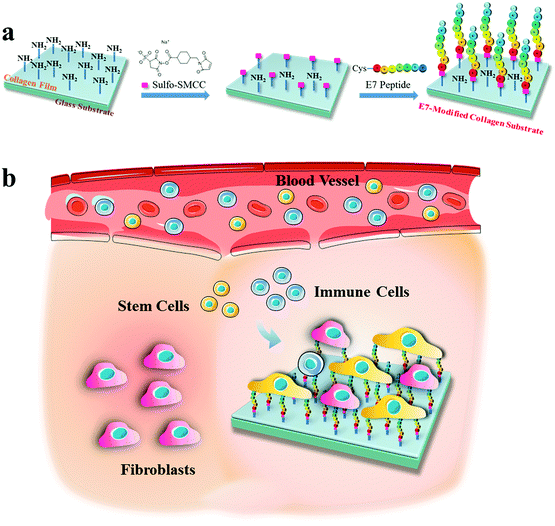
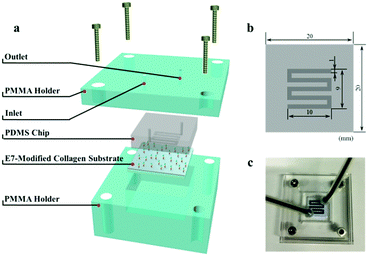
Selective capture of mesenchymal stem cells over fibroblasts and immune cells on E7-modified collagen substrates under flow circumstances†
Xiaowen
Zheng
,
Xin
Pan
,
Qian
Pang
,
Chong
Shuai
,
Lie
Ma
 * and
Changyou
Gao
* and
Changyou
Gao

MOE Key Laboratory of Macromolecular Synthesis and Functionalization, Department of Polymer Science and Engineering, Zhejiang University, Hangzhou 310027, China. E-mail: liema@zju.edu.cn
First published on 4th December 2017
Abstract
Recruitment of endogenous mesenchymal stem cells (MSCs) has become an attractive strategy for in situ tissue regeneration. However, it is of great importance to endow an implant with a specific affinity to MSCs, for many types of cells such as immune cells and fibroblasts can also be recruited. It has been demonstrated that E7 peptides have a specific affinity to MSCs, but their selectivity for MSCs when co-cultured with other cells, especially in flow conditions, has rarely been investigated. In this study, E7-modified collagen substrates were prepared using sulfosuccinimidyl 4-(N-maleimidomethyl) cyclohexane-1-carboxylate (sulfo-SMCC) as the coupling agent. The results of X-ray photoelectron spectroscopy (XPS) and quartz crystal microbalance (QCM) proved that the densities of the immobilized E7 peptides could be modulated by changing the amounts of sulfo-SMCC. The results of cell adhesion rate, adhesion area and adhesion force demonstrated that the immobilization of E7 peptides led to a significant enhancement of the adhesion of bone marrow-derived MSCs (BMSCs) compared to RAW264.7 cells and NIH3T3 cells. The selective adhesion was verified by co-culturing BMSCs with RAW264.7 cells and NIH3T3 cells, which indicated that higher proportions of BMSCs were adhered on the E7-immobilized substrates. By mimicking in vivo flow circumstances, the selective capture of BMSCs by the E7-modified substrates was revealed by a flow model. All these results suggest that E7 immobilization might be a promising strategy for an implant to achieve a better regeneration outcome by enhancing the affinity to the recruited MSCs.
Introduction
Mesenchymal stem cells (MSCs) have attracted much attention in the past several decades in the field of tissue engineering and regenerative medicine owing to their pluripotency, low immunogenicity, and expansion potential.1 However, there are many limitations in the applications of the ex vivo expanded MSCs, the expansion process of which is time-consuming and causes phenotype loss.2 Moreover, it has been manifested that more than 90% of the engrafted MSCs were dead after 4 days’ implantation,3–5 and only a few of them were directly involved in tissue remodeling. It is believed that the implanted MSCs played a role mainly in forming a microenvironment via paracrine signaling to modulate the immune response and recruit the endogenous stem cells involved in the regeneration process.6,7 Therefore, in situ tissue regeneration has been a promising approach via recruiting endogenous MSCs to tissue defects and awakening the intrinsic repair potential of the body.Although the human body has inherent mechanisms to recruit stem cells to a damaged tissue, these spontaneous recruitment processes are often inefficient to meet the requirements of tissue regeneration.8 Therefore, it has become a critical issue to recruit sufficient endogenous MSCs in an efficient way. To solve this problem, many strategies have been developed by the application of chemoattractants9,10 like substance P (SP),11–13 vascular endothelial growth factor (VEGF),14 and stromal derived factor-1 alpha (SDF-1α).15,16 Among them, SDF-1α, a member of the CXCL12 family, has a specific affinity to CXCR4 expressed by MSCs and therefore facilitates MSC recruitment and has been widely applied in in situ regeneration of many kinds of tissues.17
However, it has been shown that SDF-1α recruits not only MSCs, but also fibroblasts and immune cells (e.g. macrophages and monocytes) due to their high expression of CXCR4.18,19 The application of SDF-1α might result in fibrosis due to the unspecific recruitments of fibroblasts and monocytes.20 Therefore, it is vital to construct a surface with selective adhesion of MSCs over fibroblasts and immune cells to result in a regenerative process other than fibrosis. Collagen is a prevalently used biomaterial due to its good biocompatibility and biodegradability. However, the interactions between collagen and many kinds of cell receptors such as integrin, OSCAR, GPR56, LAIR-1, and GPVI, which are widely expressed by fibroblasts and immune cells, would activate the signaling cascades to promote fibrosis, which results in failure to restore tissue structures and functions.21 Therefore, it is of great importance to modify collagen implants to achieve specific affinity for MSCs over fibroblasts and immune cells.
Recently, a peptide with the sequence of EPLQLKM (E7) was identified to have a specific affinity to MSCs.22 Wang et al. demonstrated that an E7-immobilized collagen scaffold could accelerate the healing rate and promote angiogenesis in a porcine model, which was mainly attributed to its MSC affinity.23 However, few investigations have been conducted on the selective adhesion of MSCs over fibroblasts and immune cells by the immobilization of E7 peptides. Furthermore, for the mobilized endogenous MSCs that are mainly recruited through vessel networks,24 it is more significant to evaluate the selective capture under flow circumstances, to which little attention has been devoted so far.
In this study, mimicking the situation of a collagen implant in trauma, collagen substrates modified by E7 are constructed to realize the selective adhesion of MSCs over fibroblasts and immune cells (Scheme 1). Collagen substrates with different E7 densities were obtained by modulating the concentration of sulfosuccinimidyl-4-(N-maleimidomethyl) cyclohexane-1-carboxylate (sulfo-SMCC). The effects of the E7-modified collagen substrates on MSC adhesion rate, adhesion area and adhesion force were investigated in comparison with that of NIH3T3 cells and RAW264.7 cells. The adhesion selectivity and the selective capture under flow circumstances were evaluated by the co-culture of BMSCs, NIH3T3 cells and RAW264.7 cells in static and flow models, respectively.
Experimental
Materials
Collagen type I was isolated from fresh bovine tendon by trypsin digestion and acetic acid dissolution as described previously.25 E7 peptides (EPLQLKM, purity >98%) were customized from GL Biochem Ltd (China), and an extra cysteine was linked at the amino terminal of glutamine acid (E) to facilitate further conjugation. sulfo-SMCC was purchased from Thermo Fisher Scientific (Waltham, MA, USA).Preparation of the E7-modified collagen substrates
Collagen I was dissolved in 3% acetic acid at 37 °C for 24 h to form a 0.5% (w/v) solution. Collagen substrates were prepared by spin-coating of the collagen solution onto glass slides (1.0 cm × 1.0 cm) for further modification. The E7 conjugation was performed according to the previous literature reports22,26,27 with some modifications. The collagen substrates were washed twice with the activation buffer (0.1 M PBS containing 0.15 M NaCl, pH 7.2). In order to investigate the relationship between E7 density and cell adhesion behavior, different dosages of sulfo-SMCC of 0.25, 0.5, and 1 mg per slice were used to prepare collagen substrates with different E7 densities, which were named as CP1, CP2 and CP3 correspondingly. Accordingly, the collagen substrates were incubated in 500 μL sulfo-SMCC solution at different concentrations for 1 h at room temperature, and washed twice with the conjugation buffer (the activation buffer containing 0.1 M EDTA, pH 7.0), then soaked in the E7 solution (0.2 mg mL−1) for 2 h at 37 °C. Meanwhile a pure collagen substrate and a collagen substrate soaked in E7 solution without sulfo-SMCC were chosen as controls, named Col and Col/E7, respectively. The E7-modified collagen substrates were then washed thrice with Milli-Q (Q-Gard 1, Merck Millipore, France) and dried with nitrogen gently.Characterization of the E7-modified collagen substrates
The surface chemical compositions of the substrates were verified by X-ray photoelectron spectroscopy (XPS, ESCALAB250Xi, Thermo Scientific, USA) with a scanning intersection angle of 90° and a circular scanning radius of 250 μm. The binding energy was normalized by setting the lowest binding energy of the C 1s peak at 284.8 eV.Quartz crystal microbalance (QCM) chips spin-coated with collagen were firstly treated with the different concentrations of sulfo-SMCC at 37 °C for 2 h and then used for QCM testing. During the tests, E7 solution was slowly introduced at a rate of 45 μL min−1 at 37 °C. The mass of the conjugated E7 was quantified by QCM-D (QSense E4, Biolin Scientific, Sweden). The wettability of the E7-modified collagen substrates was analyzed in a sessile-drop model by static water contact angle measurement (DSA 100, Krüss, Germany). The volume of each droplet was 2 μL and each measurement was repeated five times.
Cell culture
Bone marrow-derived MSCs (BMSCs) were isolated from Sprague-Dawley (SD) male rats aged from 6 to 8 weeks with the methods reported previously.28 The animals for BMSC isolation were approved by the Institutional Animal Care and Use Committee of Zhejiang University and used in accordance with the guidelines for animal experimentation. Briefly, the rats were killed by cervical dislocation. Then, the animal skeleton was rinsed freely in 70% ethanol. An incision was made around the perimeter of the hind limbs, which were then dissected from the trunk of the body. After carefully removing the attached muscles and connective tissues, the bone marrow was harvested by flushing femurs and tibias with complete medium (α-MEM supplemented with 10% fetal bovine serum (FBS, Gibco, Life Technologies, New York, USA), 100 U mL−1 penicillin and 100 μg mL−1 streptomycin) into the marrow cavity. Then the harvested cells were cultured in a 9 cm cell culture dish (Corning, USA) and incubated at 37 °C under 5% CO2. After 3 days, non-adherent cells were removed by 2–3 washes with PBS and the adherent cells were further cultured in fresh medium until the cells were confluent. At about 80–90% confluence, the adherent cells were retrieved by trypsinization and then serially subcultured. The harvested BMSCs at passage 2–3 were used in this study. NIH3T3 cells and RAW264.7 cells were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). NIH3T3 cells and RAW264.7 cells were maintained in high glucose Dulbecco's modified Eagle's medium (DMEM, GENOM, China), supplemented with 10% FBS, 100 μg mL−1 streptomycin, and 100 U mL−1 penicillin.The E7-modified collagen substrates were placed in 24 well plates and sterilized in 75% ethanol for 5 min and rinsed 10 times with sterile phosphate buffered saline (PBS, pH 7.4) for subsequent cell culture.
Cell adhesion assay
BMSCs, NIH3T3 cells and RAW264.7 cells were seeded on the collagen substrates at a density of 3 × 104 cells per cm2. After being cultured for 4 h, the cells were then detached in 0.5% trypsin and 0.53 mM Ethylene Diamine Tetraacetic Acid (EDTA) for 3 min and resuspended. The cells were collected by centrifugation at 1200 rpm for 5 min and were resuspended in 60 μL DMEM and counted by a hemacytometer. The cell adhesion rate was calculated by divided the number of adhered cells by the cell seeding number. The cell adhesion force was measured by a centrifugation method following Reyes's work.29 The cell suspension was centrifuged and rinsed with serum-free medium 3 times to remove residual esterase. Then the suspension was mixed with 0.6 μg mL−1 calcein-AM (Yeasen, China) for 25 min at 37 °C and washed with serum-free medium 3 times. Each kind of cell was seeded on the E7-modified collagen substrates at a density of 1.5 × 104 cells per well, respectively. After 3 h, the substrates were washed with PBS (pH 7.4) 3 times to remove the floating cells and observed under a fluorescence microscope (IX81, Olympus, Japan) to count the number of adhered cells in each view. Then the substrates were placed upside down on the bottom of a centrifuge tube filled with PBS and centrifuged at two different speeds (400 rpm and 800 rpm for RAW264.7 cells, 500 rpm and 1000 rpm for BMSCs, and 600 rpm and 1200 rpm for NIH3T3 cells, respectively) for 5 min. Then the cell numbers were counted again under a fluorescence microscope to calculate the ratios of the adhered cells before and after centrifugation. Finally, the adhesion force was calculated according to the formula described in Reyes's work.To measure the cell adhesion areas, the cells cultured on the collagen substrates for 3 h and 24 h were stained with 0.6 μg mL−1 calcein-AM, respectively. Then the cell images were taken under a fluorescence microscope, and the cell areas were measured using Image Pro Plus software (Media Cybernetics, USA).
Cell morphology
Cell morphologies on the collagen substrates were visualized under a confocal laser scanning microscope (CLSM, LSM-510, Zeiss, Germany). BMSCs, RAW264.7 cells and NIH3T3 cells were seeded on the E7-modified substrates at a density of 5 × 103 cells per cm2 and cultured for 3 h. Then the cells were fixed with 4% paraformaldehyde (Sinopharm Chemical Reagent Co., Ltd China) at 37 °C for 15 min, washed with PBS 3 times and permeabilized using 0.5% Triton X-100 (Sigma-Aldrich, USA) for 10 min. After being rinsed with PBS thrice, the samples were incubated in a 1% bovine serum albumin (BSA, AMRESCO, USA)/PBS solution for 30 min at 37 °C, Then the cells were washed thrice and stained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich, USA) and rhodamine-labeled phalloidin (Thermo Fisher Scientific, USA) at room temperature for 30 min. After being rinsed thrice in PBS, the cells were observed under a CLSM.Selective adhesion assay
To study the selective adhesion property of the E7-modified collagen substrate, BMSCs, NIH3T3 cells and RAW264.7 cells were co-cultured by low glucose Dulbecco's Modified Eagle Medium (DMEM, Gilbco, USA) containing 10% FBS. The co-cultured cell suspensions at a density of 5 × 103 cells per cm2 for each cell were seeded onto the collagen substrates. Here, BMSCs, NIH3T3 cells and RAW264.7 cells were pre-stained with 0.6 μg mL−1 calcein-AM, 2 × 10−6 M PKH26 (Sigma-Aldrich, USA) and 0.5 μg mL−1 Hoechst 33342 (Beyotime, China) according to the manuals or literature reports,30,31 respectively. After being cultured for 3 h, the proportions of each kind of cell were calculated by the cell number counted under a fluorescence microscope.Cell selective capture in a flow model
To test the ability of cell selective capture under dynamic conditions, a flow model was designed.32 As described in Scheme 2, the flow model is composed of three parts: (i) a PDMS chip with serpentine microfluidic channels (1 mm wide, 0.2 mm high), (ii) the E7-modified collagen substrate (2.0 cm × 2.0 cm), and (iii) the upper and lower PMMA holders. The rectangular microfluidic channel was connected with two through holes at both ends of the channel for tubing connection.Prior to each test, the flow model was sterilized with 75% alcohol, and the microfluidic channel was washed with 1 mL low glucose DMEM using a syringe pump (WZS-50F2 from Zhejiang University Medical Instrument, Hangzhou, China). BMSCs, NIH3T3 cells and RAW264.7 cells were pre-stained with 0.6 μg mL−1 calcein-AM, 2 × 10−6 M PKH26 and 0.5 μg mL−1 Hoechst 33342, respectively and cultured in low glucose DMEM. Then, 2.5 mL mixed cell suspensions at a density of 2 × 104 cells per mL for each kind of cell were flowed through the microfluidic channel at a rate of 0.5 mL h−1 (0.7 × 10−3 m s−1 in average). Then, 200 μL PBS was pumped through followed by 0.5 mL of 4% paraformaldehyde to fix the captured cells. Finally, the flow model was disassembled and the substrates were observed under a fluorescence microscope. The proportions of the captured cells were calculated by image analysis with Image Pro Plus software.
Statistical analysis
At least three independent experiments were carried out to get reproducible data if not specifically stated. Experimental data were expressed as mean ± standard deviation (SD) and the significant differences were analyzed by one way ANOVA (SPSS 16.0) using SPSS software (SPSS Inc., Chicago, USA). The significance level of p < 0.05 was chosen for all of the tests.Results and discussion
Characterization of E7-modified collagen substrates
The compositions of the collagen substrates with/without E7 modification were characterized by XPS (Fig. 1a). It showed that the sulfur-to-carbon (S/C) ratio of Col was 0.589% and increased slightly to 0.610% for Col/E7, which indicated that only a small amount of E7 was physically adsorbed on the collagen substrates. As for CP, E7 peptides were covalently bound on the substrates under the crosslinking of sulfo-SMCC, resulting in a S/C ratio of 0.741%, which was significantly higher than those of Col and Col/E7. With the increasing concentrations of sulfo-SMCC, i.e. CP2 and CP3, the S/C ratios increased to 0.959% and 1.178%, respectively. According to the amino acid composition of collagen I (Table S1, ESI†) and E7 peptides, the S/C ratio of E7 peptides is 19 times higher than that of collagen I. It is supposed that the increase of the S/C ratio could be attributed to the physical adsorption (Col/E7) and covalent binding (CP) of E7. It should be noted that the sulfonic acid groups are detached after the reaction of amine and sulfo-SMCC, and the unreacted sulfo-SMCC can be flushed by plenty of Milli-Q water. Therefore, the sulfo-SMCC crosslinking treatment has no effect on the increase of the S/C ratios.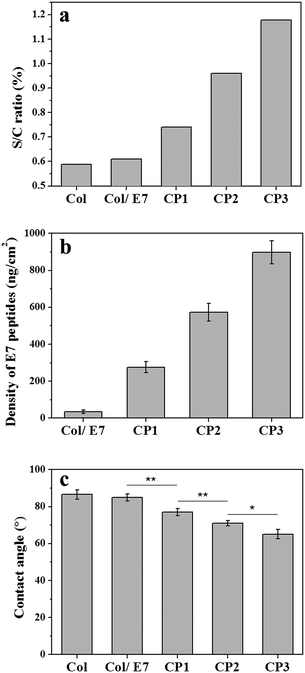 | ||
| Fig. 1 The effects of the sulfo-SMCC amount on (a) the S/C ratio, (b) the E7 densities, and (c) the static water contact angles of the E7-modified collagen substrates. | ||
The immobilized E7 was analyzed quantitatively by QCM-D (Fig. S1, ESI†). It shows that the resonance frequency continuously went down and then reached a plateau after injecting the E7 solution for 1.5 h, and then went up after washing by water and finally reached to a platform again. From the frequency decease, the amounts and thereby the densities of the immobilized E7 were calculated (Fig. 1b). It shows that the E7 densities increased along with the sulfo-SMCC concentrations (CP1, CP2, and CP3) and reached to 275, 573, and 897 ng cm−2, respectively, which are markedly higher than that of Col/E7 (34 ng cm−2). The growth trend of the E7 densities is in good agreement with the XPS data. Both XPS and QCM results indicate that the amount of immobilized E7 can be modulated by changing the sulfo-SMCC concentrations. The E7 immobilization was also proved intuitively by the fluorescence images of the FITC-E7 modified collagen substrates, which showed stronger green fluorescence with the concentration increase of sulfo-SMCC (Fig. S2, ESI†). We also investigated the static water contact angles of the E7-modified substrates (Fig. 1c). The contact angles of Col were about 87°, indicating a relatively hydrophobic surface. There was no significant difference in the contact angles between Col and Col/E7. However, the contact angles of the E7-modified substrates decreased from 77° to 64° along with the increasing concentration of sulfo-SMCC. It is known that E7 peptides are more hydrophilic than collagen I. The decrease of the contact angles should be attributed to the introduction of E7 peptides. Moreover, it is reported that the contact angle of a substrate ranging from 40° to 70° is more suitable for cell adhesion.33
Cell adhesion behaviors
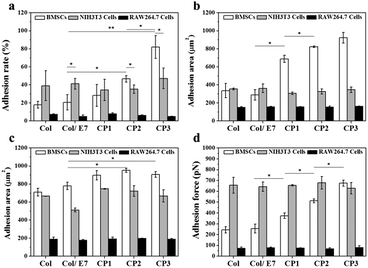
A series of cascade cell behaviors involving adhesion, spreading and proliferation occur when the interaction between cells and substrates occurs. Among these behaviors, cell adhesion lays the foundation for subsequent cell behaviors. In this study, the adhesion behaviors of BMSCs, NIH3T3 cells and RAW264.7 cells on the E7-modified collagen substrates were compared from the aspects of adhesion rate, adhesion force and adhesion area, respectively. First, the results of adhesion rates are shown in Fig. 2a. The adhesion rates of BMSCs on Col (18.0%), Col/E7 (20.7%) and CP1 (28.4%) did not show significant differences, which means that the physically adsorbed E7 and the covalently bonded E7 with low density are not sufficient to increase the adhesion rate of BMSCs. However, the adhesion rate of BMSCs on CP2 (46.7%) showed a significant increase compared to that on Col/E7 (p < 0.05). The highest adhesion rate of BMSCs was obtained on CP3 (82.0%), which is 4 times higher than that of Col/E7. In contrast, the adhesion rates of NIH3T3 cells on all the substrates fluctuated around 40% and no significant difference was observed among the different substrates. It is worth mentioning that the adhesion rates of NIH3T3 cells (38.9% and 41.2%) were higher than those of BMSCs on Col and Col/E7, but they were significantly lower than those of BMSCs on CP2 and CP3. RAW264.7 cells showed the lowest adhesion rates on all substrates when compared to BMSCs and NIH3T3 cells.The areas of the cells adhered for 3 h were investigated (Fig. 2b). On Col, BMSCs had relatively low adhesion areas (336 μm2), which had no obvious difference with those on Col/E7. However, with the increase of the E7 densities, the adhesion areas of BMSCs increased sharply to 715 μm2 (CP1), 817 μm2 (CP2), and 931 μm2 (CP3), respectively. For NIH3T3 cells, no significant difference was observed among the different substrates (ranging from 306 μm2 to 362 μm2). Although the adhesion areas of BMSCs and NIH3T3 cells did not show a significant difference in Col and Col/E7, BMSCs possessed higher adhesion areas than NIH3T3 cells on all the CP substrates. In contrast, RAW.264.7 cells showed the lowest and almost constant adhesion areas on all the substrates (ranging from 150 μm2 to 162 μm2). It was demonstrated that BMSCs on CP have significantly higher spreading velocities than NIH3T3 cells and RAW264.7 cells.
The 24 h-adhesion areas of BMSCs, NIH3T3 cells and RAW 264.7 cells were also investigated (Fig. 2c). Except for CP3, the adhesion areas of BMSCs on all the other substrates became larger when the culture time prolonged to 24 h. Although the adhesion areas of NIH3T3 cells and RAW264.7 cells also augmented with the increasing culture time, BMSCs always showed larger adhesion areas than NIH3T3 cells and RAW264.7 cells on all the substrates except for Col. Similar to the results of 3 h-adhesion areas, there was still no difference among those of RAW264.7 cells on all the substrates. Interestingly, the adhesion area of BMSCs on CP3 at 3 h was similar to that at 24 h, implying that the immobilized E7 peptides can accelerate the spreading of BMSCs significantly to reach a fully spreading state at 3 h. Comparably, no accelerating effect was observed on the spreading of NIH3T3 cells and RAW264.7 cells.
Furthermore, we studied the effects of the immobilized E7 on the adhesion forces of three kinds of cells (Fig. 2d). It shows that NIH3T3 cells had the highest adhesion forces (∼650 pN) on almost all the substrates except for those on CP3, which may be attributed to the binding sites such as RGD on collagen substrates. Comparably, RAW264.7 cells showed the lowest and constant adhesion forces on all the substrates (∼75 pN). This indicates that the introduction of E7 did not significantly enhance the adhesion force of NIH3T3 cells and RAW264.7 cells. As for the adhesion forces of BMSCs, there was no significant difference between Col and Col/E7, while they increased steadily from 244 pN (Col/E7) to 675 pN (CP3) with the increase of the E7 densities, indicating that the covalently grafted E7 can significantly enhance the adhesion forces of BMSCs. It is worth noting that on CP3, the adhesion force of BMSCs became similar to that of NIH3T3 cells. Although NIH3T3 cells showed a stronger interaction with the unmodified collagen substrates (Col), due to the specific interactions of E7 peptides to BMSCs, the CP substrates specifically provided additional recognition sites for BMSCs to increase their adhesion forces.
Cell morphologies
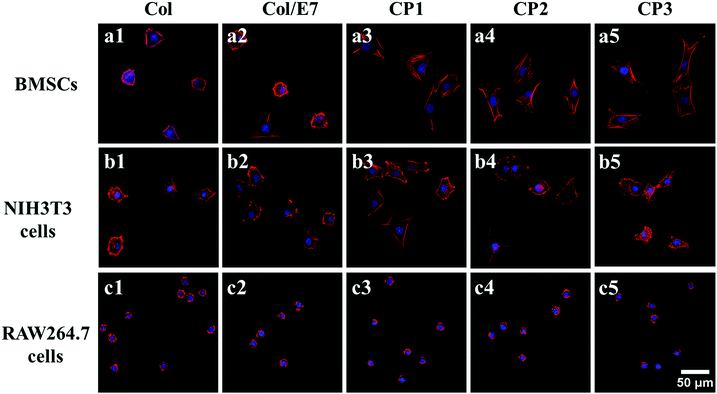
Cell morphologies visualized under CLSM are presented in Fig. 3 The BMSCs on Col showed contractive morphologies, the cytoskeletons of which were not clear. There was no obvious difference between the morphologies of BMSCs on Col and Col/E7 (Fig. 3(a1 and a2)). In contrast, BMSCs on CP substrates were spindle-shaped and exhibited good spreading morphologies with clear and aligned cytoskeletons. No significant effect of the E7 densities was observed on the morphologies of BMSCs (Fig. 3(a3–a5)). As for NIH3T3 cells, their morphologies remained almost unchanged, and there was no obvious association with the E7 density. On all the substrates, RAW264.7 cells kept similar, i.e. round, morphologies, indicating that RAW264.77 cells did not spread well on all the collagen-based substrates. The above results proved that E7 peptides could selectively promote the spreading of BMSCs, but had no significant effects on RAW264.7 cells and NIH3T3 cells. This is due to the fact that spreading area and adhesion force are associated with focal adhesions, which are formed by the specific interaction between cell integrins and substrate ligands.34Selective adhesion of BMSCs
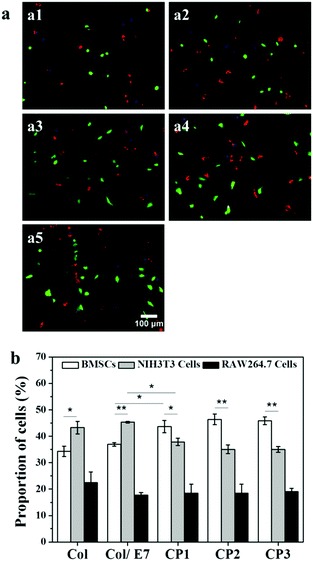
The selective adhesion properties were further characterized by co-culturing BMSCs, RAW264.7 cells and NIH3T3 cells on the substrates (Fig. 4). It shows that more NIH3T3 cells (red) could be observed than BMSCs (green) and RAW264.7 cells (blue) on Col and Col/E7. However, on the CP substrates, more green cells and fewer red cells could be observed, indicating the enhanced selectivity to BMSC adhesion. RAW264.7 cells kept almost the same number regardless of the substrate types. Moreover, BMSCs on CP showed more spreading morphologies than those on Col and Col/E7, which was consistent with the results of the separately cultured BMSCs. Accordingly, the proportions of the three kinds of cells on each substrate were statistically analyzed (Fig. 4b). The proportions of BMSCs, NIH3T3 cells and RAW264.7 cells on Col were 34.3%, 43.3% and 22.4%, respectively. When these cells were cultured on Col/E7, no significant changes were found for all types of cells compared to those on Col. This means that the physical adsorption of E7 does not change the properties of collagen substrates too much. As for the CP substrates, the proportions of BMSCs increased to 43.7% (CP1), 46.4% (CP2) and 46.0% (CP3), respectively. Meanwhile, the proportions of NIH3T3 cells decreased to 37.9%, 35.1% and 35.0%, correspondingly. One could notice that the proportions of BMSCs became higher than those of NIH3T3 cells when the substrates were changed from Col to CP, although there was no significant difference among the different CP substrates. It is worth mentioning that the increase of the BMSC proportion only reduced the proportion of NIH3T3 cells, and did not affect the proportion of RAW264.7 cells, which almost kept at ∼20%. We tentatively attribute this to the existence of space competition between NIH3T3 cells and BMSCs with similar spreading area. The above results lead us to conclude that selective adhesion of BMSCs is significantly enhanced as compared with RAW264.7 cells and NIH3T3 cells by E7 immobilization.Selective capture of BMSCs in a flow model
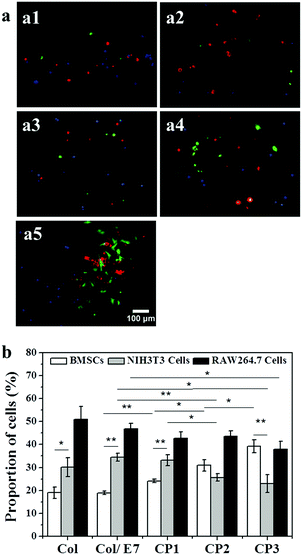
The selective capture of BMSCs on the E7-immobilized substrates was investigated using a flow model. As shown in Fig. 5(a1–a5), BMSCs (green) could be hardly found on Col, Col/E7 and even CP1, on which more RAW264.7 cells were observed. However, with the increase of E7 density, more BMSCs could be observed on CP2 and CP3. Most of the cells show round morphologies except for the BMSCs captured on CP3, which showed more spreading morphologies (Fig. 5(a5)). The statistical results are further presented in Fig. 5b. The BMSC proportions of the total captured cells were lower than those of NIH3T3 cells and RAW264.7 cells on Col and Col/E7, on which RAW264.7 cells showed the highest proportions. However, on the substrates with E7 modification, the proportion of BMSCs increased to 24% (CP1), 31% (CP2) and 39% (CP3), respectively with the increase of E7 density. The proportion of BMSCs became higher than that of NIH3T3 cells on CP2 and similar to that of RAW264.7 cells on CP3, proving that the E7-modified substrates possess an obvious selectivity for capturing BMSCs. However, the highest proportions of RAW264.7 cells in the flow model were not consistent with the results of the static adhesion assays, in which the proportions of RAW264.7 cells on each substrate were always the lowest. This conflicted phenomenon might be attributed to the fast adhesion of RAW264.7 cells, as well as the weaker shear force of the fluids in the flow channel compared to the washing force in the static adhesion tests. Although the flow model could simulate the in vivo dynamic conditions of cell capture and provide reproducible results, it was reported that the shear force can be a crucial regulator of MSC differentiation.35Therefore, it is not yet determined whether the force generated by the fluids would induce BMSC differentiation and thereby affect the adhesion behavior. In addition, we preliminarily focused on the short-term capture capabilities in this study, for the continuous CO2 supply in the channels could not be ensured. Notwithstanding its limitations, this study suggests that the substrates modified by E7 peptides could selectively enhance the capture of BMSCs from circulating cells.
Conclusions
E7-modified collagen substrates were prepared by using sulfo-SMCC as the coupling agent, the amounts of which were modulated to obtain substrates with different E7 densities. With the increase of E7 density, the adhesion rate of BMSCs increased significantly accompanied by a larger adhesion area and a higher adhesion force compared to those of NIH3T3 cells and RAW264.7 cells. It was further validated that the E7-modified substrates selectively promoted the adhesion of BMSCs in both static and flow models. This indicates that conjugating E7 peptides might be a promising strategy for designing advanced implants for in situ tissue regeneration.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors acknowledge the financial support by the National Key Research Program of China (2017YFA0104901), the National Natural Science Foundation of China (51673167 and 21434006) and the Fundamental Research Funds for the Central Universities (2017XZZX008-05).References
- S. Yamanaka, Cell Stem Cell, 2007, 1, 39–49 CrossRef CAS PubMed.
- K. Andreas, M. Sittinger and J. Ringe, Trends Biotechnol., 2014, 32, 483–492 CrossRef CAS PubMed.
- A. Y. Sheikh, S. Lin, F. Cao, Y. Cao, K. E. A. van der Bogt, P. Chu, C. Chang, C. H. Contag, R. C. Robbins and J. C. Wu, Stem Cells, 2007, 25, 2677–2684 CrossRef PubMed.
- C. Toma, M. F. Pittenger, K. S. Cahill, B. J. Byrne and P. D. Kessler, Circulation, 2002, 105, 93–98 CrossRef PubMed.
- E. Abdelwahid, A. Kalvelyte, A. Stulpinas, K. A. T. de Carvalho, L. C. Guarita-Souza and G. Foldes, Apoptosis, 2016, 21, 252–268 CrossRef CAS PubMed.
- C. Balbi, M. Piccoli, L. Barile, A. Papait, A. Armirotti, E. Principi, D. Reverberi, L. Pascucci, P. Becherini, L. Varesio, M. Mogni, D. Coviello, T. Bandiera, M. Pozzobon, R. Cancedda and S. Bollini, Stem Cells Transl. Med., 2017, 6, 1340–1355 CrossRef CAS PubMed.
- C. Cunha, C. R. Almeida, M. I. Almeida, A. M. Silva, M. Molinos, S. Lamas, C. L. Pereira, G. Q. Teixeira, A. T. Monteiro, S. G. Santos, R. M. Goncalves and M. A. Barbosa, Stem Cells Transl. Med., 2017, 6, 1029–1039 CrossRef CAS PubMed.
- H. Du, H. Naqvi and H. S. Taylor, Stem Cells Dev., 2012, 21, 3324–3331 CrossRef CAS PubMed.
- P. Chen, J. Tao, S. Zhu, Y. Cai, Q. Mao, D. Yu, J. Dai and H. Ouyang, Biomaterials, 2015, 39, 114–123 CrossRef CAS PubMed.
- Y. Dai, Z. Gao, L. Ma, D. Wang and C. Gao, Macromol. Biosci., 2016, 16, 1632–1642 CrossRef CAS PubMed.
- M. Shafiq, Y. Jung and S. H. Kim, J. Biomed. Mater. Res., Part A, 2016, 104, 1352–1371 CrossRef CAS PubMed.
- M. Shafiq, Y. Jung and S. H. Kim, J. Biomed. Mater. Res., Part A, 2015, 103, 2673–2688 CrossRef CAS PubMed.
- M. Shafiq, Y. Jung and S. H. Kim, Eur. Cells Mater., 2015, 30, 282–302 CrossRef CAS PubMed.
- H. Okuyama, B. Krishnamachary, Y. F. Zhou, H. Nagasawa, M. Bosch-Marce and G. L. Semenza, J. Biol. Chem., 2006, 281, 15554–15563 CrossRef CAS PubMed.
- B. P. Purcell, J. A. Elser, A. Mu, K. B. Margulies and J. A. Burdick, Biomaterials, 2012, 33, 7849–7857 CrossRef CAS PubMed.
- M. Zamani, M. P. Prabhakaran, E. S. Thian and S. Ramakrishna, J. Colloid Interface Sci., 2015, 451, 144–152 CrossRef CAS PubMed.
- Y. Won, A. N. Patel and D. A. Bull, Biomaterials, 2014, 35, 5627–5635 CrossRef CAS PubMed.
- J. Ding, K. Hori, R. Zhang, Y. Marcoux, D. Honardoust, H. A. Shankowsky and E. E. Tredget, Wound Repair Regen., 2011, 19, 568–578 CrossRef PubMed.
- D. Pilling, T. Fan, D. Huang, B. Kaul and R. H. Gomer, PLoS One, 2009, 4, e7475 Search PubMed.
- Y. Tabata, J. R. Soc., Interface, 2009, 6(Suppl 3), S311–S324 CrossRef CAS PubMed.
- N. M. Coelho and C. A. McCulloch, Cell Tissue Res., 2016, 365, 521–538 CrossRef CAS PubMed.
- Z. Shao, X. Zhang, Y. Pi, X. Wang, Z. Jia, J. Zhu, L. Dai, W. Chen, L. Yin, H. Chen, C. Zhou and Y. Ao, Biomaterials, 2012, 33, 3375–3387 CrossRef CAS PubMed.
- Q. Tan, J. Dai, X. Yan, Y. Yang, H. Wang, L. Shen, S. Li, Y. Lin, S. Wang, X. Hou and C. Shi, Int. J. Burns Trauma, 2014, 2, 181 CrossRef PubMed.
- F. Chen, L. Wu, M. Zhang, R. Zhang and H. Sun, Biomaterials, 2011, 32, 3189–3209 CrossRef CAS PubMed.
- L. Ma, C. Y. Gao, Z. W. Mao, J. Zhou, J. C. Shen, X. Q. Hu and C. M. Han, Biomaterials, 2003, 24, 4833–4841 CrossRef CAS PubMed.
- Z. Man, L. Yin, Z. Shao, X. Zhang, X. Hu, J. Zhu, L. Dai, H. Huang, L. Yuan, C. Zhou, H. Chen and Y. Ao, Biomaterials, 2014, 35, 5250–5260 CrossRef CAS PubMed.
- Q. Meng, Z. Man, L. Dai, H. Huang, X. Zhang, X. Hu, Z. Shao, J. Zhu, J. Zhang, X. Fu, X. Duan and Y. Ao, Sci. Rep., 2015, 5, 17802 CrossRef CAS PubMed.
- S. Huang, L. Xu, Y. Sun, T. Wu, K. Wang and G. Li, J Orthop. Translat., 2015, 3, 26–33 CrossRef.
- C. D. Reyes and A. J. Garcia, J. Biomed. Mater. Res., Part A, 2003, 67, 328–333 CrossRef PubMed.
- Y. Peng, Y. He and S. Zhu, Chin. J. Tissue Eng. Res., 2012, 2487–2490 CAS.
- Y. Fu, S. Gong, Q. Xue, Y. Liu and K. Yan, J. Clin. Otorhinolaryngol. Head Neck Surg., 2007, 21, 172–175 Search PubMed.
- Y. Lu, L. Zhao, Q. Shen, M. A. Garcia, D. Wu, S. Hou, M. Song, X. Xu, W. OuYang, W. W. L. OuYang, J. Lichterman, Z. Luo, X. Xuan, J. Huang, L. W. K. Chung, M. Rettig, H. Tseng, C. Shao and E. M. Posadas, Methods, 2013, 64, 144–152 CrossRef CAS PubMed.
- Y. Arima and H. Iwata, Biomaterials, 2007, 28, 3074–3082 CrossRef CAS PubMed.
- N. Q. Balaban, U. S. Schwarz, D. Riveline, P. Goichberg, G. Tzur, I. Sabanay, D. Mahalu, S. Safran, A. Bershadsky, L. Addadi and B. Geiger, Nat. Cell Biol., 2001, 3, 466–472 CrossRef CAS PubMed.
- J. Hao, Y. Zhang, D. Jing, Y. Shen, G. Tang, S. Huang and Z. Zhao, Acta Biomater., 2015, 20, 1–9 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7tb02812a |
| This journal is © The Royal Society of Chemistry 2018 |