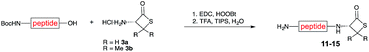Open Access Article
Open Access ArticleCoupling of sterically demanding peptides by β-thiolactone-mediated native chemical ligation†
Huan
Chen
a,
Yunxian
Xiao
a,
Ning
Yuan
c,
Jiaping
Weng
a,
Pengcheng
Gao
a,
Leonard
Breindel
b,
Alexander
Shekhtman
b and
Qiang
Zhang
 *a
*a
aDepartment of Chemistry, University at Albany, State University of New York, 1400 Washington Avenue, Albany, NY 12222, USA. E-mail: qzhang5@albany.edu
bDepartment of Chemistry, University at Albany, State University of New York, 1400 Washington Avenue, Albany, NY 12222, USA
cState Key Laboratory of Natural and Biomimetic Drugs, School of Pharmaceutical Sciences, Peking University, 38 Xueyuan Road, Beijing 100191, China
First published on 23rd January 2018
Abstract
The ligation of sterically demanding peptidyl sites such as those involving Val–Val and Val–Pro linkages has proven to be extremely challenging with conventional NCL methods that rely on exogenous thiol additives. Herein, we report an efficient β-thiolactone-mediated additive-free NCL protocol that enables the establishment of these connections in good yield. The rapid NCL was followed by in situ desulfurization. Reaction rates between β-thiolactones and conventional thioesters towards NCL were also investigated, and direct aminolysis was ruled out as a possible pathway. Finally, the potent cytotoxic cyclic-peptide axinastatin 1 has been prepared using the developed methodology.
Introduction
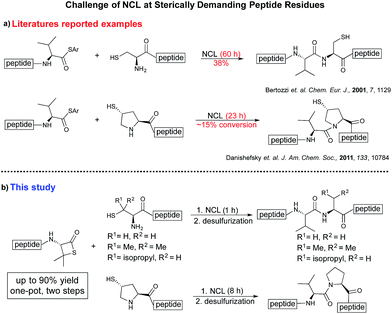
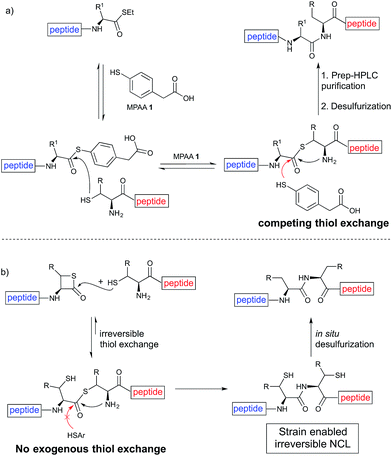
Despite the major advances in the synthetic coupling of peptides and proteins such as the use of Native Chemical Ligation (NCL) procedures1 and metal free desulfurization,2 the accomplishment of ligations3 involving amino acids with sterically demanding side chains continues to be problematic (Fig. 1a). In the previous reports by Bertozzi,4 Danishefsky4 and others,4 the ligation sites involving valine or proline were furnished in low yield and required an extended reaction time. This challenge is particularly relevant for the synthesis of membrane proteins in which amino acids such as Val, Ile, Leu, and Ala constitute more than one-third of the transmembrane domains.5 Thus, the need to develop methodologies for the ligation of these residues is tremendous. Although considerable efforts have been directed towards improving the ligation efficiency at sterically hindered sites,6,7 such as the elegant report by Dong8 on the internal activation of thioesters promoting the occurrence of ligation at proline sites, the problem remains and continues to place severe limitations on the ability to access membrane proteins by NCL.9,10 Herein we describe a protocol that utilizes readily available β-thiolactones to facilitate the challenging peptide ligation at sterically demanding Val–Ala, Val–Leu, Val–Val, and Val–Pro sites (Fig. 1b). This allows the connection of two bulky peptidyl residues, through a one-pot additive-free NCL followed by desulfurization, and provides a significant advance in protein synthesis.NCL is typically initiated by adding an exogenous thiol additive such as 4-mercaptophenylacetic acid11 (MPAA 1) (Fig. 2a); a reversible transthioesterification between an activated thioester and cysteine provides a tethered peptidyl adduct, which then undergoes a facile N–S shift to form a new amide bond. There are two critical drawbacks associated with the use of 1. When the coupling partners involve bulky sidechains, the MPAA thiol exchange outcompetes the forward N–S shift, resulting in the disintegration of the tethered peptide and low yields. In addition, it necessitates the inclusion of a step to remove the thiol additive prior to the desulfurization. Although there are multiple procedures for simplified MPAA removal,7,11 reverse phase preparative HPLC purification remains the most effective technique. The additional purification and subsequent requisite lyophilization further lower the reaction yield and efficiency.
Results and discussion
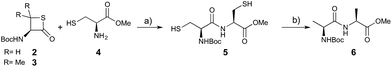
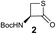
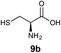
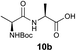
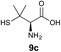
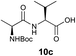
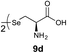
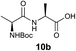
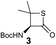
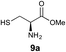
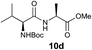
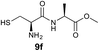
We envisioned that the constraints mentioned above could be overcome if β-thiolactones12 were used as thioester surrogates13 (Fig. 2b). Considering that the concept of using macrothiolactones as thioesters has been elegantly illustrated by the Nishiuchi group,14 the relief of the β-thiolactone ring strain might promote ligation without pre-activation. Furthermore, difficulty of regenerating the thietane could circumvent the need for the thiol additive, which might not only enable in situ desulfurization, but it might also prevent the reversible transthioesterification and improve the reactivity at the sterically hindered sites. This accessible methodology could provide a significant advance in protein synthesis and eliminate the additional steps typically required for peptide ligation, while achieving the synthesis of sterically challenging proteins and peptides with high efficiency. Pioneering work,15 elegantly illustrated by Crich and co-workers, has demonstrated the connections of single amino acids in the presence of exogenous thiol activators, however the investigation of β-thiolactones in the context of polypeptides as well as at sterically hindered sites has been elusive.16 Our approach commenced with the cyclization of commercially available Boc-L-cysteine and Boc-L-penicillamine (Boc-Cys and Boc-Pen respectively)15,17 to furnish thiolactones 2 and 3(ref. 18) in high yield (Table 1). Under the standard NCL conditions with the presence of MPAA, the mixture of thietan 2 and cysteine ester 4 produced the dipeptide 5 in 82% yield. The ligation between 2 and 4 in the absence of 1 was completed within 30 min, and the dipeptide 5 was obtained. Subsequent in situ desulfurization cleanly provided Ala–Ala dipeptide 6. Although β-thiolactones are known to undergo reactions at their carbon sites,19 we did not observe any by-products. After screening, the use of Na2HPO4 buffer provided the best result, furnishing the dipeptide 6 in 84% yield over two steps. The addition of Gn·HCl lowered the reaction yield, and the presence of TCEP did not change the ligation outcome. Under the optimized reaction conditions, the scope of the reaction was investigated. It was found that L-cysteine boosted the reaction yield to 90% when coupled to thiolactone 2 (Table 1, entry 1). The reaction between 2 and L-Pen209c smoothly yielded the Ala–Val dipeptide 10c. The coupling of thiolactone 2 and selenocystine 9d generated the desired product 10b in two steps.21 Interestingly, the mass spectrum of the initial ligated dipeptide was messy, and no meaningful mass peaks could be identified; however, after the desulfurization, the mass spectrum shows that dipeptide 10b can be detected as the major product. We believed that our observation was caused by the poor solubility of the intermediate dipeptide. The penicillamine-derived thiolactone 3 was next investigated. It was reacted with cysteine to provide the corresponding dipeptide 10d in 81% yield. The thiolactone-bearing dipeptide 7, prepared using a literature reported procedure,15 was mixed with dipeptide 9g in the NCL buffer to produce tetrapeptide 10l in excellent yield (80%). Finally, the Pen dipeptide 8 was examined. The tripeptide 10m and tetrapeptides 10n and 10o could be prepared in good yield. To further explore the reaction scope and limitations, we embarked on studies of more extended peptides (Table 2). The thiolactone bearing peptides were prepared using an EDC/HOOBt coupling method23 followed by global sidechain deprotection. The peptides 11–15 were generated without epimerization.24 Ligation between peptidyl fragments 11 and 16a provided polypeptide 17a in good yield. The reaction between 12 and 16a afforded 17b in 86% yield after two steps. The reaction between thiolactone 11 and a 3-mercapto-leucine-bearing peptide2516b provided the desired product 17c in high yield. Interestingly, considering the notorious difficulty of amide bond generation between a valine and valine or proline residues,4 we attempted to construct Val–Val and Val–Pro bonds using our protocol. The coupling between 12 and 16c occurred smoothly and the overall Val–Val linked polypeptide 17e was produced in good yield over two steps (76%). Finally, the possibility of ligating valine and proline constructs was examined. The linkage of 12 and 16d was successfully achieved in 57% yield in a one-pot fashion. A few more examples of coupling between thiolactones and 4-mercapto-prolines were probed, and the yields varied based on the sequences (42% for 17g and 32% for 17h). The β-thiolactone mediated ligation has a wide functional group tolerance; a variety of amino acid residues have been investigated, and the side-chains of Asp, Lys, Arg, His, Thr, Glu, Gln, and Tyr do not interfere with the NCL (Table 2 entries 9 and 10). It is worth noting that among all the ligations described in Table 2, the initial NCL conversions were quantitative or near-quantitative;26 it was the subsequent desulfurization that significantly reduced the overall reaction yield. More specifically, regarding the sequence 17h in entry 8 which gave the lowest yield, the precursor product (bis-thiol) was isolated in 54% yield prior to desulfurization. Considering the yield reduction from prep-HPLC purification and the known obstacle of thiol removal required for hydrophobic residues,27 this method still provides a fundamental improvement over current protocols.| Entry | Thiolactones | Ligating amino acids/peptides | Products | Yield (%) |
|---|---|---|---|---|
| a Reagents and conditions: (a) buffer (0.1 M Na2HPO4, 0.01 M TCEP, pH = 7.8), rt, 1–8 h; (b) desulfurization buffer (0.5 M TCEP pH = 7.2, 0.1 M VA-044, t-BuSH), 37 °C, 2 h. | ||||
| 1 |
|
|
|
90% |
| 2 | 2 |
|
|
53% |
| 3 | 2 |
|
|
80% |
| 4 |
|
|
|
81% |
| 5 | 2 |
|
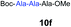
|
77% |
| 6 | 3 | 9f |
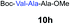
|
67% |
| 7 | 3 |
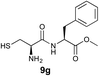
|
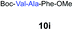
|
69% |
| 8 |
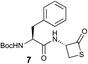
|
4 |
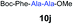
|
71% |
| 9 | 7 | 9f |

|
79% |
| 10 | 7 | 9g |
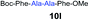
|
80% |
| 11 |
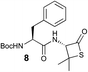
|
4 |
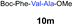
|
62% |
| 12 | 8 | 9f |
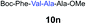
|
72% |
| 13 | 8 | 9g |
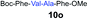
|
77% |
| Entry | Thiolactones | Ligating peptides | Products | Yield (%) |
|---|---|---|---|---|
| a Reagents and conditions: (a) EDC (2.5 equiv.), HOOBt (2.5 equiv.), CH2Cl2, −20 °C, (b) TFA/H2O/TIPS = 95/2.5/2.5; NCL conditions: 0.1 M Na2HPO4, 0.01 M TCEP, pH = 7.8, rt, 1–8 h; desulfurization: buffer (0.5 M TCEP pH = 7.2, 0.1 M VA-044, t-BuSH), 37 °C, 2–3 h. * = isolated yield after the initial NCL shown in parentheses. | ||||
| 1 |

|
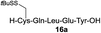
|
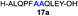
|
84% |
| 2 |

|
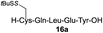
|
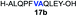
|
86% |
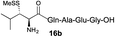
| 3 |
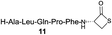
|
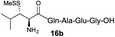
|
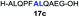
|
73% |
| 4 |
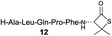
|
|
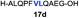
|
81% |
| 5 |
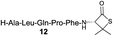
|
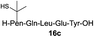
|
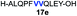
|
76% |
| 6 |
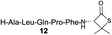
|
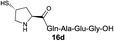
|
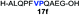
|
57% |
| 7 |

|
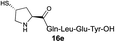
|
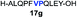
|
42% |
| 8 |
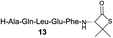
|
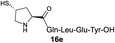
|
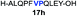
|
32% (54%) |
| 9 |

|
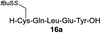
|
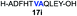
|
82% |
| 10 |
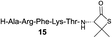
|
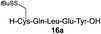
|
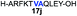
|
83% |
Intrigued by the facile β-thiolactone reactivity, we next investigated the reaction mechanism and rate. Reaction rates of the thiolactone and other known activated thioesters10,28 were compared (Fig. 3a). By monitoring with LC-MS, the ligation of the L-cysteine and thiolactone-bearing peptide 13 was determined to be completed within 5 min. Analogously, peptides that possessed phenyl selenoesters 13a/13c and phenyl thioesters 13b/13d were subjected to identical reaction conditions. After 5 min, alanine phenyl selenoester 13a was consumed by 55% and alanine phenyl thioester 13b was converted by 4%. More intriguingly, the analogous valine phenyl selenoester 13c and thioester 13d were merely consumed by 4% and 1% respectively during the same period. This experiment illustrated that β-thiolactone was a superior thioester compared to existing activated thioesters regarding the reaction rate. Using a cross-over experiment, we probed whether the reaction proceeded through an NCL process or an undesired aminolysis pathway (Fig. 3b). A combination of peptides 13, 16a and 16f were mixed in the NCL buffer. After 60 min, two major peaks appeared in the LC trace. One peak corresponded to peptide 17i, which is the desired NCL product, and another was identified as the unreacted peptide 16f. At 120 min, the trace amounts of 13 and 16a were consumed, furnishing 17i exclusively. The direct aminolysis product 17j was not observed, which suggested that thietan-mediated peptide ligation did indeed proceed by the NCL pathway.29
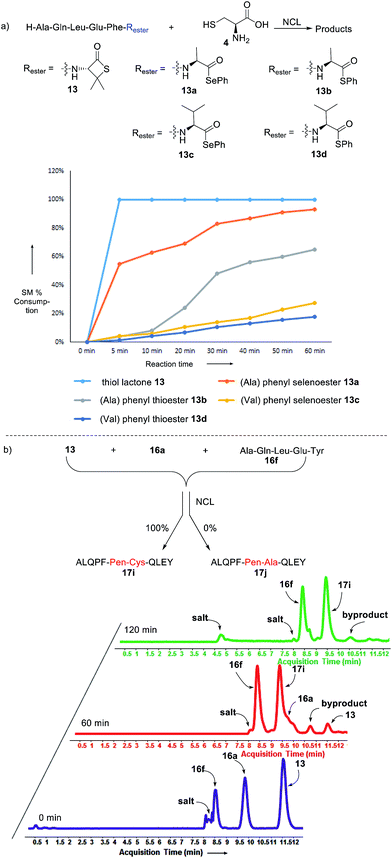 | ||
| Fig. 3 Reaction rate and mechanistic investigation. Reagents and conditions: NCL condition for (a) and (b): 0.1 M Na2HPO4, 0.01 M TCEP, pH = 7.8, rt. | ||
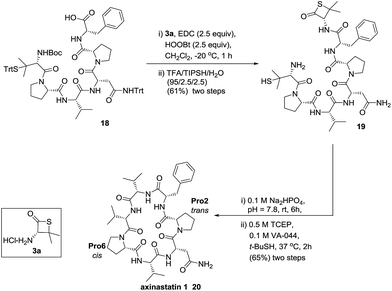
With our enhanced understanding of the strain facilitated peptide ligation, we explored an expansion of the reaction scope and application by pursuing the synthesis of a cyclic peptide axinastatin 1 (20) with our methodology (Scheme 1). Axinastatin 1 is a naturally occurring cyclic heptapeptide which was isolated in minute quantities from a marine sponge (4.5 × 10−5% yield).30 It shows potent cytotoxicity against a leukemia cancer cell line (ED50 = 0.21 μg mL−1). Structurally, 20 contains three valines and two prolines in the seven amino acid backbone. Importantly, Pro2 and Pro6 of the naturally occurring axinastatin 1 are in trans and cis configurations, respectively. The synthesis herein commenced with the preparation of 18 using an Fmoc solid phase peptide synthesis with a TGT resin, and the thiolactone 19 was produced through the coupling of 3a and the peptide 18 without noticeable epimerization.23 The thiolactone 19 was subjected to a one-pot NCL followed by desulfurization to successfully produce the cyclic peptide 20 intramolecularly in good yield. The high resolution MS, 1H, and 13C NMR data of synthesized axinastatin 1 are identical to the reported ones.31 The proper proline conformations were confirmed by 2D NMR experiments.32
Conclusions
In summary, we have successfully developed a novel one-pot protocol for peptide ligations at sterically demanding sites utilizing the strained β-thiolactone. The strain not only greatly accelerates the reaction and enables in situ thiol removal, but also provides a practical method for the otherwise challenging ligation linking sterically hindered peptidyl residues. The reaction scopes and limitations have been evaluated and discussed, and the β-thiolactones proved to be a much more rapid thioester surrogate compared to conventional thioesters. Furthermore, the mode of the reaction has been verified to be an NCL pathway. The versatility of the method was demonstrated in part by the synthesis of axinastatin 1. Overall, this strain-driven thietan-mediated ligation provides a powerful tool for the synthesis of peptides and proteins. In particular, our method could make the hydrophobic regions of membrane proteins33 more synthetically accessible. Studies towards the synthesis of complex membrane proteins using our approach will be reported in due course.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Support for this work was provided by: the National Science Foundation (CHE 1710174), the University at Albany-SUNY to Q. Zhang, and a China Research Council Scholarship to P. Gao. Thanks are extended to Prof. Rabi Musah (SUNY Albany) and Prof. Paramjit Arora (NYU) for helpful suggestions. The early work performed by Kyle McHugh is acknowledged.Notes and references
- P. Dawson, T. Muir, I. Clark-Lewis and S. Kent, Science, 1994, 266, 776–779 CAS.
- L. Z. Yan and P. E. Dawson, J. Am. Chem. Soc., 2001, 123, 526–533 CrossRef CAS PubMed; Q. Wan and S. Danishefsky, Angew. Chem., Int. Ed., 2007, 46, 9248–9252 CrossRef PubMed.
- C. Unverzagt and Y. Kajihara, Chem. Soc. Rev., 2013, 42, 4408–4420 RSC; A. Tejada, J. Brailsford, Q. Zhang, J. Shieh, M. Moore and S. Danishefsky, Top. Curr. Chem., 2014, 362, 1–26 CrossRef CAS; C. Hackenberger and D. Schwarzer, Angew. Chem., Int. Ed., 2008, 47, 10030–10074 CrossRef PubMed; L. Raibaut, N. Ollivier and O. Melnyk, Chem. Soc. Rev., 2012, 41, 7001–7015 RSC.
- L. Marcaurelle, L. Mizoue, J. Wilken, L. Oldham, S. Kent, T. Handel and C. Bertozzi, Chem.–Eur. J., 2001, 7, 1129–1132 CrossRef CAS; S. Shang, Z. Tan, S. Dong and S. Danishefsky, J. Am. Chem. Soc., 2011, 133, 10784–10786 CrossRef PubMed; H. Ding, A. Shigenaga, K. Sato, K. Morishita and A. Otaka, Org. Lett., 2011, 13, 5588–5591 CrossRef PubMed.
- T. Sato, Biopolymers, 2016, 106, 613–621 CrossRef CAS PubMed.
- M. Ulmschneider and M. Sansom, Biochim. Biophys. Acta, Biomembr., 2001, 1512, 1–14 CrossRef CAS; B. Nilsson, L. Kiessling and R. Raines, Org. Lett., 2000, 2, 1939–1941 CrossRef PubMed; R. Hondal, B. Nilsson and R. Raines, J. Am. Chem. Soc., 2001, 123, 5140–5141 CrossRef PubMed; M. Gieselman, L. Xie and W. Donk, Org. Lett., 2001, 3, 1331–1334 CrossRef PubMed; J. Bode, R. Fox and K. Baucom, Angew. Chem., Int. Ed., 2006, 45, 1248–1252 CrossRef PubMed; J. Canosa and P. Dawson, Angew. Chem., Int. Ed., 2008, 47, 6851–6855 CrossRef PubMed; X. Li, H. Lam, Y. Zhang and C. Chan, Org. Lett., 2010, 12, 1724–1727 CrossRef PubMed; T. Durek and P. Alewood, Angew. Chem., Int. Ed., 2011, 50, 12042–12045 CrossRef PubMed; N. Ollivier, J. Vicogne, A. Vallin, H. Drobecq, R. Desmet, O. Mahdi, B. Leclercq, G. Goormachtigh, V. Fafeur and O. Melnyk, Angew. Chem., Int. Ed., 2012, 51, 209–213 CrossRef PubMed; R. Thompson, X. Liu, N. García, P. Pereira, K. Jolliffe and R. Payne, J. Am. Chem. Soc., 2014, 136, 8161–8164 CrossRef PubMed; M. Jbara, M. Seenaiaha and A. Brik, Chem. Commun., 2014, 50, 12534–12537 RSC; J. Tailhades, N. Patil, M. Hossain and J. Wade, J. Pept. Sci., 2015, 21, 139–147 CrossRef PubMed; M. Raj, H. Wu, S. Blosser, M. Vittoria and P. Arora, J. Am. Chem. Soc., 2015, 137, 6932–6940 CrossRef PubMed; F. Burlina, G. Papageorgiou, C. Morris, P. D. Whitee and J. Offer, Chem. Sci., 2014, 5, 766 RSC; J. Blanco-Canosa, B. Nardone, F. Albericio and P. Dawson, J. Am. Chem. Soc., 2015, 137, 7197–7209 CrossRef PubMed; P. M. Shelton, C. Weller and C. Chatterjee, J. Am. Chem. Soc., 2017, 139, 3946–3949 CrossRef PubMed; C. Rao and C. Liu, Org. Biomol. Chem., 2017, 15, 2491–2496 Search PubMed; Y. Asahina, T. Kawakami and H. Hojo, Chem. Commun., 2017, 53, 2114–2117 RSC.
- N. Mitchell, L. Malins, X. Liu, R. Thompson, B. Chan, L. Radom and R. Payne, J. Am. Chem. Soc., 2015, 137, 14011–14014 CrossRef CAS PubMed; O. Reimann, C. Smet-Nocca and C. Hackenberger, Angew. Chem., Int. Ed., 2015, 54, 306–310 CrossRef PubMed.
- Y. Gui, L. Qiu, Y. Li, H. Li and S. Dong, J. Am. Chem. Soc., 2016, 138, 4890–4899 CrossRef CAS PubMed.
- J. Li, S. Tang, J. Zheng, C. Tian and L. Liu, Acc. Chem. Res., 2017, 50, 1143–1153 CrossRef CAS PubMed; S. Kent, Bioorg. Med. Chem., 2017, 25, 4926–4937 CrossRef PubMed; H. Burke, L. McSweeney and E. Scanlan, Nat. Commun., 2017, 8, 15655 CrossRef PubMed.
- D. Bang, B. L. Pentelute and S. Kent, Angew. Chem., Int. Ed., 2006, 45, 3985–3988 CrossRef CAS PubMed; J. Zheng, H. Cui, G. Fang, W. Xi and L. Liu, ChemBioChem, 2010, 11, 511–515 CrossRef PubMed.
- T. Moyal, H. Hemantha, P. Siman, M. Refua and A. Brik, Chem. Sci., 2013, 4, 2496–2501 RSC; J. Shimko, C. Howard, M. Poirier and J. Ottesen, Methods Mol. Biol, 2013, 981, 177–192 Search PubMed.
- S. Aubry, G. Aubert, T. Cresteil and D. Crich, Org. Biomol. Chem., 2012, 10, 2629–2632 Search PubMed; A. Noel, B. Delpech and D. Crich, J. Sulfur Chem., 2013, 34, 104–141 CrossRef CAS; A. Noel, B. Delpech and D. Crich, J. Org. Chem., 2014, 79, 4068–4077 CrossRef PubMed; M. Michael, T. Brandon, Y. Wang, C. Geoffrey and G. Doherty, J. Am. Chem. Soc., 2014, 136, 10814–10820 CrossRef PubMed; X. Liu, Y. Wang and G. Doherty, Asian J. Org. Chem., 2015, 4, 994–1009 CrossRef.
- G. Kenner, W. McDermott and R. Sheppard, J. Chem. Soc. D, 1971, 636–637 RSC; B. Backes and J. Ellman, J. Am. Chem. Soc., 1994, 116, 11171–11172 CrossRef CAS; E. George, R. Novick and T. Muir, J. Am. Chem. Soc., 2008, 130, 4914–4924 CrossRef PubMed; G. Fang, J. Wang and L. Liu, Angew. Chem., Int. Ed., 2012, 51, 10347–10350 CrossRef PubMed; S. Panda, C. Hall, A. Oliferenko and A. Katritzky, Acc. Chem. Res., 2014, 47, 1076–1087 CrossRef PubMed.
- S. Tsuda, T. Yoshiya, M. Mochizuki and Y. Nishiuchi, Org. Lett., 2015, 17, 1806–1809 CrossRef CAS PubMed.
- D. Crich and S. Kasinath, J. Org. Chem., 2009, 74, 3389–3393 CrossRef CAS PubMed; Aubry, K. Sasaki, L. Eloy, G. Aubert, P. Retailleau, T. Cresteil and D. Crich, Org. Biomol. Chem., 2011, 9, 7134–7143 Search PubMed.
- The homocysteine derived γ-thiolactone was also investigated in the preparation of polymers, see: Z. Fan, Y. Zhang, J. Jia and X. Li, RSC Adv., 2015, 5, 16740–16747 RSC.
- M. Suzuki, K. Makimura and S. Matsuoka, Biomacromolecules, 2016, 17, 1135–1141 CrossRef CAS PubMed.
- Thiolactone 2 can be stored at 0 °C for at least one month, and pen-thiolactone 3 can be stored indefinitely in the refrigerator.
- A. Noel, B. Delpech and D. Crich, Org. Biomol. Chem., 2012, 10, 6480–6483 CAS.
- C. Haase, H. Rohde and O. Seitz, Angew. Chem., Int. Ed., 2008, 47, 6807–6810 CrossRef CAS PubMed.
- N. Metanis, E. Keinan and P. E. Dawson, Angew. Chem., Int. Ed., 2010, 49, 7049–7053 CrossRef CAS PubMed; K. Cergol, R. Thompson, L. Malins, P. Turner and R. Payne, Org. Lett., 2014, 16, 290–293 CrossRef PubMed.
- Reactions were generally carried out at 3.0 mM. Reactions carried out in diluted buffer required a longer reaction time (0.2 mM, 20 h), yet the yields are nearly identical compared to the ones in more concentrated buffers. See the ESI† for experiment details.
- S. Sakakibara, Biopolymers, 1995, 37, 17–28 CrossRef CAS PubMed.
- The D-Phe incorporating polypeptide 12a was prepared, and its LC-MS trace does not match with the one from corresponding L-Phe bearing peptide 12 produced via the EDC coupling protocol. This observation further validated that EDC/HOOBt coupling conditions did not lead to epimerization. See the ESI† section VI for details.
- Z. Harpaz, P. Siman, K. Kumar and A. Brik, ChemBioChem, 2010, 11, 1232–1235 CrossRef CAS PubMed; Z. Tan, S. Shang and S. Danishefsky, Angew. Chem., Int. Ed., 2010, 49, 9500–9503 CrossRef PubMed.
- See the ESI† for selected NCL crude LC-MS traces.
- B. Fierz, S. Kilic, A. Hieb, K. Luger and T. Muir, J. Am. Chem. Soc., 2012, 134, 19548–19551 CrossRef CAS PubMed.
- N. Mitchell, S. Kulkarni, L. Malins, S. Wang and R. Payne, Chem.–Eur. J., 2017, 23, 946–952 CrossRef CAS PubMed; L. Malins, N. Mitchell and R. Payne, J. Pept. Sci., 2014, 64–77 CrossRef PubMed; L. Raibaut, M. Cargoet, N. Ollivier, Y. Chang, H. Drobecq, E. Boll, R. Desmet, J. Monbaliu and O. Melnyk, Chem. Sci., 2016, 7, 2657–2665 RSC.
- No reaction was observed when mixing 13 and alanine bearing 16f in NCL buffer for 4 h.
- G. Pettit, C. Herald, M. Boyd, J. Leet, C. Dufresne, D. Doubek, J. Schmidt, R. Cerny, J. Hooper and K. Rützler, J. Med. Chem., 1991, 34, 3339–3340 CrossRef CAS PubMed.
- R. Konat, B. Mathä, J. Winkler and H. Kessler, Liebigs Ann./Recl., 1995, 765–774 CrossRef CAS.
- See the ESI† for structure determination details.
- R. Ignotz, B. Kelly, R. Davis and J. Massagué, Proc. Natl. Acad. Sci. U. S. A., 1986, 6307–6363 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7sc04744d |
| This journal is © The Royal Society of Chemistry 2018 |