Bacterial and archaeal community structure involved in biofuels production using hydrothermal- and enzymatic-pretreated sugarcane bagasse for an improvement in hydrogen and methane production
Juliana K.
Braga
 *,
Fabrício
Motteran
*,
Fabrício
Motteran
 ,
Isabel K.
Sakamoto
,
Isabel K.
Sakamoto
 and
Maria Bernadete A.
Varesche
and
Maria Bernadete A.
Varesche
 *
*
University of São Paulo, School of Engineering of São Carlos, Department of Hydraulics and Sanitation, Av. Trabalhador Sãocarlense, 400, 13566-590 São Carlos, SP, Brazil. E-mail: jukawanishi@gmail.com; varesche@sc.usp.br
First published on 6th September 2018
Abstract
Sugarcane bagasse (SCB) was used as a lignocellulosic substrate, combining the co-production of H2 (Stage I) and CH4 (Stage II) by a dark fermentation process in batch reactors. Hydrothermally- and enzymatic (Aspergillus niger)-pretreated SCB were applied as substrate sources. Two fermentative inocula (In1 and In2) were used in Stage I and a methanogenic inoculum in Stage II (In3), comprising in total three experimental series in relation to Stage I: A (In1), B (In1 plus In2), and C (In2). The final metabolites (solid, liquid, and gaseous fractions) from Stage I were used for CH4 production (Stage II). The SCB pretreatment employed was favorable for biogas and organic acids production. Higher H2 and CH4 yields were obtained in C (4.3 and 6.3 mmol g−1 SCB, respectively). For all conditions, the H2 production occurred primarily via an acetic acid route. The predominance of cellulolytic enzyme producers (Enterococcus and Clostridium) may have favored the H2 and subsequent CH4 production; this last was produced mainly from members of the Methanoregulaceae and Methanosaetaceae families. Furthermore, homoacetogenic bacteria (Acetobacterium, Clostridium, Eubacterium, Holophaga) were also identified in both stages. The synergistic action of these microbial groups promoted the hydrolysis of SCB as well as hydrogen and methane production.
1 Introduction
Agricultural biomasses are important platforms for the production of biofuels from renewable raw materials.1 Hydrogen (H2) and methane (CH4) are important energy sources, which can be produced by the anaerobic digestion of different organic wastes.2 Given that the dark fermentation for hydrogen production from organic substrates only occurs partially, the combination with methane production can benefit energy recovery.3 In addition, compared to liquid fuels, such as ethanol, hydrogen and methane can be readily separated from the liquid phase, which can reduce the process costs.2Two-stage processes have previously already been employed for CH4 production,4 for the separation of the hydrolysis/acidogenesis phase from the methanogenesis phase, and for improving each process independently.5 Studies into hydrogen and methane production using two-stage fermentation processes have been carried out using a variety of substrates, such as food waste,3 solid household waste,4 and sludge from a wastewater treatment plant.6
However, few studies have been carried out on the co-production of H2 and CH4 from sugarcane bagasse (SCB). Some of the most studied raw materials are sorghum,7 corn cob,8 olive pulp,9 mushroom,10,11 and potato waste.12 SCB is a carbohydrate-rich substrate that has been used for hydrogen production.13,14 The biotechnology processes used in biomass conversion are more ecologically correct and capable of generating energy as well as the thermochemical and electrochemical processes.15 In this way, the use of SCB for energy generation through biological processes can reduce the environmental impacts caused by the combustion, thus reducing the CO2 emission indices.
The design and optimization of hydrogen production from hydrothermally (200 °C for 10 min at 16 bar)-pretreated SCB was evaluated using a response surface methodology and anaerobic consortium from an upflow anaerobic sludge blanket reactor (UASB) treating vinasse.16 The authors observed a maximum hydrogen production of 17.7 mmol L−1 at 60 °C with 3.0 g L−1 of yeast extract, with a predominance of acetic and butyric acids.
Among the microorganisms involved in the H2 production were Clostridium genus, like Clostridium butyricum, C. acetobutylicum, C. saccharoperbutylacetonicum, C. pasteurianum, and C. cellulolyticum.17–21 Hydrogen and organic acids production were evaluated with autochthonous (Clostridium and Paenibacillus from SCB) and allochthonous (Clostridium, Bacillus and Enterobacter) bacteria from SCB (pretreated in an autoclave at 121 °C, 1.5 kg cm−2 for 15 min) in batch reactors.22 The authors observed a maximum production of 23.1 mmoL H2 L−1 under optimized conditions of 7.0 g SCB per L and pH 7.2 at 37 °C through the acetic acid (1.57 g L−1) pathway. A higher hydrogen production of 50 mmol L−1 was observed when a co-culture of Clostridium thermocellum and Thermoanaerobacterium aotearoense was employed with 40 g L−1 of alkali-pretreated SCB (3% NaOH, 55 °C for 3 h).23
Meanwhile, due the complexity of SCB components, it is evident that this lignocellulosic biomass is difficult to be fully utilized by only a single bacterial strain. A microbial community, such as anaerobic sludge, is thus usually used as an inoculum source for hydrogen or methane production.3,4 However, it is still difficult to control and optimize the hydrogen-producing microbial consortium, as the hydrogen production rate and yield are not always high. It is therefore essential to obtain a stable and efficient microbial community that is able to improve the hydrolysis of biomass and hydrogen production.
SCB is composed of hemicellulose, lignin, and cellulose, as well as other minor components. Hemicellulose and lignin fractions protect cellulose fibers from microbial cellulases,24 which are essential in the conversion of complex carbohydrates to fermentable glucose. However, the conversion of lignocellulosic substrates into H2 requires many steps, including substrate pretreatment, enzymatic hydrolysis, and fermentation.
In a study of hydrogen production from mushroom compost by Clostridium thermocellum, the authors supplemented the culture medium with recombinant β-glucosidases and obtained an increase in hydrogen production of 37%.10 In contrast, in the present study, commercial enzymes were not used for the hydrolysis of SCB, which makes the biogas production process more attractive due to its reduced costs since the cellulolytic enzymes were obtained from both, the A. niger and the inoculum.
The purpose of the pretreatment of recalcitrant substrates is to change or remove the lignin and/or hemicellulose structure, decrease the crystallinity of cellulose, and increase the substrate surface area in order to benefit the enzymes accessing the structure of cellulose for the better recovery of fermentable sugars.25
According to da Cruz et al.,26 hydrothermal pretreatment eliminates these barriers by solubilizing hemicellulose and breaking the lignin structure. Furthermore, cellulose is also decrystallized, thereby allowing the cellulase enzymes to access those microfibers. This technology for SCB pretreatment is of particular interest because fibers are heated in water, the only solvent used, which eliminates the cost of a catalyst and reduces the waste generation process.27
Pretreatment employing a hydrothermal system consists of keeping SBC in pressurized liquid hot water and promoting rapid depressurization, after achieving a severity parameter, for the purpose of breaking down the lignocellulosic structure to supply the cellulolytic and fermenting microorganism with access to the cellulose structure.28 Therefore, the hydrothermal pretreatment of SCB could solubilize more than 80% of the hemicellulose and more than 40% of the lignin.29 In a study into the hydrothermal pretreatment of SCB (reaction temperature and time between 160 °C and 200 °C and 5–20 min, respectively) for ethanol production, the authors observed in the solid fraction about 5–22% xylan, 22% Klason lignin, and 44–55% glucan.26
Usually, the process of the enzymatic treatment of lignocellulose consists of two steps: primarily, the lignocellulosic biomass is pretreated to destroy its complex structure; and then, the substrate is converted by the enzymes to fermentable sugars.30
Accordingly, this conversion of the lignocellulosic substrate to glucose requires the presence of cellulolytic enzymes. Since fungal pretreatment is a low cost and environmentally friendly pretreatment method, it has attracted increased importance in recent years.31 Furthermore, enzymatic pretreatment is conducted under mild conditions and several inhibitory compounds are not generated.32 Therefore, combining hydrothermal pretreatment with enzymatic pretreatment is a very interesting proposition.
The main disadvantages of biological pretreatment are the substantial loss of carbohydrates that may occur, long residence time, necessity for careful control of the growth conditions, the need to avoid contamination, and the space required, which limit its applications. To overcome these limitations, biological treatments are usually used in combination with other treatments.33 The combination of fungal treatment with liquid hot water was studied to evaluate the hydrolysis of the lignocellulosic biomass Populus tomentosa.34
Among the microorganisms applied in the biological treatment of lignocellulosic substrates, fungi have gained distinction, especially the filamentous fungi that adapt more easily because they are less demanding and do not require large amounts of available water and as their growth is stimulated by the formation of hyphae. Among these, the genus Aspergillus is considered superior to other genera in relation to the production of cellulolytic and hemicellulolytic complexes.35
Although the association of SCB pretreatments has already been studied,23,36–38 knowledge of the association of the hydrothermal and enzymatic pretreatment of SCB using A. niger as well as the effect of the inoculum source and phylogenetic knowledge of the community are still lacking for H2 and CH4 production in a two-stage anaerobic digestion process from lignocellulosic biomass.
Therefore, the purpose of this research was to study the feasibility of the co-production of H2 (Stage I) and CH4 (Stage II) from pretreated sugarcane bagasse (SCB) using a two-stage anaerobic digestion process. The metabolites (organic acids and H2) generated at the end of Stage I served as substrates in Stage II, for CH4 production. The pretreatment of SCB was conducted in a hydrothermal system combining enzymatic pretreatment with the fungus Aspergillus niger. The resulting solid fraction from both pretreatments was used as a substrate in the H2/CH4 assays. Moreover, the microbial community structure in the H2 and CH4 production reactors was assessed, using the Illumina MiSeq platform for the bacteria and archaea domains, as well as the relation with the metabolites produced in both operational stages.
2 Material and methods
2.1 Sugarcane bagasse pretreatment
The SCB used in this study was supplied by São Martinho sugar mill (Pradópolis, SP, Brazil). The SCB composition was 31.4% cellulose, 36.6% xylan, 5.0% arabinan, 24.8% lignin, and 1.2% ash.39 The SCB (5 g) was pretreated using a 100 mL capacity hydrothermal system, 20 bar pressure at 200 °C for 10 min,14 followed by enzymatic pretreatment with the fungus Aspergillus niger. At the end of both pretreatments, the solid fraction was employed as a substrate in the co-production of H2 and CH4.In this study, A. niger strain (ATCC 10577) acquired from the André Tosello Foundation (Campinas, SP, Brazil) was used. The lyophilized strain was reactivated by adding 0.5 mL sterile distilled water to the tube (30 min); then the sample was inoculated in solid medium (20 g L−1 malt extract, 20 g L−1 glucose, 5 g L−1 peptone, 15 g L−1 agar, and pH 7) and incubated at room temperature for approximately 7 days or until total growth on the plate for subsequent application in the SCB pretreatment.
For the enzymatic pretreatment, the hydrothermally pretreated SCB (solid fraction) was used as a carbon source in Mandels and Weber culture media.40 For this, 1 mL L−1 of Tween 80 and 5 g of SCB was added into an Erlenmeyer flask. This substrate was autoclaved at 121 °C for 20 min and then 5 mL of sterile culture medium was added to it. Subsequently, a spore suspension with 1 × 108 spores per mL of A. niger was added to the Erlenmeyer flask and incubated at 37 °C with periodic shaking for 7 days. At the end of this period, the SCB pretreated hydrothermally and enzymatically by the fungus was used as a substrate in the H2 and CH4 production assays.
2.2 Anaerobic mixed consortia
The first inoculum (In1) was obtained from an anaerobic lagoon of a pulp and paper mill's wastewater treatment plant (WWTP) in Suzano (São Paulo, Brazil). To obtain the cellulolytic and fermentative consortium, 10% (v/v) of the sludge was inoculated in cellulose anaerobe medium41 containing 2 g L−1 cellulose for 24 h. The second inoculum (In2) was a fermentative and cellulolytic consortia (Clostridium, Bacillus, Bacteroides, and Paenibacillus) obtained in the Biological Processes Laboratory, University of São Paulo (Brazil), using cellulose as the substrate.42 This consortia was inoculated in cellulose anaerobe medium41 containing 2 g L−1 cellulose for 24 h.The methanogenic inoculum (In3) was a granular sludge obtained from the upflow anaerobic sludge blanket reactor (UASB) used in the treatment of poultry slaughterhouse wastewater operated in mesophilic conditions. To obtain the methanogenic consortium, 10% (v/v) of the sludge was inoculated in cellulose anaerobe medium41 containing 2 g L−1 cellulose for 72 h. After the incubation period, the inocula were centrifuged at 6000 rpm, 27 °C for 10 min, the supernatant were discarded and the concentrated biomasses were resuspended in cellulose anaerobe medium,41 which were the inocula of the H2 and CH4 production assays.
2.3 Culture media and biogas production assays
Batch reactors (Duran® flasks of 250 mL) were operated in triplicate. The reactors contained 90 mL of cellulose anaerobe medium,41 10 mL of inoculum (10% v/v), and 2 g L−1 of pretreated SCB as the substrate. All the assays were performed in mesophilic conditions (37 °C), pH 6.0, and under stirring at 100 rpm. The total fermentation times were 196 h and 464 h for Stages I and II, respectively.The cellulose anaerobe medium41 used for the cultivation of the microbial consortia was supplemented with yeast extract (1 g L−1). This medium contained the following compounds: NaHCO3 (2.1 g L−1); NH4Cl (0.68 g L−1); KH2PO4 (0.18 g L−1); (NH4)2SO4 (0.15 g L−1); MgSO4·7H2O (0.12 g L−1); K2HPO4 (296 mg L−1); CaCl·2H2O (61 mg L−1); FeSO4·7H2O (21 mg L−1); nitriloacetic acid (15 mg L−1); NaCl (10 mg L−1); MnSO4·H2O (5 mg L−1); CoCl2·6H2O (1 mg L−1); ZnSO4·7H2O (1 mg L−1); CuSO4·5H2O (0.1 mg L−1); KAl(SO4)2·12H2O (0.1 mg L−1); H3BO3 (0.1 mg L−1); Na2MoO4·2H2O (0.1 mg L−1); and vitamin solution (5 mL L−1). The vitamin solution was composed of: pyridoxine HCl (10 mg L−1); calcium DL-pantothenate (5 mg L−1); lipoic acid (5 mg L−1); nicotinic acid (5 mg L−1); p-amino benzoic acid (5 mg L−1); riboflavin (5 mg L−1); thiamine HCl (5 mg L−1); biotin (2 mg L−1); folic acid (2 mg L−1); and vitamin B12 (0.1 mg L−1).
The solid fraction of pretreated SCB (2 g L−1) was used as the substrate source in all reactors instead of 2 g L−1 cellulose. Three experimental series were performed: A (10% of In1), B (5% of In1 + 5% of In2), and C (10% of In2) (Table 1). The reactor headspace was purged with nitrogen (N2 100%) for 10 min in order to provide an anaerobic environment. The final metabolites (solid, liquid, and gaseous fractions) of Stage I (H2 production) were used for the subsequent CH4 production stage (Stage II). Therefore, the reactor of Stage I was inoculated with 10% (v/v) of inoculum 3 (In3 – methanogenic) using a syringe so that there was no loss of biogas, to form the reactor from Stage II. The initial pH of the methanogenic stage was adjusted to pH 7 with NaOH (10% w/v). For this, an aliquot of the liquid fraction at the end of Stage I was withdrawn with a syringe and the final pH was measured and then NaOH (10% w/v) was added to the reactors, also with a syringe.
| Experimental series | Stage I | Stage II |
|---|---|---|
| a In – inoculum; In1 – anaerobic lagoon of a wastewater treatment plant; In2 – fermentative inoculum; In3 granular ludge from UASB reactor used in the treatment of poultry slaughterhouse wastewater. | ||
| A | In1 | In3 |
| B | In1 + In2 | In3 |
| C | In2 | In3 |
2.4 Physicochemical and chromatographic analyses
The accumulated hydrogen and methane production were monitored by gas chromatography (GC) 2010 (Shimadzu, Japan) equipped with a thermal conductivity detector (TCD) and a Carboxen 1010 PLOT column (30 m × 0.53 mm) according to Motteran et al.43 H2 and CH4 yields were calculated by the amount of these biogas accumulations in the headspace per amount of carbohydrate consumed.The quantification of volatile organic acids (VOA) was carried out for all reactor conditions at the end of each stage (Stage I and Stage II), by high-performance liquid chromatography (HPLC), equipped with a refraction index detector (RID-10A), UV diode array detector (SPD-M10Avp), LC-10ADvp Pump, CTO-20A oven, SCL 10 Avp controller, and Aminex HPX-87H column, 300 mm × 7.8 mm (BioRad). The mobile phase consisted of H2SO4 (0.01 N) at a 0.5 mL min−1 flow rate.44 Alcohols were determined by gas chromatography (GC) equipped with a HPINNOWAX column and flame ionization detector using H2 as the carrier gas with synthetic air and N2 as the auxiliary gas.45
Determination of the total volatile solids (TVS) and pH were conducted following standard methods for the examination of water and wastewater,46 whereas carbohydrates determination was carried out indirectly by the phenol–sulfuric acid method using glucose as the standard.47
2.5 Kinetic analysis
The experimental data (H2 and CH4 values) were adjusted to the average values obtained from triplicates using the Origin® 9.0 software (OriginLab, Northampton, MA). The data of the accumulated H2 and CH4 production were adjusted using the Gompertz equation, modified by Zwietering (eqn (1)).48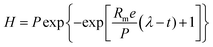 | (1) |
2.6 Microbial community analysis
The analyzes of molecular biology were processed by Illumina MiSeq, and for that, the biomass samples collected from the batch reactors fed with SCB at the end of the operation (Stages I and II) were washed in a phosphate buffer and centrifuged at 6000 rpm for 10 min. The pellets were stored at −20 °C. DNA was extracted according to Griffiths et al.49 Polymerase chain reaction (PCR) of the 16S rRNA gene fragments employed the primers 968F and 1401R for the Bacteria domain,50 while for the archaea domain, the primers 1100F and 1400R51 were used.The phylogenetic diversity of the microbial consortium (A I, C I, A II, and C II) was analyzed by Illumina MiSeq sequencing.52 Primers 515F (5′-barcode-GTGCCAGCMGCCGCGG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) were used to amplify the V4 regions of the bacteria 16S ribosomal RNA gene using a GeneAmp PCR System (ABI company, USA). Amplicons were purified using calibrated Ampure XP beads. Then, the pooled and purified PCR product was used to prepare the Illumina DNA library. Sequencing was performed at MR DNA (https://www.mrdnalab.com, Shallowater, Texas, USA) on a MiSeq following the manufacturer's instructions. Sequence data were processed using an MR DNA analysis pipeline (MR DNA, Shallowater, Texas, USA).
The sequences were joined, depleted of barcodes, then sequences <150 bp and sequences with ambiguous base calls were removed. Sequences were de-noised, OTUs generated, and chimeras removed. Operational taxonomic units (OTU) were defined by clustering at 97% similarity cutoff. Final OTUs were taxonomically classified using Ribosomal Database Project (RDP-II) Classifier and the National Center for Biotechnology Information (NCBI) (https://rdp.cme.msu.edu; https://www.ncbi.nlm.nih.gov). Finally, the raw reads were deposited into the NCBI as a Sequence Read Archive (SRA) database, under Bioproject number PRJNA387459, BioSample SAMN07152251, SRX2841069 (Experiment), SRR5583156 (Run).
3 Results and discussion
3.1 H2 and CH4 production from pretreated SCB
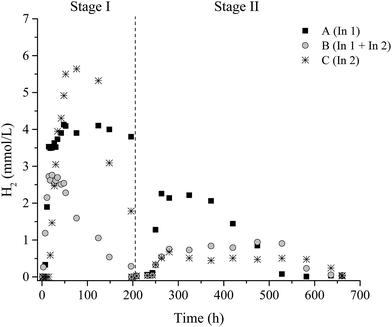
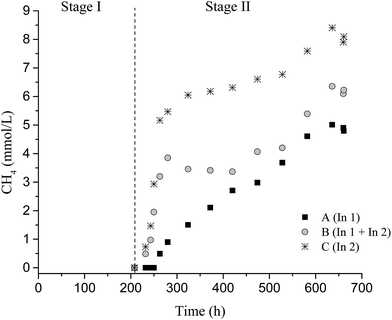
In the degradation of lignocellulosic biomass, H2 and CH4 production are influenced by several environmental conditions, mainly temperature and pH.4,6,53,54 A temperature of 37 °C and initial pH 6.0 were applied for all conditions. Fig. 1 and 2 show the variations of H2 and CH4 production from 2 g L−1 pretreated SCB in the two-stage dark fermentation process. The reactors were operated for 660 h and different behaviors with respect to H2 and CH4 production were verified for the batch reactors studied.For the conversion of lignocellulosic material (such as SCB) to fuel, the cellulose and hemicellulose must be broken down into their equivalent monomer carbohydrates so that microorganisms can use them in their biological route.55 In this study, the H2 production suggested that the pretreated SCB by hydrothermal plus enzymatic processes facilitated the access and use of sugars by the fermentative bacteria. According to Taherzadeh and Karimi,56 the hydrothermal pretreatment of lignocellulosic biomass improves the available and susceptible surface area of cellulose molecules and makes them more accessible to hydrolytic enzymes. In this way, the pretreatment methods applied in this study were effective in breaking and/or disrupting the SBC structures in order to increase the access of cellulolytic enzymes and fermenting-bacteria, thereby aiding the hydrolysis of sugars for acids conversion and H2 production.
The presence of microorganisms in WWTP inoculum (In1), as well as the presence of the cellulolytic-fermentative consortium (In2), slowly promoted the cellulose degradation from SCB producing H2 in all stages. This fact was evidenced by H2 observed in all inoculum conditions A (In1), B (In1 + In2), and C (In2) and CH4 only in Stage II.
In Stage I, H2 consumption was observed for conditions B I and C I. The development of homoacetogenic bacteria favored the consumption of H2 under these conditions since CH4 production was not observed in this stage. Nine genera with homoacetogenic representatives were identified (Acetobacterium, Clostridium, Eubacterium, Holophaga, Moorella, Ruminococcus, Sporomusa, Thermoanaerobacter, and Treponema). According to Drake et al.,57 members from these genera may have both a H2-producing and consuming behavior. Thus, it is likely that these microbial groups are homoacetogens that play an essential role in acetate formation under the operational conditions employed in this study.
The homoacetogens produce specific enzymes that catalyze the formation of acetyl-CoA, which is then transformed to acetate in catabolism or to cell carbon in anabolism.58 In the present study, higher concentrations of acetic acid (Table 6) were observed in conditions with higher H2 consumption (B I and C I), thus confirming the presence of homoacetogenic metabolism.
In Stage II, H2 consumption was also verified, however, with the presence of CH4 for all conditions. In this case, the hydrogenotrophic methanogens were probably the most likely responsible for this decrease. After all, under mesophilic or thermophilic conditions, studies have revealed that homoacetogenesis cannot strongly compete with hydrogenotrophic methanogenesis since the latter produces more energy.59,60 Even though the occurrence of OTUs related to H2-producing bacteria, such as Clostridium, Enterobacter and Bacillus in this study, lower H2 production was observed under A II, B II, and C II, compared to the previous stage (Stage I). It is, thus, likely that H2 consumption by homoacetogens (Table 6) and/or methanogens (Table 8) could explain this finding.
Table 2 shows the kinetic parameters for H2 and CH4, such as potential production (P), maximum production rate (Rm), and λ. The mixture of inoculum 1 and inoculum 2 (condition B) did not favor higher H2 production compared to the assays using the inocula separately (conditions A and C); probably, due to the lower amount (5%) employed of each inoculum, which may have promoted slower growth of the interest microorganisms.
| Condition | H2 | CH4 | ||||||
|---|---|---|---|---|---|---|---|---|
| P (mmol L−1) | R m (mmol L−1 h−1) | λ (h) | R 2 | P (mmol L−1) | R m (mmol L−1 h−1) | λ (h) | R 2 | |
| a A – inoculum 1, B – inoculum 1 + inoculum 2; C – inoculum 2; I – Stage I and II – Stage II. * Maximum values not adjusted in modified Gompertz equation. | ||||||||
| A I | 3.54 ± 0.2 | 0.59 ± 0.6 | 7.5 ± 3.9 | 0.95 | — | — | — | — |
| A II | 2.0 ± 0.1 | 0.57 ± 0.2 | 36.7 ± 3.1 | 0.89 | 4.8 ± 0.1 | 0.04 ± 0.004 | 698.4 ± 5.7 | 0.98 |
| B I | 2.64 ± 0.04 | 0.32 ± 0.04 | 3.1 ± 0.6 | 0.98 | — | — | — | — |
| B II | 0.8 ± 0.05 | 0.29 ± 0.04 | 37 ± 3.7 | 0.87 | 6.1 ± 0.3 | 0.03 ± 0.007 | 634.99 ± 20.2 | 0.89 |
| C I | 5.47 ± 0.1 | 0.22 ± 0.01 | 16 ± 0.8 | 0.99 | — | — | — | — |
| C II | 0.52 ± 0.03 | 0.21 ± 1.8 | 37 ± 2.4 | 0.88 | 7.6 ± 0.2 | 0.1 ± 0.01 | 668.6 ± 4.5 | 0.97 |
In Stage I, 3.5 ± 0.2 mmol L−1, 2.6 ± 0.04 mmol L−1, and 5.5 ± 0.1 mmol L−1 of H2 were observed for conditions A (In1), B (In1 + In2), and C (In 2), respectively (Table 2). Moreover, in Stage II, the maximum H2 production was 2.0 ± 0.1 mmol L−1, 0.8 ± 0.05 mmol L−1, and 0.5 ± 0.03 mmol L−1 for the same conditions, respectively. H2 consumption was observed in all the studied conditions of both stages, except in A I. The presence of homoacetogenic bacteria and/or methanogenic archaea contributed to this reduction on H2 concentration. The homoacetogenic bacteria are very versatile organisms and are able to convert a variety of substrates to acetic acid as the main end product observed.58 Acetic acid was the main end product here with a little production of other metabolites for all conditions studied (Table 4).
| Condition | Parameters | |||
|---|---|---|---|---|
| H2 yield (mmol H2 g−1 substrate) | CH4 yield (mmol CH4 g−1 substrate) | Specific production H2 (mmol L−1 g−1 TVS) | Specific production CH4 (mmol L−1 g−1 TVS) | |
| a A – inoculum 1, B – inoculum 1 + inoculum 2; C – inoculum 2; I – Stage I and II – Stage II. | ||||
| A I | 3.12 | — | 0.94 | — |
| A II | 1.67 | 3.85 | 0.70 | 1.66 |
| B I | 2.06 | — | 0.62 | — |
| B II | 0.71 | 4.75 | 0.28 | 2.20 |
| C I | 4.26 | — | 1.81 | — |
| C II | 0.51 | 6.30 | 0.20 | 2.94 |
| Condition | VOA (mg L−1) | Alcohol (mg L−1) | ||||
|---|---|---|---|---|---|---|
| Acetic acid | Butyric acid | Propionic acid | Acetic acid (%) | Butanol | Ethanol | |
| a VOA – volatile organic acid; SCB – sugarcane bagasse; A – inoculum 1, B – inoculum 1 + inoculum 2; C – inoculum 2 I – Stage I and II – Stage II. | ||||||
| A I | 460 | 50 | 50 | 68 | 24.4 | 19.3 |
| A II | 760 | 70 | 100 | 75 | — | — |
| B I | 5880 | 520 | 50 | 84 | — | — |
| B II | 4500 | 620 | 1260 | 68 | 23.8 | — |
| C I | 9270 | 480 | 700 | 87 | 24.1 | — |
| C II | 30 | 10 | — | 36 | — | — |
It was reported that the H2 produced from butyrate oxidation reacts rapidly with CO2 to form acetate by homoacetogenesis.61 In the present study, the higher concentrations of acetic acid to the detriment of butyric acid in all the studied conditions is consistent with this statement.
No CH4 was detected in Stage I; however, 4.8 ± 0.1 mmol L−1, 6.1 ± 0.3 mmol L−1, and 7.6 ± 0.2 mmol L−1 CH4 were observed in Stage II for conditions A, B, and C, respectively. The methane produced, under conditions A and B, originated mainly from hydrogenotrophic methanogenic archaea, due to the decrease in H2 production and low consumption of acetic acid (Table 4). In this study, representatives of methanogenic archaea were distributed in four orders (Methanobacteriales, Methanomassiliicoccales, Methanomicrobiales, and Methanosarcinales), totaling 82%, 55.8%, 62%, and 64.6% of the sequences for samples from conditions A I, C I, A II, and C II, respectively.
Condition C (In2) provided a higher CH4 potential production (25.3 ± 0.9 mmol L−1) in Stage II. Probably, In2 (fermentative and cellulolytic consortia) enabled a greater conversion of soluble sugars to H2 and organic acids, which promoted higher CH4 production. Thus, in this condition, the CH4 was produced mainly by methanogenic acetoclastic archaea since the acetic acid produced in Stage I was practically all consumed in CH4 formation (Table 4).
Values of λ below those observed by Lay62 were observed in this study (Table 2). The authors evaluated H2 production in a batch reactor operated at 37 °C, pH 7, with cellulose as the substrate (12.5 g L−1 and 50 g L−1). These authors used anaerobic sludge as the inoculum and solid residues as the substrate and observed λ values ranging from 3.74 to 4.26 days.
In the present study, λ ranged from 3.1 h in condition B I (In1 + In2; Stage I) to 37 h in B II (In1 + In2; Stage II). The condition that used In2 (fermentative and cellulolytic consortia) as the sole inoculum source was the best strategy for H2 (Stage I) and CH4 (Stage II) production, although the rates were lower compared to conditions A (In1 – anaerobic lagoon of WWTP) and B (In1 + In2). Since In2 is less diverse than In1, related to the microbial community, the production and consumption of metabolites were slower, leading to a lower Rm and higher P.
In a study of H2 production, Soares et al.37 employed 2 g L−1 of hydrothermally pretreated SCB and the same culture medium of this study (cellulose anaerobe). The authors observed a λ of 17.5 h and 4.44 mmol H2 per L when a thermophilic sludge was employed as the inoculum and the reactors were operated at 55 °C. According to these authors, the higher λ verified for the SCB assays when compared to the glucose assay (9.4 h) was due to the lower availability of sugars in the SCB. In the present study, the association of enzymatic pretreatment to the hydrothermally pretreated SCB may have improved the availability of sugars in the lignocellulosic biomass; thus, promoting lower values of λ in Stage I.
The H2 production rates (Rm) in this study were similar to those observed by Ren et al.,63 who evaluated the effects of cellulose concentration on H2 production rates using cow manure as the inoculum. The authors observed production rates of 0.42 mmol L−1 h−1 (5 g L−1 cellulose) and 0.65 mmol L−1 h−1 (10 g L−1 cellulose), while in the present study, the highest values for 2 g L−1 SCB were 0.59 mmol L−1 h−1 (A I), 0.57 mmol L−1 h−1 (A II), and 0.3 mmol L−1 h−1 (B I) (Table 2).
Although Ren et al.63 presented similar Rm values, the substrate conversion values were 0.084 and 0.065 mmol H2 L−1 h−1 for 5 and 10 g L−1 of cellulose. In this study, these values were much higher, namely 0.29, 0.28, and 0.15 mmol H2 L−1 h−1 for the reduced applied substrate (2 g L−1). The inocula employed in this study were more efficient in converting a more complex substrate to H2, and also the combination of both pretreatments (hydrothermal and enzymatic) had an effective effect on biogas production.
Rabelo64 evaluated H2 production using the same inoculum as in the present study (In1) and observed a much lower H2 production rate (0.05 mmol L−1 h−1) when 2 g L−1 cellulose without enzyme addition was used as the substrate. The enzymatic and hydrothermal pretreatment of SCB in the present study may have contributed to a higher H2 production rate (0.59 mmol L−1 h−1), compared to that observed by Rabelo.64
The maximum H2 (4.26 mmol g−1 SCB) and CH4 (6.30 mmol g−1 SCB) yields were obtained for condition C (In 2) in stages I and II, respectively (Table 3). Liu et al.4 observed a similar H2 yield (43 mL g−1 VS added or 5 mmol g−1 VS) in a study of H2 and CH4 production from household solid waste (HSW) in a two-stage fermentation process. The HSW consisted mainly of food residue, paper and garden waste. Instead, the CH4 yield achieved by these authors was slightly lower (500 mL g−1 VS or 4 mmol g−1 VS).
The specific H2 and CH4 productions are shown in Table 3. The highest specific H2 production was observed for condition C I (1.81 mmol L−1 g−1 TVS). A decrease in H2 production was observed with increasing stages (Stage I to II), with a reduction of 25.7%, 54%, and 88.8% for conditions A, B, and C, respectively. The highest specific CH4 production (2.94 mmol L−1 g−1 TVS) was also observed for condition C (Stage II) (Table 3).
Rabelo64 evaluated H2 and CH4 production using the same inoculum as in the present study (In1) and observed a lower specific CH4 production (0.25 mmol L−1 g−1 TVS) when 2 g L−1 cellulose (without enzyme addition) was used as the substrate. In this study, the addition of another inoculum source, In3 (granular sludge obtained from the UASB reactor used in the treatment of poultry slaughterhouse wastewater), in Stage II may have favored the higher specific CH4 production. In addition, it is noteworthy that the use of physical (hydrothermal) and biological (enzymatic) pretreatments conjugated in SCB may have increased the availability of sugars in Stage I, which were converted to acids, generating more CH4 at the end of Stage II, compared to in ref. 65
In this study, the pretreatment with A. niger provided the necessary enzymes for the SCB breakdown, different from that carried out by ref. 66, in which external enzymes were added directly in to the culture medium. Xie et al.66 evaluated H2 and CH4 production from potatoes by a two-phase anaerobic fermentation. Better performances of 271 mL H2 per g TVS (22 mmol L−1 g−1 TVS) and 158 mL CH4 per g TVS (14 mmol L−1 g−1 TVS) were achieved when the residue was pretreated by α-amylase and glucoamylase. Furthermore, the H2 production process was more interesting due to its reduced costs since the microorganisms that produce these enzymes were acquired from the inocula and the biological pretreatment.
Ratti et al.67,68 also used commercial enzymes (cellulose, Sigma-Aldrich®) for the hydrolysis of cellulose in H2 employing batch reactors assays. The authors used as an inoculum source pretreated (acid and heat shock) and leached rumen fluid. The authors observed that the inoculum pretreatment may have inhibited the cellulolytic microorganisms, since H2 was detected only in the presence of the enzyme. On the other hand, the pretreatment applied in the present study did not inhibit the cellulolytic-fermentative microorganisms, making this process more attractive for the generation of biogas (H2 and CH4) in two stages. The biological process in two or more stages for sequential H2 and CH4 production has been considered as an alternative to improve the economic viability of waste treatment.69
3.2 Production of soluble microbial products
At an initial pH of 6.0, acetic acid was the main metabolite formed in all studied conditions (Table 4), suggesting that fermentation occurred mainly through this route. The organic acids produced (such as acetic, butyric, and propionic) at the end of Stage I were available for use in Stage II, thus favoring the development of acetoclastic methanogens represented by members of the genus Methanosaeta (Methanosarcinales order) (A II: 21.8% and C II: 21.5%, respectively) and, according to Whitman et al.,70 archaea belonging to the family Methanosaetace when using only acetate as a catabolic substrate. In this study, the proportion of acetic acid varied from 62% to 81%.A higher acetic acid concentration at the end of Stage I was observed for conditions B I (5880 mg L−1) and C I (9270 mg L−1). For condition A I (In1), H2 production was stable and the production of acetic acid was lower (460 mg L−1). In addition, decreases of 8%, 90%, and 68% in the concentration of H2 (headspace) were observed under conditions A I, B I, and C I, respectively, without CH4 production. At the end of Stage II, there was a total consumption of the H2 produced. It is likely that in all cases there was the development of homoacetogenic bacteria. According to Lay (2001), this group may consume 11–43% of the H2 (in liquid medium) present in the reactors, since they produce acetic acid from CO2 and H2.62 Acetic acid-producing bacteria, which may also consume the H2, were identified in this study; for example sequences similar to Acetobacterium, Clostridium, and Holophaga.
The higher concentration of acetic acid availability at the end of Stage I provided in Stage II a potential CH4 production of 4.8 ± 0.1 mmol L−1, 6.07 ± 0.3 mmol L−1, and 7.6 ± 0.2 mmol L−1 for A II, B II, and C II, respectively. The lowest acetic acid concentration in C II (30 mg L−1) corresponded to a higher CH4 yield (6.3 mmol g−1 SCB) (Table 3).
The pretreatment (hydrothermal plus enzymatic) applied on SCB was feasible for breaking down the complex molecules of SCB, thereby generating fermentable sugars, which were then converted to organic acids. The acetic acid route for H2 production was the most evident in the present study. Some other short chain organic acids may have been generated in Stage I, such as lactic acid, although this was not detected in the present study, since its conversion to other acids may have occurred. According to Kapdan et al.,15 this acid may follow the conversion pathway to produce acetic, butyric, and propionic acids. Thus, the prevalence of these acids as the main end products occurred in this stage (Fig. 3).
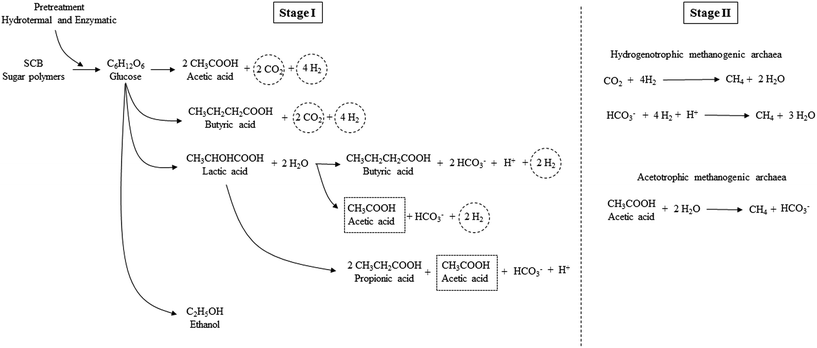 | ||
| Fig. 3 Scheme of soluble metabolites production in Stages I and II for H2 and CH4 production from pretreated SCB. | ||
Thus, lactic acid may be related to another H2 production pathway by microorganisms such as lactic acid bacteria (LAB), since these organisms are commonly present in the autochthonous community of sugarcane.71,72 Members from LAB were identified in this study. The most representative LAB was the genus Enterococcus (0.5–28.9% of total relative abundance), while in a reduced proportion (<1% of total relative abundance), the genera Granulicatella, Lactobacillus, and Streptococcus were also identified.
According to Moreira et al.,73 the diversity of the produced metabolites (acids) in the sugars fermentation may be due to competition among the strains present in the more complex microbial consortium (condition B). The glucose consumption from the polysaccharides from the SCB breakdown (hydrothermal plus enzymatic pretreatment) and their processing routes in acids and H2 in Stage I and for CH4 in Stage II can be verified in Fig. 3.
Rabelo64 identified Desulfovibrio sp. (sulfate reducing bactéria-SRB) when using the same inoculum as in the present study (In1) for H2 production from cellulose. According to these authors, the sulfate present in the culture medium and the acetic acid generated may have favored the growth and maintenance of these groups in the H2 production reactors.
By using Illumina MiSeq sequencing 1.13% (A I), 0.08% (C I), 1.97% (A II), and 3.89% (C II) of the sequences were related to SRB, totaling 19 genera. Among these, the most representative genera were Desulfitobacterium (0.1–0.95%), Desulfosporosinus (0.03–3.28%), Desulfotomaculum (0.02–0.14%), and Desulfovibrio (0.01–0.42%). These genera were also identified as SBR in other studies.74–76
In all conditions, butyric and propionic acids were also observed, but in smaller proportions (Table 4). Butanol and/or ethanol were observed at low concentrations compared to acetic acid in conditions A I, B II, and C I (Table 4).
3.3 Illumina sequencing analysis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 027
027![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 023 sequences from the four samples (Table 5). After trimming, singletons were removed, and the resulting sequences, with an average length of 272 bp, were used to determine a total of 40 and 2128 OTUs (operational taxonomic units) used in the taxonomical classification for archaea and bacteria domains, respectively.
023 sequences from the four samples (Table 5). After trimming, singletons were removed, and the resulting sequences, with an average length of 272 bp, were used to determine a total of 40 and 2128 OTUs (operational taxonomic units) used in the taxonomical classification for archaea and bacteria domains, respectively.
| Sequencing result analysis | A I | C I | A II | C II | ||||
|---|---|---|---|---|---|---|---|---|
| Arch | Bact | Arch | Bact | Arch | Bact | Arch | Bact | |
| a bp–bases pair; OTU – operation taxonomic units; A – inoculum 1; B – inoculum 1 + inoculum 2; C – inoculum 2; Arch – archaea domain; Bact – bacteria domain. | ||||||||
| Good's estimated coverage (%) | 0.919 | 0.999 | 1.000 | 0.999 | 0.999 | 0.999 | 0.999 | 0.998 |
| Sequence length (bp) | 271 | 271 | 272 | 273 | ||||
| Total sequences (trimming data) | 111 | 278![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 605 605 |
52 | 308![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 665 665 |
4813 | 235![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 537 537 |
3910 | 135![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 336 336 |
| Total of OTUs (taxonomical classification) | 27 | 1131 | 16 | 843 | 38 | 1740 | 36 | 1537 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||||
| Richness estimation | ||||||||
| Chao1 | 36.17 | 1432 | 25.33 | 1243 | 38.5 | 1863 | 37 | 1749 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||||
| Diversity index | ||||||||
| Shannon (H) | 2.64 | 3.84 | 2.38 | 2.72 | 2.19 | 4.35 | 2.37 | 4.53 |
| Simpson (D) | 0.88 | 0.93 | 0.88 | 0.87 | 0.82 | 0.94 | 0.87 | 0.97 |
Sequences were classified in two phyla for the archaea domain (Crenarchaeota and Euryarchaeota) present in all the samples studied. A total of 25 phyla were observed in the bacteria domain, including 22 in A I, 19 in C I, 25 in A II, and 24 in C II. In the two conditions of the H2 stage process (A I and C I), the most prevalent phylum was Firmicutes (79.47% and 55.65%, respectively). According to Ratti et al.79 and Santos et al.,80 this phylum contains the main microorganisms that produce H2. Moreover, the anaerobic members of the phylum Firmicutes are also known to be producers of organic acids with short chains of 3 and 4 carbons, like propionic acid and butyric acid,81 which justifies the metabolic routes found in this study (Fig. 3 and Table 4).
The Shannon–Weiner (H) and Simpson (D) indices of the microbial consortium were analyzed (Table 5). Comparing the samples from the H2 stage process (Stage I), the microbial community in A I (3.84) had a higher diversity than C I (0.93). The bacterial community was composed of 209 OTUs with a predominance of the families Enterococcaceae (28.8%) and Clostridiaceae (23.5%), both from phylum Firmicutes. Enterococcaceae was also observed in C I with a similar relative abundance (24.1%), but there was a predominance of Porphyromonadaceae from phylum Bacteroidetes (28.8%).
Bacteria genera belonging to the family Enterococcaceae are Gram-positive and catalase-negative cocci, being also ovoid or spherical in shape, and the cell wall comprises the diamino acid lysine. These organisms are non-endospore-forming and have a low G + C content (33–46%) in the DNA. They are facultative anaerobic and chemo-organotrophic.82
Members of the family Porphyromonadaceae have a variety of cellular morphologies and chemotaxonomic behaviors. Many species of this family are indigenous of the human and animal gastrointestinal tract and oral cavity, nonetheless some species are usually related with a variety of human and animal infections.83
Comparing the samples from CH4 stage process (Stage II), the microbial community had similar values of diversity indices (Table 5), the bacterial community from A II and C II was composed of 296 and 298 OTUs with a predominance of the family Rikenellaceae (phylum Bacteroidetes) (19.9%) and Thermotogaceae (phylum Thermotogae) (11.9%), respectively.
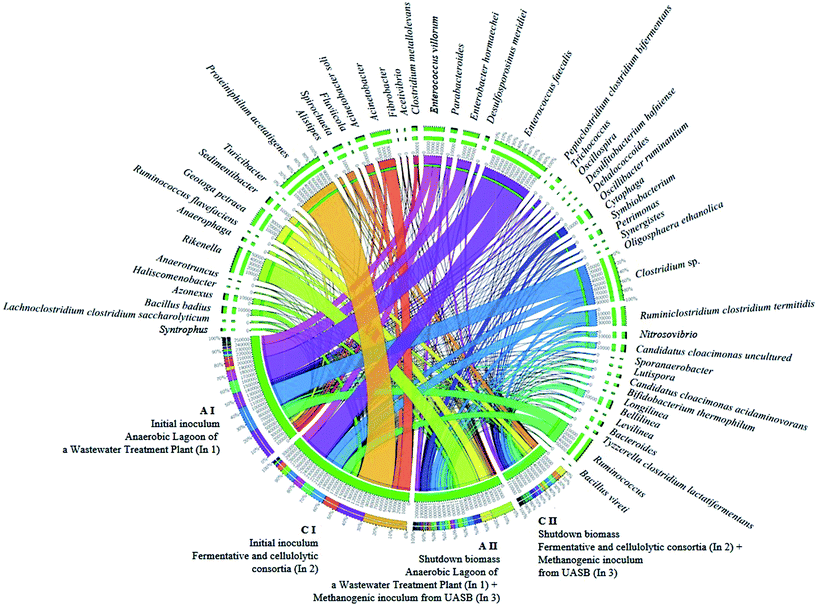 | ||
| Fig. 4 Interaction of the main bacterial genera involved in the two-stage biofuel production from sugarcane bagasse by dark fermentation. | ||
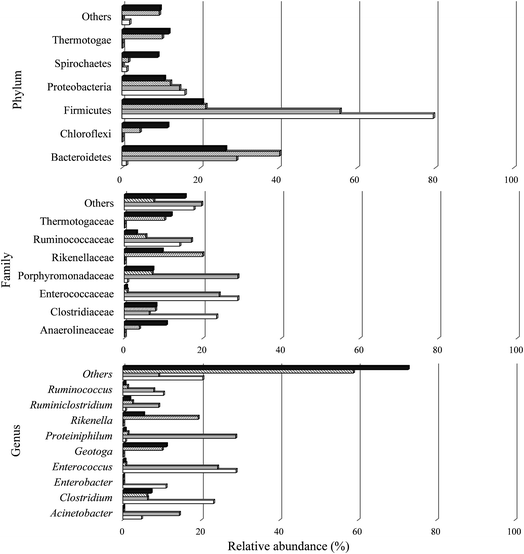 | ||
| Fig. 5 Relative abundance of bacterial phylum, family and genus, in reactors from conditions A I (white), C I (gray), A I (cross stripes), and C II (black). | ||
The Firmicutes phylum comprises some microorganisms able to degrade lignocellulosic residues,84 and according to Bergquist et al.,85 the Firmicutes together with Actinobacteria phyla comprise 80% of the cellulolytic bacteria. The Actinobacteria and Bacteroidetes phyla include microorganisms able to degrade lignocellulosic substrates.86 These microbial groups show potential cellulolytic activity, producing enzymes that are capable of degrading or initiating the breakdown of complex carbon compounds, such as cellulose and hemicelluloses (mainly in soils), thus representing a key step in the carbon cycle,85,87 which justifies their considerable presence in terms of the relative abundance of these phyla in this study.
Streptomyces was the genera belonging to the Actinobacteria phylum with a greater relative abundance (A-II 0.06% and C-II 0.05%) and Bifidobacterium thermophilum (A-I 1.71%; –I 0.02%; A-II 0.03%; C-II 0.02%) as well as representatives of the Micromonosporaceae family (A-II 0.03%). Both of these genera may have played an important role in the ecology of the reactors in the biogas production. Streptomyces genus can produce antibiotics,88 restricting the growth of other microorganisms, Micromonospora species can produce several hydrolytic enzymes, thus playing an active role in the degradation of organic matter.89
Within the Clostridiaceae family (Firmicutes phylum), 23.1% (A I), 6.3% (C I), 6.1% (A II), and 7.2% (C II) of the reads were related to the Clostridium genus. Many anaerobic bacteria can produce H2 from carbohydrate-rich organic wastes. Species such as C. buytricum,90C. thermolacticum,91C. pasteurianum,20C. paraputrificum,92 and C. bifermentants93 are obligate anaerobes and form spores. Clostrida members produce H2 during the exponential growth phase.
Rabelo64 also observed sequences related to Clostridium (67 clones) when the author used the same inoculum as in this study (In1) for H2 and CH4 production using cellulose as a substrate in anaerobic batch reactors. Furthermore, sequences similar to Desulfovibrio (6 clones), Raoultella (2 clones), and Klebsiella (2 clones) were observed to a lesser extent.
The higher diversity observed in A I (H′: 3.84) may be behind the occurrence of H2-consuming bacteria, which may explain the lower H2 yield observed when compared with C I (Table 5). The presence of members from the genera with a homoacetogenic behavior was greater in A I (33.72% of the sequences) compared with C I (14.26% of the sequences) (Table 6). However, the higher H2 production rate in A I (0.59 mmol L−1 h−1) may have provided a stable H2 level during Stage I, although H2 consumption by these microorganism may also have occurred. On the other hand, the lower H2 production rate in C I (0.22 mmol L−1 h−1) promoted a sudden decrease in H2 production, although this production reached higher values (Fig. 1). This diversity of members with possible homoacetogenic behavior favored the low values of H2 yield compared to the theoretical one.
| Genus | A I (%) | C I (%) | A II (%) | C II (%) |
|---|---|---|---|---|
| Acetobacterium | 0.00 | 0.00 | 0.01 | 0.00 |
| Clostridium | 23.13 | 6.26 | 6.13 | 7.20 |
| Eubacterium | 0.02 | 0.01 | 0.08 | 0.13 |
| Holophaga | 0.00 | 0.00 | 0.02 | 0.03 |
| Moorella | 0.00 | 0.00 | 0.03 | 0.06 |
| Ruminococcus | 10.33 | 7.85 | 1.19 | 0.54 |
| Sporomusa | 0.23 | 0.14 | 0.06 | 0.07 |
| Thermoanaerobacter | 0.00 | 0.00 | 0.00 | 0.01 |
| Treponema | 0.00 | 0.00 | 0.23 | 0.26 |
| Total | 33.72 | 14.26 | 7.74 | 8.31 |
The presence of homoacetogens in Stage II was less evident than in Stage I (Table 6), contributing to the hypothesis that the methanogens were favored at this Stage and were the main organisms responsible for the decrease in H2 production as well as in its consumption. In this study, homoacetogens were probably unable to compete effectively for H2, as they show quite poor growth kinetics compared to methanogens.94,95
Even though the bacterial community of condition B was not phylogenetically assessed, it was possible to infer that there was also the presence of these homoacetogens organisms, since the inoculum source consisted of the mixture of In1 (used in condition A I) and In2 (used in condition C I). The remarkable presence of the genus Clostridium, in condition A I may be related mainly to H2 production and not to homoacetogenesis, since representatives of this genus may have both behaviors. Some genera from the Clostridiacea family may have had a direct influence on the biogas production, either for the direct H2 production, or for the degradation of sugars, such as cellulose, hexoses, and glucose, generating organic acids (acetic acid and formic acid) as substrates for the methanogenic archaea (Fig. 4).
The genera Brassicibacter (A I 0.06%, C I 0.01%, A II 0.29%, and C II 0.14%) and Anaerosporobacter are capable of producing acetic acid and formic acid, which may have occurred in Stage II, as well as propionic acid, ethanol,96 and H2 by Anaerosporobacter.97 The species of these genera are also capable of using fructose, glucose, and sucrose as a carbon source, which may have been generated by the enzymatic hydrolysis in the biological pretreatment and by the enzymatic action of other microorganisms from SCB.96
However, although the genus Lutispira found in this study is capable of using a few carbohydrates, it cannot directly ferment arabinose, cellobiose, cellulose, fructose, galactose, glucose, glycerol, lactose, maltose, mannose, ribose, starch, sucrose, xylose, among others, and thus members of this group are restricted to the availability of other energy sources.98 Likewise the genus Oxobacter are not able to use sugars, amino acids, organic acids and alcohols as energy sources; however, the fermentation of pyruvate by these microorganisms generates acetic acid and CO2. When CO2 is catabolically used, acetic acid and butyric acid are generated as metabolic products.99
In the same way, the genus Caloramator has ethanol and acetic acid as end products in the glucose degradation.100 In the present study, the acetic acid produced will contribute to the methane production in Stage II. The differential of the genera Fonticella and Geosporobacter is that these microorganisms are able to use cellobiose, besides fructose, sucrose, and glucose as electrons donors,101 with formic acid, acetic acid, ethanol, and CO2 as the main end products from the fermentation of these sugars.102
The genus Fervidicella (family Clostidiacea) is capable of fermenting various types of carbohydrates, among them glucose, and produces ethanol, acetic acid, CO2, and H2. However, it does not use organic acids as a source of carbon and energy.103 In the same way, members of the genus Saccharofermentans are able to use hexoses, polysaccharides, glycoses, and alcohols, generating acetic acid, lactic acid, fumaric acid, CO2, and H2. However, in cellulose degradation, the representatives of this microbial group do not generate organic acids, being therefore dependents on the action of the enzymatic treatment of other microorganisms.104 Probably, this group benefited from the enzymatic treatment of SCB used in this study.
Furthermore, a higher relative abundance of the hydrogenotrophic methanogenic Methanobacterium was observed in this condition, thereby allowing higher H2 consumption (40.4% – A I and 7.7% – C I). Another genus identified mainly in samples from Stage I was Enterobacter (Firmicutes phylum; Enterobacteriacea family). Members from this genus are facultative anaerobes and the quantity of H2 produced is equivalent to that from Clostridum members.90,105 Both aforementioned genera were observed primarily in condition A I (Fig. 5).
A high H2 yield was observed when Enterobacter sp. was used as the inoculum with 16.5 g L−1 xylose in batch reactors (2.0 ± 0.05 mol H2 per mol substrate).106 The higher value obtained by these authors was probably due to the simplicity of the substrate and to the higher concentration used when compared to that used in the present study (2 g L−1 g−1 SCB). Some bacteria from this genus have the ability to degrade cellulosic substrates, since cellulase activity has been observed in some members of the Entrobacteriaceae family.107
With their higher relative abundance in this study, members from Enterococcus genera (Firmicutes phylum) were observed mainly in the H2 production stage (A I and C I). In the present study, two species belonging to Enterococcus may be related to SCB degradation and H2 production in stage I, with E. faecalis (21.2% and 17.9%) and E. villorum (7.6% and 6.1%) identified in samples from A I and C I, respectively.
The potential of Enterococcus gallinarum G1 to degrade cellulose and produce H2 under mesophilic conditions (37 °C) and pH 6.5 was evaluated.108 The authors noted that cellulose hydrolysis and H2 production occurred with 5 h of λ time and H2 continued to be produced in the following 60 h. The H2 yield observed was 2.38 mmol H2 g−1 cellulose and the end liquid products were primarily acetate, propionate, and butyrate. Hu and Zhu109 and Hu et al.110 also reported that acetic acid and butyric acid were the predominant metabolites.
In the present study, acetate, propionate and butyrate were also the main metabolites generated. Moreover, a higher H2 yield was observed for both conditions (A I and C I). Although the substrate employed in this study was more complex (SCB), the higher microbial diversity used as the inoculum may have favored the conversion of SCB to easily degradable carbohydrates, thereby promoting the higher H2 yield (A I – 3.12 mmol H2 per g SCB added and C I – 4.26 mmol H2 per g SCB added).
A bacterium able to degrade cellulose was isolated from the gut of Microcerotermes diversus.111 According to the authors, three cellulose degrading bacteria isolated belonged to the genera Acinetobacter, Pseudomonas and Staphylococcus. Members from the genus Acinetobacter were also observed in this study, mainly in samples from Stage I, 4.8% and 14.4% for A I and C I, respectively.
Furthermore, some members from this genus have the ability to degrade hemicellulose, an important component of SCB. According to Lo et al.,112 xylanase is the main enzyme to hydrolyze hemicellulose and to originate fermentable sugars (five-carbon) for H2 production. A cellulolytic bacterium, Acinetobacter junii F6-02, was isolated from soil in Taiwan and this strain was able to produce xylanase and cellulase extracellularly with high efficiency.
Within this genus, two species were identified, Acinetobacter genomo sp. 3 (2.5% – A I and 10.2% – C I) and Acinetobacter soli (1.9% – A I and 4% – C I). The high proportion of this genus on C I may have led to the higher degradation of cellulose and hemicellulose from SCB, which provided a higher H2 yield in this condition.
Many cellulolytic microorganisms have been described by their capacity to hydrolyze lignocellulosic substrates for energy production. Those cellulolytic organisms include the bacteria Ruminococcus, Bacillus, Streptomyces, and Bacteroides.113 These genera were also observed in the present study. Among them, Ruminococcus had a higher relative abundance (10.3%, 7.8%, 19.2%, and 5.3%), followed by Bacillus (0.6%, 5%, 0.3%, and 0.3%), Bacterioides (0.1%, 0.1%, 2.5%, and 2.3%), and finally, Streptomyces (0%, 0%, 0.06%, and 0.05%) for samples from A I, C I, A II, and C II, respectively. Consequently, these microbial groups may have used the cellulose from the sugarcane bagasse as an energy and catabolic source.
Another group, probably involved in cellulose degradation, was found at a high proportion in the CH4 stage. The Geotoga genus (Thermotogales order) was observed mainly in A II (10.1%) and C II (11.1%). Thermotogales are very important due to their ability to metabolize simple and complex carbohydrates, such as sucrose, cellulose, maltose, and xylose.114 The SCB used in this study served as a cellulose source for this bacteria group.
Ruminiclostridium is an anaerobic and cellulolytic bacteria observed mainly in C I (9.1%). This mesophilic bacterium metabolizes cellulose and some hemicellulosic polysaccharides into fermentable sugars.115 The substrate employed in this study (SCB) may have favored the development of members from this genus. More noticeable in the samples from CH4 production (A II and C II), the genus Rikenella, members of the genus Bacteroides, were found in several different digestive tracts and probably indigenous from the inoculum used in the CH4 stage.
The characteristics of the culture medium may have favored some microbial groups. In fact, approximately, 0.7% (A I), 28.8% (C I), 1.3% (A II), and 0.6% (C II) of the sequences found were associated with Proteiniphilum genus. Proteiniphilum is an obligately anaerobic Gram-negative bacterium. Members from this genus grow better under mesophilic conditions and the yeast extract (YE) and peptone can be used as energy sources. P. acetatigenes was first observed and isolated from an upflow anaerobic sludge blanket-type anaerobic digester.116 The presence of YE (1 g L−1) in the culture medium, in this study, may have caused the development of this species. Furthermore, higher concentrations of acetic acid (9262.3 mg L−1) and propionic acid (700.1 mg L−1) were observed for the condition with a higher relative abundance of P. acetatigenes (C I).
A total of 250 bacterial species were classified in the biomass of the reactors. In this study, 13 of the most common species (>1% of the classified sequences) accounted for 51.6%, 80.6%, 16.1%, and 17.8% of the classified sequences for reactors from conditions A I, C I, A II, and C II, respectively (Table 7). In the four samples, the difference in the bacterial community composition was evident with a higher abundance of Enterococcus faecalis and Proteiniphilum acetatigenes in the H2 production stage (A I and C I, respectively) and Geotoga petraea in the CH4 production stage (A II and C II).
| Species | A I (%) | C I (%) | A II (%) | C II (%) |
|---|---|---|---|---|
| Acinetobacter genomo sp. 3 | 2.52 | 10.16 | 0.07 | 0.09 |
| Acinetobacter soli | 1.91 | 4.05 | 0.04 | 0.07 |
| Bacillus badius | 0.10 | 4.17 | 0.03 | 0.04 |
| Bifidobacterium thermophilum | 1.71 | 0.02 | 0.03 | 0.02 |
| Candidatus cloacimonas acidaminovorans | 0.01 | 0.02 | 0.52 | 3.39 |
| Clostridium lactatifermentans | 1.46 | 0.07 | 0.26 | 0.06 |
| Clostridium metallolevans | 2.86 | 0.03 | 0.82 | 0.33 |
| Clostridium termitidis | 0.46 | 9.08 | 2.42 | 1.74 |
| Enterobacter hormaechei | 10.99 | 0.13 | 0.04 | 0.08 |
| Enterococcus faecalis | 21.25 | 17.93 | 0.51 | 0.35 |
| Enterococcus villorum | 7.61 | 6.15 | 0.23 | 0.17 |
| Geotoga petraea | 0.09 | 0.10 | 10.06 | 11.09 |
| Proteiniphilum acetatigenes | 0.67 | 28.75 | 1.07 | 0.38 |
| Others | 48.35 | 19.36 | 83.89 | 82.19 |
| Total | 100 | 100 | 100 | 100 |
All known methanogens belong to the Euryarchaeota phylum.118 In this study, the phylum Euryarchaeota represented a large majority of the sequences identified in the biomass under all conditions. There was a predominance of four archaeal families (Fig. 6). The majority of representatives in all samples were Methanobacteriaceae (7.9–55.8%), Methanoregulaceae (22.5–52.9%), and Methanosaetaceae (4.5–25%).
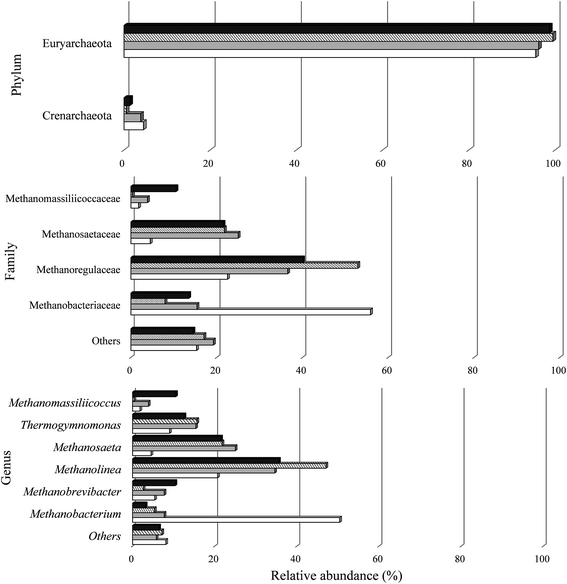 | ||
| Fig. 6 Relative abundance of archaeal phylum, family and genus in the reactors under conditions A I (white), C I (gray), A II (cross stripes), and C II (black). | ||
In the four samples studied, 11 genera were classified. The biomass from condition A I contained higher populations of Methanobacterium (50.4%) and from C I, A II, and C II higher populations of Methanolinea (34.6%, 47.1% and 35.6%, respectively).
Methanogens belonging to the genus Methanobrevibacter were also observed in this study. Similar to both genera, Methanobacterium sp. and Methanolinea, they can also use the end products of bacterial fermentation to produce CH4.119,120 All these genera produce CH4 from formate and H2/CO2, but not from acetate. In this study, it is possible that the H2 produced in the last stage (A II: 2 ± 0.1 mmol H2 L−1 and C II: 0.5 ± 0.03 mmol H2 L−1) served as substrates for the development of those genera in the reactors (Fig. 5).
The genus Methanosaeta are specialized in metabolizing acetate, reflecting a high affinity for the compound.121 This genus has been observed to dominate the methanogenic community in anaerobic reactors.122 In this study, representatives of this genus were uniformly distributed between samples A II (21.8%) and C II (21.5%). This genus was responsible for the conversion of acetic acid to CH4 from Stage I to II with a reduction of 55.7% and 99.8% for A II and C II, respectively.
A total of nine archaeal species were classified in the biomass of the reactors (Table 8), which accounted for 14.4%, 11.5%, 8.9%, and 17.7% of the classified sequences for the reactors under conditions A I, C I, A II, and C II, respectively. Differences in the composition of the archaeal community in the four samples were evident with the predominance of Methanosphaerula palustris, Methanobacterium subterraneum and Candidatus methanomassiliicoccus intestinalis in the CH4 production stage (A II and C II).
| Species | A I (%) | C I (%) | A II (%) | C II (%) |
|---|---|---|---|---|
| Candidatus methanomassiliicoccus intestinalis | 1.80 | 3.85 | 0.35 | 10.51 |
| Candidatus methanoregula boonei | 0.00 | 0.00 | 0.17 | 0.20 |
| Candidatus nitrosocaldus | 4.50 | 1.92 | 0.50 | 1.20 |
| Candidatus nitrosocaldus yellowstonii | 0.00 | 1.92 | 0.02 | 0.13 |
| Methanobacterium bryantii | 1.80 | 0.00 | 0.12 | 0.05 |
| Methanobacterium subterraneum | 2.70 | 1.92 | 2.80 | 1.51 |
| Methanosaeta concilii | 0.90 | 0.00 | 1.06 | 1.05 |
| Methanosaeta pelagica | 0.90 | 0.00 | 0.08 | 0.00 |
| Methanosphaerula palustris | 1.80 | 1.92 | 3.84 | 3.09 |
| Others | 85.6 | 88.5 | 91.0 | 82.3 |
| Total | 100 | 100 | 100 | 100 |
Methanosphaerula palustris is strictly anaerobic, mesophilic, and mildly acidophilic.123Methanobacterium subterraneum is an autotrophic, halotolerant methanogen.124 Both species use H2/CO2 for CH4 formation and do not use acetate. The affiliation of Ca. Methanomassiliicoccus intestinalis to a large cluster of sequences retrieved from paddy soils, freshwater, and marine sediments suggest their recent adaptation to gut environments.125
The generation of biofuels from renewable resources, mainly lignocellulosic biomass, is possible from the majority of low-priced and abundant non-food materials that can be obtained from plants. Thus, lignocellulosic materials have the potential to provide second-generation biofuels.126 The production of H2, bio-oils, biogas, alcohols, and biodiesel from renewable biomass, such as SCB, have been a major research focus around the world in order to enhance petroleum fuels and reduce environmental pollution.55
The application of two-stage anaerobic digestion and hydrothermal and enzymatic pretreatment, is an excellent strategy to improve H2 and CH4 production, since the metabolites produced in one stage could subsequently be used in the next operational stages. It was also observed that the microbial community adapted to the conditions imposed, thus favoring each operational stage, optimizing the SCB degradation system in a two-stage process. Integrating H2 and CH4 production in the same reactor, in different stages, provides a higher potential to use the SCB to produce bioenergy. The highly diversified microbial consortia used in this study allowed high yields of both H2 and methane production.
4. Conclusion
The anaerobic conversion of SCB to H2 and CH4 is an attractive alternative to renewable energy. This is a rare study that investigated the combination of the hydrothermal and enzymatic pretreatment of SCB through the activity of A. niger, with biogas measurements, by-products analyses, and phylogenetic characterization of the microorganisms involved. The combination of both pretreatments was adequate in the hydrolysis of SCB, since H2 was detected in all the studied conditions. The mixed culture obtained from wastewater treatment plant sludge (inoculum 1) was a favorable source for H2 production using SCB, as well as the cellulolytic and fermentative culture (inoculum 2). However, the mixture of inoculum 1 and inoculum 2 did not favor H2 production. The highest H2 and CH4 potential production was obtained when inoculum 2 (fermentative and cellulolytic consortia) was employed in Stage I (C I), obtaining 5.47 mmol L−1 and 7.60 mmol L−1, respectively. The metabolite generated in the first stage fermentation contributed to methane production in the second one. Acetic acid was the major metabolite formed under all the conditions studied, suggesting that fermentation occurred via this route. Microbial consortium able to produce cellulase and xylanase enzymes, such as Enterobacter, Enterococcus, Acinetobacter, Clostridium, and Geotoga, were identified in this study. In the methanogenic stage, three genera of hydrogenotrophic methanogens (Methanolinea, Methanobrevibacter, and Methanobacterium) and one genus of acetoclastic methanogens (Methanosaeta) were identified. Furthermore, homoacetogenic bacterias, such as Acetobacterium, Clostridium, Eubacterium, Holophaga, Moorella, Ruminococcus, Sporomusa, Thermoanaerobacter, and Treponema were also identified. This microbial group played an essential role in Stage I, consuming a large part of the H2 produced with acetic acid generation. These microorganisms act in synergy for the hydrolysis of SCB and H2 and CH4 production. Methanogenesis and homoacetogenesis play decisive roles in the efficiency of H2 production; thus, further studies are needed to elucidate the interaction of these microbial groups in the degradation of cellulose wastes and in bioenergy production. Therefore, the employment of a consortium containing members from Clostridium, Bacillus, Bacteroides, and Paenibacillus genera (inoculum 2) for hydrogen production and a methanogenic consortium for subsequent methane production represents a promising insight into the anaerobic digestion of hydrothermal- and enzymatic-pretreated SCB, which provides an example of the production of biogas and value-added by-products from lignocellulosic biomass.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This project was funded by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) process number 2013/20196-7 and 2009/15984-0.References
- B. Kamm and M. Kamm, Appl. Microbiol. Biotechnol., 2004, 64, 137–145 CrossRef PubMed
.
- Y. Lu, Q. Lai, C. Zhang, H. Zhao, K. Ma, X. Zhao, H. Chen, D. Liu and X. H. Xing, Bioresour. Technol., 2009, 100, 2889–2895 CrossRef PubMed
.
- S. K. Han and H. S. Shin, J. Air Waste Manage. Assoc., 2004, 54, 242–249 CrossRef
.
- D. Liu, D. Liu, R. J. Zeng and I. Angelidaki, Water Res., 2006, 40, 2230–2236 CrossRef PubMed
.
- V. Blonskaja, A. Menert and R. Vilu, Adv. Environ. Res., 2003, 7, 671–678 CrossRef
.
- C. Ting and D. Lee, Int. J. Hydrogen Energy, 2007, 32, 677–682 CrossRef
.
- G. Antonopoulou, H. N. Gavala, I. V. Skiadas, K. Angelopoulos and G. Lyberatos, Bioresour. Technol., 2008, 99, 110–119 CrossRef PubMed
.
- Y. C. Lo, G. D. Saratale, W. M. Chen, M. Der Bai and J. S. Chang, Enzyme Microb. Technol., 2009, 44, 417–425 CrossRef
.
- E. C. Koutrouli, H. Kalfas, H. N. Gavala, I. V. Skiadas, K. Stamatelatou and G. Lyberatos, Bioresour. Technol., 2009, 100, 3718–3723 CrossRef PubMed
.
- B.-B. Hu and M.-J. Zhu, Int. J. Hydrogen Energy, 2017, 42, 7866–7874 CrossRef
.
- H.-N. Lin, Y.-T. Wang and M.-J. Zhu, Int. J. Hydrogen Energy, 2017, 42, 26687–26694 CrossRef
.
- H. Zhu, A. Stadnyk, M. Béland and P. Seto, Bioresour. Technol., 2008, 99, 5078–5084 CrossRef PubMed
.
- R. P. Ratti, T. P. Delforno, I. K. Sakamoto and M. B. A. Varesche, Int. J. Hydrogen Energy, 2015, 40, 6296–6306 CrossRef
.
- J. K. Braga, L. A. Soares, F. Motteran, I. K. Sakamoto and M. B. A. Varesche, Int. J. Hydrogen Energy, 2016, 41(48), 22812–22823 CrossRef
.
- I. K. Kapdan and F. Kargi, Enzyme Microb. Technol., 2006, 38, 569–582 CrossRef
.
- L. A. Soares, J. K. Braga, F. Motteran, I. K. Sakamoto, E. L. Silva and M. B. A. Varesche, Water Sci. Technol., 2017, 76(1–2), 95–105 CrossRef PubMed
.
- H. Yokoi, S. Mori, J. Hirose, S. Hayashi and Y. Takasaki, Biotechnol. Lett., 1998, 20, 895–899 CrossRef
.
- H. L. Chin, Z. S. Chen and C. P. Chou, Biotechnol. Prog., 2003, 19, 383–388 CrossRef PubMed
.
- M. Ferchichi, E. Crabbe, G.-H. Gil, W. Hintz and A. Almadidy, J. Biotechnol., 2005, 120, 402–409 CrossRef PubMed
.
- G. Liu and J. Shen, J. Biosci. Bioeng., 2004, 98, 251–256 CrossRef PubMed
.
- J. K. Braga, A. A. Abreu, F. Motteran, M. A. Pereira, M. M. Alves and M. B. A. Varesche, Waste Biomass Valorization, 2017, 1–11 Search PubMed
.
- C. A. B. S. Rabelo, L. A. Soares, I. K. Sakamoto, E. L. Silva and M. B. A. Varesche, J. Environ. Manage., 2018, 223, 952–963 CrossRef PubMed
.
- J. Cheng and M. Zhu, Bioresour. Technol., 2013, 144, 623–631 CrossRef PubMed
.
- N. Reddy and Y. Yang, Trends Biotechnol., 2005, 23, 22–27 CrossRef PubMed
.
- M. Balat, H. Balat and C. Öz, Prog. Energy Combust. Sci., 2008, 34, 551–573 CrossRef
.
- S. H. da Cruz, B. S. Dien, N. N. Nichols, B. C. Saha and M. A. Cotta, J. Ind. Microbiol. Biotechnol., 2012, 39, 439–447 CrossRef PubMed
.
- S. Kumar, U. Kothari, L. Kong, Y. Y. Lee and R. B. Gupta, Biomass Bioenergy, 2011, 35, 956–968 CrossRef
.
- S. G. Allen, D. Schulman, J. Lichwa, M. J. Antal, M. Laser and L. R. Lynd, Ind. Eng. Chem. Res., 2001, 40, 2934–2941 CrossRef
.
- Y. Gao, J. Xu, Y. Zhang, Q. Yu, Z. Yuan and Y. Liu, Bioresour. Technol., 2013, 144, 396–400 CrossRef PubMed
.
- H. Jørgensen, J. B. Kristensen and C. Felby, Biofuels, Bioprod. Biorefin., 2007, 1, 119–134 CrossRef
.
- B. C. Saha, N. Qureshi, G. J. Kennedy and M. A. Cotta, Int. Biodeterior. Biodegrad., 2016, 109, 29–35 CrossRef
.
- M. J. Taherzadeh and K. Karimi, BioResources, 2007, 2, 707–738 Search PubMed
.
- C. N. Hamelinck, G. van Hooijdonk and A. P. Faaij, Biomass Bioenergy, 2005, 28, 384–410 CrossRef
.
- W. Wang, T. Yuan, K. Wang, B. Cui and Y. Dai, Bioresour. Technol., 2012, 107, 282–286 CrossRef PubMed
.
- C. L. Aguiar and T. J. B. Menezes, Bol. Cent. Pesqui. Process. Aliment., 2000, 18, 57–70 Search PubMed
.
- T. Zhang and M.-J. Zhu, Bioresour. Technol., 2016, 214, 769–777 CrossRef PubMed
.
- L. A. Soares, J. K. Braga, F. Motteran, I. K. Sakamoto, P. A. S. Monteiro, P. Seleghim and M. B. A. Varesche, Waste Biomass Valorization, 2018, 1–14 Search PubMed
.
- Z. Zhu, M. Zhu and Z. Wu, Bioresour. Technol., 2012, 119, 199–207 CrossRef PubMed
.
-
F. Ahmad, Methane Production in Response to Sulfuric Acid and Hydrogen Peroxide Assisted Hydrothermal Pretreatment of Sugarcane Bagasse, São Carlos, 2017 Search PubMed
.
- M. Mandels and J. Weber, Cellulases and Their Applications, Advances in Chemistry, 1969, ch. 23, vol. 95, pp. 391–414 Search PubMed.
-
R. Atlas, Handbook of Microbiological Media, Boca Raton, 4th edn, 2010 Search PubMed
.
- C. A. B. S. Rabelo, L. A. Soares and M. B. A. Varesche, Austin Clinical Microbiology, 2017, 2, 1–6 Search PubMed
.
- F. Motteran, J. K. Braga, E. L. Silva and M. B. A. Varesche, J. Environ. Sci. Health, Part A: Environ. Sci. Eng., 2016, 51, 1288–1302 CrossRef PubMed
.
- E. D. Penteado, C. Z. Lazaro, I. K. Sakamoto and M. Zaiat, Int. J. Hydrogen Energy, 2013, 38, 6137–6145 CrossRef
.
- J. K. Braga and M. B. A. Varesche, Am. J. Anal. Chem., 2014, 05, 8–16 CrossRef
.
-
APHA, Standard Methods for the Examination of Water and Wastewater, 20th edn, 2005 Search PubMed
.
- M. Dubois, K. A. Gilles, J. K. Hamilton, P. A. Rebers and F. Smith, Anal. Chem., 1956, 28, 350–356 CrossRef
.
- M. Zwietering, I. Jongenburger, F. Rombouts and K. Van’t, Riet, 1990, 56, 1875–1881 Search PubMed
.
- R. I. Griffiths, A. S. Whiteley, A. G. Oapos Donnell and M. J. Bailey, Appl. Environ. Microbiol., 2000, 66, 5488–5491 CrossRef PubMed
.
- U. Nübel, B. Engelen, A. Felske, J. Snaidr, A. Wieshuber, R. I. Amann, W. Ludwig and H. Backhaus, J. Bacteriol., 1996, 178, 5636–5643 CrossRef
.
- Y. Kudo, FEMS Microbiol. Ecol., 1997, 22, 39–48 CrossRef
.
- B. Si, Z. Liu, Y. Zhang, J. Li, X.-H. Xing, B. Li, N. Duan and H. Lu, Int. J. Hydrogen Energy, 2015, 40, 3191–3200 CrossRef
.
- Y. Y. Li, H. Sasaki, K. Yamashita, K. Seki and I. Kamigochi, Water Sci. Technol., 2002, 45, 143–150 CrossRef PubMed
.
- Y. Ueno, H. Fukui and M. Goto, Environ. Sci. Technol., 2007, 41, 1413–1419 CrossRef PubMed
.
- R. Chandra, H. Takeuchi and T. Hasegawa, Renewable Sustainable Energy Rev., 2012, 16, 1462–1476 CrossRef
.
- M. J. Taherzadeh and K. Karimi, Int. J. Mol. Sci., 2008, 9, 1621–1651 CrossRef PubMed
.
-
L. Drake, H. L. Kusel and C. Matthies, in The Prokaryotes, New York, 2006, pp. 354–420 Search PubMed
.
- G. Diekert and G. Wohlfarth, Antonie van Leeuwenhoek, 1994, 66, 209–221 CrossRef PubMed
.
- R. Conrad and M. Klose, FEMS Microbiol. Ecol., 1999, 30, 147–155 CrossRef PubMed
.
- H. H. Fang and X.-S. Jia, Water Res., 1999, 33, 1791–1798 CrossRef
.
- V. Siriwongrungson, R. J. Zeng and I. Angelidaki, Water Res., 2007, 41, 4204–4210 CrossRef PubMed
.
- J. J. Lay, Biotechnol. Bioeng., 2001, 74, 280–287 CrossRef PubMed
.
- N. Q. Ren, J. F. Xu, L. F. Gao, L. Xin, J. Qiu and D. X. Su, Int. J. Hydrogen Energy, 2010, 35, 2742–2746 CrossRef
.
- C. A. B. S. Rabelo, Avaliação de consórcio microbiano obtido a partir de lodo de estação de tratamento de efluentes de indústria de papel e celulose visando à produção de H2 a partir de celulose, UNIVERSIDADE DE SÃO PAULO, 2014.
-
C. A. B. S. Rabelo, L. S. Botta and M. B. A. Varesche, Proc. XI DAAL - Lat. Am. Work. Symp. Anaerob. Dig, Havana, Cuba, 2014 Search PubMed
.
- B. Xie, J. Cheng, J. Zhou, W. Song and K. Cen, Int. J. Hydrogen Energy, 2008, 33, 5006–5011 CrossRef
.
- R. P. Ratti, L. S. Botta, I. K. Sakamoto and M. B. A. Varesche, Int. J. Hydrogen Energy, 2013, 38, 9707–9717 CrossRef
.
- R. P. Ratti, L. S. Botta, I. K. Sakamoto, E. L. Silva and M. B. A. Varesche, Biotechnol. Lett., 2014, 36, 537–546 CrossRef PubMed
.
- M. Kim, C. Liu, J. Noh, Y. Yang, S. Oh, K. Shimizu, D. Lee and Z. Zhang, Int. J. Hydrogen Energy, 2013, 38, 8648–8656 CrossRef
.
-
W. B. Whitman, D. R. Boone, Y. Koga and J. Keswani, in Bergey's manual of systematic bacteriology, ed. G. M. G. D. R. Boone and R. W. Castenholz, New York, 2nd edn, 2001, pp. 211–213 Search PubMed
.
- Y. Sobrun, A. Bhaw-Luximon, D. Jhurry and D. Puchooa, Adv. Biosci. Biotechnol., 2012, 03, 398–407 CrossRef
.
- F. C. O. Gomes, C. L. C. Silva, C. R. Vianna, I. C. A. Lacerda, B. M. Borelli, Á. C. Nunes, G. R. Franco, M. M. Mourão and C. A. Rosa, Braz. J. Microbiol., 2010, 41, 486–492 CrossRef PubMed
.
- F. S. Moreira, R. G. Machado, B. B. Romão, F. R. X. Batista, J. S. Ferreira and V. L. Cardoso, Process Biochem., 2017, 58, 60–68 CrossRef
.
- H. J. Laanbroek, H. J. Geerligs, L. Sijtsma and H. Veldkamp, Appl. Environ. Microbiol., 1984, 47, 329–334 Search PubMed
.
- M. Pester, N. Bittner, P. Deevong, M. Wagner and A. Loy, ISME J., 2010, 4, 1591–1602 CrossRef PubMed
.
- T. Aüllo, A. Ranchou-Peyruse, B. Ollivier and M. Magot, Front. Microbiol., 2013, 4, 362 Search PubMed
.
- F. E. Mosey, Water Sci. Technol., 1983, 15, 209–232 CrossRef
.
-
S. R. Harper and F. G. Pohland, in Microbiology and Biochemistry of Strict Anaerobes Involved in Interspecies Hydrogen Transfer, Springer US, Boston, MA, 1990, pp. 387–390 Search PubMed
.
- R. P. Ratti, L. S. Botta, I. K. Sakamoto, E. L. Silva and M. B. A. Varesche, Biotechnol. Lett., 2014, 36, 537–546 CrossRef PubMed
.
- S. C. Santos, P. R. F. Rosa, I. K. Sakamoto, M. B. Amâncio Varesche and E. L. Silva, Int. J. Hydrogen Energy, 2014, 39, 9599–9610 CrossRef
.
- S. Venkata Mohan, L. Agarwal, G. Mohanakrishna, S. Srikanth, A. Kapley, H. J. Purohit and P. N. Sarma, Int. J. Hydrogen Energy, 2011, 36, 8234–8242 CrossRef
.
-
J. M. Whiley and R. A. Hardie, in Bergey's Manual of Systematic Bacteriology The Firmicutes, ed. D. Jones, P. De Vos and G. M. Garrity, Springer US, New York, USA, 2nd edn, 2009, pp. 655–711 Search PubMed
.
-
M. Sakamoto, in The Prokaryotes, Springer Berlin Heidelberg, Berlin, Heidelberg, 2014, pp. 811–824 Search PubMed
.
- C. R. Carere, R. Sparling, N. Cicek and D. B. Levin, Int. J. Mol. Sci., 2008, 9, 1342–1360 CrossRef PubMed
.
- P. L. Bergquist, M. D. Gibbs, D. D. Morris, V. S. J. Te’o, D. J. Saul and H. W. Morgan, FEMS Microbiol. Ecol., 1999, 28, 99–110 CrossRef
.
- C. Guevara and M. M. Zambrano, FEMS Microbiol. Lett., 2006, 255, 52–58 CrossRef PubMed
.
- A. Pourcher, L. Sutra, I. I. Hébé, G. Moguedet, C. Bollet, P. Simoneau and L. Gardan, FEMS Microbiol. Ecol., 2001, 34, 229–241 CrossRef PubMed
.
- J. Bérdy, J. Antibiot., 2005, 58, 1–26 CrossRef PubMed
.
- E. A. Barka, P. Vatsa, L. Sanchez, N. Gaveau-Vaillant, C. Jacquard, H.-P. Klenk, C. Clément, Y. Ouhdouch and G. P. van Wezel, Microbiol. Mol. Biol. Rev., 2016, 80, 1–43 CrossRef PubMed
.
- H. Yokoi, A. Saitsu, H. Uchida, J. Hirose, S. Hayashi and Y. Takasaki, J. Biosci. Bioeng., 2001, 91, 58–63 CrossRef PubMed
.
- C. Collet, Int. J. Hydrogen Energy, 2004, 29, 1479–1485 CrossRef
.
- D. Evvyernie, K. Morimoto, S. Karita, T. Kimura, K. Sakka and K. Ohmiya, J. Biosci. Bioeng., 2001, 91, 339–343 CrossRef PubMed
.
- C. C. Wang, C. W. Chang, C. P. Chu, D. J. Lee, B.-V. Chang, C. S. Liao and J. H. Tay, Water Res., 2003, 37, 2789–2793 CrossRef PubMed
.
- V. Peters, P. Janssen and R. Conrad, FEMS Microbiol. Ecol., 1998, 26, 317–324 CrossRef
.
- A. Stams, Antonie van Leeuwenhoek, 1994, 66, 271–294 CrossRef PubMed
.
- M. X. Fang, W. W. Zhang, Y. Z. Zhang, H. Q. Tan, X. Q. Zhang, M. Wu and X. F. Zhu, Int. J. Syst. Evol. Microbiol., 2012, 62, 3018–3023 CrossRef PubMed
.
- H. Jeong, Y. W. Lim, H. Yi, Y. Sekiguchi, Y. Kamagata and J. Chun, Int. J. Syst. Evol. Microbiol., 2007, 57, 1784–1787 CrossRef PubMed
.
- H. Shiratori, H. Ohiwa, H. Ikeno, S. Ayame, N. Kataoka, A. Miya, T. Beppu and K. Ueda, Int. J. Syst. Evol. Microbiol., 2008, 58, 964–969 CrossRef PubMed
.
-
F. A. Rainey, in Bergey's Manual of Systematics of Archaea and Bacteria, John Wiley & Sons, Ltd, Chichester, UK, 2015, pp. 1–4 Search PubMed
.
- C. D. Ogg and B. K. C. Patel, Int. J. Syst. Evol. Microbiol., 2009, 59, 95–101 CrossRef PubMed
.
- N. Klouche, M.-L. Fardeau, J.-F. Lascourreges, J.-L. Cayol, H. Hacene, P. Thomas and M. Magot, Int. J. Syst. Evol. Microbiol., 2007, 57, 1757–1761 CrossRef PubMed
.
- B. Fraj, W. Ben Hania, A. Postec, M. Hamdi, B. Ollivier and M.-L. Fardeau, Int. J. Syst. Evol. Microbiol., 2013, 63, 1947–1950 CrossRef PubMed
.
- C. D. Ogg and B. K. C. Patel, Int. J. Syst. Evol. Microbiol., 2010, 60, 1394–1400 CrossRef PubMed
.
- S. Chen, L. Niu and Y. Zhang, Int. J. Syst. Evol. Microbiol., 2010, 60, 2735–2738 CrossRef PubMed
.
- Y. Nakashimada, M. A. Rachman, T. Kakizono and N. Nishio, Int. J. Hydrogen Energy, 2002, 27, 1399–1405 CrossRef
.
- C. Long, J. Cui, Z. Liu, Y. Liu, M. Long and Z. Hu, Int. J. Hydrogen Energy, 2010, 35, 6657–6664 CrossRef
.
- A. Chatterjee, Y. Cui, Y. Liu, C. K. Dumenyo and A. K. Chatterjee, Appl. Environ. Microbiol., 1995, 61, 1959–1967 Search PubMed
.
- A. Wang, L. Gao, N. Ren, J. Xu and C. Liu, Biotechnol. Lett., 2009, 31, 1321–1326 CrossRef PubMed
.
- B.-B. Hu and M.-J. Zhu, Microb. Cell Fact., 2017, 16, 77 CrossRef PubMed
.
- B.-B. Hu, M.-Y. Li, Y.-T. Wang and M.-J. Zhu, Chem. Eng. J., 2018, 335, 979–987 CrossRef
.
- Z. Pourramezan, G. R. Ghezelbash, B. Romani, S. Ziaei and A. Hedayatkhah, Microbiology, 2012, 81, 736–742 Search PubMed
.
- C. M. Lo, Q. Zhang, N. V. Callow and L. K. Ju, Bioresour. Technol., 2010, 101, 717–723 CrossRef PubMed
.
- P. Rapp and F. Wagner, Appl. Environ. Microbiol., 1986, 51, 746–752 Search PubMed
.
- S. A. Van Ooteghem, S. K. Beer and P. C. Yue, Appl.
Biochem. Biotechnol., 2002, 98–100, 177–189 CrossRef PubMed
.
- J. Ravachol, R. Borne, I. Meynial-Salles, P. Soucaille, S. Pagès, C. Tardif and H.-P. Fierobe, Biotechnol. Biofuels, 2015, 8, 114 CrossRef PubMed
.
- S. Chen and X. Dong, Int. J. Syst. Evol. Microbiol., 2005, 55, 2257–2261 CrossRef PubMed
.
- B. Schink, Microbiol. Mol. Biol. Rev., 1997, 61, 262–280 Search PubMed
.
- A. Söllinger, C. Schwab, T. Weinmaier, A. Loy, A. T. Tveit, C. Schleper and T. Urich, FEMS Microbiol. Ecol., 2016, 92, fiv149 CrossRef PubMed
.
- G. N. Jarvis, C. Strömpl, D. M. Burgess, L. C. Skillman, E. R. Moore and K. N. Joblin, Curr. Microbiol., 2000, 40, 327–332 CrossRef PubMed
.
- H. Imachi, S. Sakai, Y. Sekiguchi, S. Hanada, Y. Kamagata, A. Ohashi and H. Harada, Int. J. Syst. Evol. Microbiol., 2008, 58, 294–301 CrossRef PubMed
.
- M. S. M. Jetten, A. J. M. Stams and A. J. B. Zehnder, FEMS Microbiol. Lett., 1992, 88, 181–197 CrossRef
.
- L. Raskin, D. Zheng, M. E. Griffin, P. G. Stroot and P. Misra, Antonie van Leeuwenhoek, 1995, 68, 297–308 CrossRef PubMed
.
- H. Cadillo-Quiroz, P. Browne, N. Kyrpides, T. Woyke, L. Goodwin, C. Detter, J. B. Yavitt and S. H. Zinder, Genome Announcements, 2015, 3, e01280-1–5 CrossRef PubMed
.
- S. Kotelnikova, A. J. L. Macario and K. Pedersen, Int. J. Syst. Bacteriol., 1998, 48, 357–367 CrossRef PubMed
.
- G. Borrel, H. M. B. Harris, N. Parisot, N. Gaci, W. Tottey, A. Mihajlovski, J. Deane, S. Gribaldo, O. Bardot, E. Peyretaillade, P. Peyret, P. W. O'Toole and J.-F. Brugere, Genome Announcements, 2013, 1, e00453-13 CrossRef PubMed
.
- M. Simpson-Holley, A. Higson and G. Evans, J. Chem. Eng., 2007, 163, 46–59 Search PubMed
.
| This journal is © The Royal Society of Chemistry 2018 |