip platform with enzymatic glycan biosynthesis without any additional protection/deprotection, purification, and immobilization processes to directly investigate the biological function of complex glycans in a simple and efficient manner. In this work, we synthesized GM1-related complex glycans, including GM1 pentasaccharide, GM2 tetrasaccharide, and GM3 trisaccharide, on lactose disaccharide-immobilized surfaces through sequential enzymatic glycosylation processes; these complex glycans are impossible to biosynthesize using commercially available glycosyltransferases due to the absence of β-1,4-N-acetylgalactosaminyltransferase with GM3 trisaccharide (3-sialyllactose) as an acceptor. These target complex glycans are sialic acid-containing glycan moieties of gangliosides that are involved in adhesion/recognition and signal transduction, and their important functions have resulted in a concerted effort to synthesize them as biological probes.22–24 Then, we analyzed interactions between whole cholera toxin (ctxAB5) and the on-chip-biosynthesized GM1 pentasaccharide-related complex glycans. To the best of our knowledge, this is the first application of an on-chip enzymatic glycosylation approach to biosynthesize GM1 pentasaccharide-related complex glycans on chip and analyze their interactions with ctxAB5 at the same time.
| Glycan | Saccharide | Sequence | Symbola |
|---|---|---|---|
| a Blue circle, glucose (Glc); yellow circle, galactose (Gal); yellow square, N-acetylgalactosamine (GalNAc); purple square, N-acetylneuramic acid (Neu5Ac). | |||
| GM1 | Pentasaccharide | Galβ1-3GalNAcβ1-4(Neu5Acα2-3)Galβ1-4Glc |
|
| GM2 | Tetrasaccharide | GalNAcβ1-4(Neu5Acα2-3)Galβ1-4Glc |
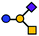
|
| GM3 | Trisaccharide | Neu5Acα2-3Galβ1-4Glc |
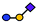
|
| Lactose | Disaccharide | Galβ1-4Glc |
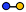
|
Three glycosyltransferases, Pasteurella multocida sialyltransferase (PmST), Campylobacter jejuni N-acetylgalactosaminyltransferase (CgtA), and C. jejuni galactosyltransferase (CgtB), were selected to prepare GM1-related complex glycans directly on chip through enzymatic glycosylation. It was reported that PmST catalyses the transfer of N-acetylneuramic acid (Neu5Ac) from CMP-Neu5Ac to lactose, CgtA transfers an N-acetylgalactosamine (GalNAc) residue from UDP-GalNAc to GM3 trisaccharide, and CgtB contributes to the formation of GM1 pentasaccharide from GM2 tetrasaccharide by transferring a galactose (Gal) residue from UDP-Gal.25,26 The PmST enzyme was purified from cell lysates by using immobilized metal affinity chromatography (see Fig. S1A in ESI†). We obtained highly purified PmST (approx. 99%) with a molecular weight of ∼40 kDa. To produce recombinant CgtA and CgtB enzymes in Escherichia coli, each coded gene was cloned into an expression vector containing a hexa-histidine (His6) affinity tag sequence and an inducible T7 promoter. The expression levels of CgtA and CgtB were not clearly detected on Coomassie blue-stained gel (data not shown), but the enzymes were successfully identified by western blotting (see Fig. S1B and C in ESI†), indicating that the expression levels of these glycosyltransferases are extremely low. However, purification of CgtA and CgtB failed due to their weak binding to Ni2+-NTA resin.
Thus, each soluble fraction from both cell lysates was directly used as a catalyst. Before enzymatic synthesis of GM1-related glycans on chip, we first analysed the activities of recombinant glycosyltransferases through high-performance liquid chromatography (HPLC). All obtained recombinant glycosyltransferases showed expected activities, indicating that the three enzymes were produced as active forms (see Fig. S2 in ESI†).
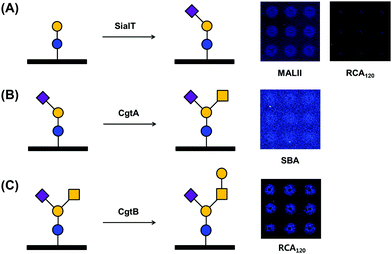
Successful enzymatic glycosylation on glass surfaces enables the direct use of a variety of glycan sources for analyzing glycan–protein interactions. To examine the feasibility of the technique, we performed glycosylation on a glass chip using three glycosyltransferases and checked the glycosylation products by using fluorescence dye-labeled lectins (Fig. 2). We applied Tris-HCl buffer (pH 8.5) containing PmST, CMP-Neu5Ac, and MgCl2 to a lactose disaccharide-immobilized chip and incubated the chip for 12 h at 37 °C. The chip was then washed with buffer and incubated with rhodamine-labeled Maackia amurensis lectin II (MALII) and Ricinus communis agglutinin I (RCA120) lectins. Fluorescence images showed that Neu5Ac-specific MALII bound to the enzymatic product on the chip, while Gal-specific RCA120 did not (Fig. 2A), indicating that GM3 trisaccharide was successfully synthesized on the lactose disaccharide-immobilized chip by PmST glycosyltransferase. Previous studies with PmST have shown a strong preference for Gal residues on N-acetyllactosamine and lactose disaccharide along with high enzyme activity (6000 U L−1).26 This could help to explain the fluorescence intensities of MALII and RCA120 binding after sialylation by PmST. Next, a GM3 trisaccharide-immobilized chip was incubated with HEPES buffer (pH 7.0) containing CgtA, UDP-GalNAc, and MnCl2. Due to the low enzyme activity (67 U L−1) of CgtA,25 the reaction was performed for 4–6 days at 37 °C. The reaction product was reacted with rhodamine-conjugated soybean agglutinin (SBA) lectin that specifically binds to GalNAc residues. We verified that GalNAc residues were successfully transferred to Gal residues on the GM3 trisaccharide-immobilized surface by CgtA glycosyltransferase (Fig. 2B). However, the fluorescence intensities of the microspots were not strong. This result seems to be due to the masking of GalNAc residues by the presence of Neu5Ac residues that are linked α-(2,3) to Gal residues of GM2 tetrasaccharide.27 Finally, CgtB-based glycosylation was performed by incubating CgtB on a GM2 tetrasaccharide-immobilized chip surface in MES buffer (pH 6.0) containing UDP-Gal and MnCl2 for 4–6 days at 37 °C. Rhodamine-labeled RCA120 interacted with the microspots where immobilized GM2 tetrasaccharide had reacted with CgtB glycosyltransferase, indicating that Gal residues were successfully transferred onto GalNAc residues on immobilized GM2 tetrasaccharides (Fig. 2C). Thus, we successfully demonstrated the possibility of using our chip platform to enzymatically synthesize complex glycans on the surface. We also checked that our glycan chip platform could be used as an analytical tool for assaying the substrate specificity of glycosyltransferase.
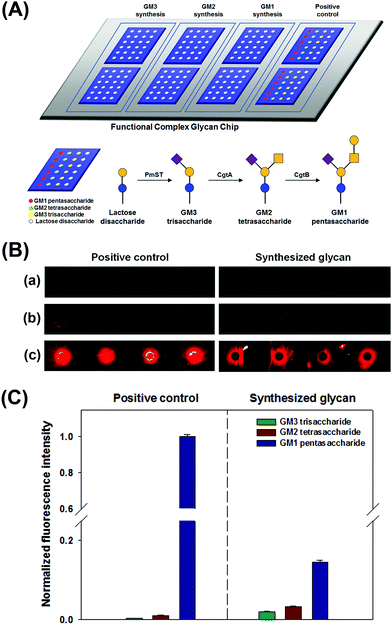
We performed simultaneous syntheses of GM3 trisaccharide, GM2 tetrasaccharide, and GM1 pentasaccharide on a lactose disaccharide-immobilized surface through a series of glycosyltransferase-based reactions. To prepare GM1 pentasaccharide-related glycans on a surface, lactose disaccharide-immobilized surfaces were separated into three blocks using Geneframe® (Fig. 3A). To confirm their syntheses, positive controls (commercially available GM3, GM2, and GM1 glycans) were also immobilized onto the same chip and separated with immobilized lactose disaccharide by using Geneframe® (Fig. 3A). Three blocks were treated with glycosyltransferase(s) to construct GM3, GM2, and GM1 glycans (Fig. 3A).
Then, we analyzed the interactions between ctxAB5 and the GM1-related glycans that were enzymatically synthesized in each block under the same conditions. We checked that the intrinsic selectivity of ctxAB5 to the biosynthesized glycans was identical to that of the positive controls as follows: GM1 pentasaccharide ≫ GM2 tetrasaccharide > GM3 trisaccharide (Fig. 3B and C). This result also indicated that GM1 pentasaccharide and GM2 tetrasaccharide were successfully biosynthesized on lactose disaccharide-immobilized surfaces. The on-chip-biosynthesized GM1 pentasaccharide showed much higher fluorescence intensity than the biosynthesized GM2 tetrasaccharide (Fig. 3B and C), indicating that terminal Gal residues significantly contribute to the interaction between ctxAB5 and GM1 pentasaccharide. However, the difference in fluorescence intensities between the on-chip biosynthesized GM1 pentasaccharide and GM2 tetrasaccharide was smaller than that between positive GM1 pentasaccharide and GM2 tetrasaccharide (Fig. 3B and C). This seems to be due to the low biosynthesis yield of GM1 pentasaccharide, which might originate from low enzyme activity (17 U L−1) and/or decreased accessibility of CgtB.18,25 The biosynthesized GM3–ctxAB5 interaction resulted in very low fluorescence intensity (Fig. 3B and C), which is identical to that of positive GM3 trisaccharide.8 The comparison of the ctxAB5-biosynthesized GM2 interaction with the ctxAB5-biosynthesized GM3 interaction also indicated that the GalNAc residue and two-finger grip formation play important roles in the ctxAB5–glycan interaction; these findings are supported by our previous report that the structural conformation of glycan and constituent terminal monosaccharides of GM1 pentasaccharide significantly affected the interaction.8,28,29
Collectively, we demonstrated the feasibility and validity of an on-chip glycosylation approach that is capable of biosynthesizing complex glycans and analyzing the interactions between directly synthesized glycans and proteins simultaneously. Our platform could mitigate the drawbacks of current glycan synthesis methods that still limit the investigation of many roles of glycans in biology, including expensive and complicated processes. We hope that the new combination of on-chip enzymatic synthesis and analysis of glycan-related interactions demonstrated herein will open a way for the understanding of glycan-related biomolecular interactions. However, this study has limitations in glycan biosynthesis yield and quantitative analysis of glycosylation yield. Increasing synthesis yield remains a major challenge in the field of glycan synthesis. Thus, we are working on further studies to devise a solution for increasing and quantitatively measuring the amount of glycan biosynthesized on a solid surface by engineering glycosyltransferases to enhance their amounts and activities and combining with a microfluidic system to easily collect the synthesized glycan and recycle the enzymes. These studies will have a significant impact on the on-chip biosynthesis of complex glycans and applications of glycan chips.
Financial support was provided by the Basic Core Technology Development Program for the Oceans and the Polar Regions (NRF-2015M1A5A1037055) of the National Research Foundation funded by the Ministry of Science and ICT (to H. J. Cha & C. S. Kim) and the Basic Science Research Program (NRF-2016R1D1A3B03933824) of the National Research Foundation funded by the Ministry of Education, Korea (to C. S. Kim). We thank Dr Y. K. Jo (POSTECH) for helping with the illustrations.
Conflicts of interest
There are no conflicts to declare.Notes and references
- K. Ohtsubo and J. D. Marth, Cell, 2006, 126, 855–867 CrossRef CAS PubMed.
- N. Sharon, Biochim. Biophys. Acta, Gen. Subj., 2006, 1760, 527–537 CrossRef CAS PubMed.
- J. D. Marth and P. K. Grewal, Nat. Rev. Immunol., 2008, 8, 874–887 CrossRef CAS PubMed.
- P. H. Seeberger and D. B. Werz, Nature, 2007, 446, 1046–1051 CrossRef CAS PubMed.
- A. Bartolozzi and P. H. Seeberger, Curr. Opin. Struct. Biol., 2001, 11, 587–592 CrossRef CAS PubMed.
- K. C. Nicolaou and H. J. Mitchell, Angew. Chem., Int. Ed., 2001, 40, 1576–1624 CrossRef CAS PubMed.
- P. H. Liang, C. Y. Wu, W. A. Greenberg and C. H. Wong, Curr. Opin. Chem. Biol., 2008, 12, 86–92 CrossRef CAS PubMed.
- C. S. Kim, J. H. Seo and H. J. Cha, Anal. Chem., 2012, 84, 6884–6890 CrossRef CAS PubMed.
- O. Blixt, K. Allin, O. Bohorov, X. F. Liu, H. Andersson-Sand, J. Hoffmann and N. Razi, Glycoconjugate J., 2008, 25, 59–68 CrossRef CAS PubMed.
- H. Seo, C. S. Kim, B. H. Hwang and H. J. Cha, Nanotechnology, 2010, 21, 215101–215108 CrossRef PubMed.
- S. P. Fodor, J. L. Read, M. C. Pirrung, L. Stryer, A. T. Lu and D. Solas, Science, 1991, 251, 767–773 CrossRef CAS PubMed.
- R. Frank, Tetrahedron, 1992, 48, 9217–9232 CrossRef CAS.
- L. Ban and M. Mrksich, Angew. Chem., Int. Ed., 2008, 47, 3396–3399 CrossRef CAS PubMed.
- C. Breton, L. Snajdrova, C. Jeanneau, J. Koca and A. Imberty, Glycobiology, 2006, 16, 29r–37r CrossRef CAS PubMed.
- A. Trincone and A. Giordano, Curr. Org. Chem., 2006, 10, 1163–1193 CrossRef CAS.
- M. Meldal, Biopolymers, 2002, 66, 93–100 CrossRef CAS PubMed.
- J. Kress, R. Zanaletti, A. Amour, M. Ladlow, J. G. Frey and M. Bradley, Chem. – Eur. J., 2002, 8, 3769–3772 CrossRef CAS.
- N. Laurent, R. Haddoub and S. L. Flitsch, Trends Biotechnol., 2008, 26, 328–337 CrossRef CAS PubMed.
- S. Park and I. Shin, Org. Lett., 2007, 9, 1675–1678 CrossRef CAS PubMed.
- S. Park, M. R. Lee, S. J. Pyo and I. Shin, J. Am. Chem. Soc., 2004, 126, 4812–4819 CrossRef CAS.
- J. H. Seo, K. Adachi, B. K. Lee, D. G. Kang, Y. K. Kim, K. R. Kim, H. Y. Lee, T. Kawai and H. J. Cha, Bioconjugate Chem., 2007, 18, 2197–2201 CrossRef CAS.
- S. K. O. Lloyd and K. Furukawa, Glycoconjugate J., 1998, 15, 627–636 CrossRef.
- S. I. Hakomori, Glycoconjugate J., 2001, 17, 143–151 CrossRef.
- D. K. Ress and R. J. Linhardt, Curr. Org. Chem., 2004, 1, 31–46 CAS.
- O. Blixt, D. Vasiliu, K. Allin, N. Jacobsen, D. Warnock, N. Razi, J. C. Paulson, S. Bernatchez, M. Gilbert and W. Wakarchuk, Carbohydr. Res., 2005, 340, 1963–1972 CrossRef CAS PubMed.
- H. Yu, H. Chokhawala, R. Karpel, H. Yu, B. Wu, J. Zhang, Y. Zhang, Q. Jia and X. Chen, J. Am. Chem. Soc., 2005, 127, 17618–17619 CrossRef CAS.
- T. K. Dam, T. A. Gerken, B. S. Cavada, K. S. Nascimento, T. R. Moura and C. F. Brewer, J. Biol. Chem., 2007, 282, 28256–28263 CrossRef CAS.
- J. H. Seo, C. S. Kim and H. J. Cha, Analyst, 2013, 138, 6924–6929 RSC.
- J. H. Seo, C. S. Kim, H. Y. Lee, T. Kawai and H. J. Cha, Anal. Chem., 2011, 83, 6011–6017 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8cc06526h |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |