Open Access Article
Open Access ArticleDesign of a fluorogenic PNA probe capable of simultaneous recognition of 3′-overhang and double-stranded sequences of small interfering RNAs†
Takaaki Tanabe,
Takaya Sato,
Yusuke Sato * and
Seiichi Nishizawa
* and
Seiichi Nishizawa *
*
Department of Chemistry, Graduate School of Science, Tohoku University, Sendai 980-8578, Japan. E-mail: satoyuu@m.tohoku.ac.jp; nishi@m.tohoku.ac.jp; Fax: +81-22-795-6549; Tel: +81-22-795-6552
First published on 18th December 2018
Abstract
We developed a new fluorescent peptide nucleic acid (PNA) probe, COT probe, capable of simultaneous recognition of 3′-overhang and double stranded sequences of target small interfering RNA (siRNA).
Much attention has been paid to the design of RNA-binding molecules capable of binding to non-coding RNAs that can regulate gene expression.1 This class of molecules have great potential as drug candidates by modulating the expression of genes related to various diseases.2 In addition, the molecules that show the fluorescence response upon binding to non-coding RNAs have been useful probes for the analysis of non-coding RNAs.3–5 For instance, the fluorescent intercalators were shown to be useful for detection of small interfering RNAs (siRNAs), typically 21-mer double-stranded RNAs (dsRNAs) containing 3′-overhanging dinucleotides,6 as well as the analysis of siRNA delivery into the cells by means of carriers.4,5 They can provide simple and easy-to-use analysis for target siRNAs with a view toward the development of siRNA therapeutics.
In this context, we have recently reported on new class of peptide nucleic acid (PNA)-based fluorescent probes for siRNA analysis.7 PNA dinucleotide that can recognize 3′-overhanging dinucleotide through Watson–Crick base pairing was conjugated with a fluorescent intercalator, thiazole orange (TO).8 Our probe thus targeted both 3′-overhanging nucleotides and the dsRNA region near the overhangs of target siRNAs, which is characteristic compared to other siRNA-binding fluorescent probes that simply aim dsRNA region. The PNA–TO conjugate was further attached with pyrene unit at C-termini for enhancing the binding ability and selectivity to target siRNAs. The resulting probe, Py-AA-TO, exhibited light-up response of the TO unit upon selective binding to the target siRNAs containing the overhanging nucleotides that are complementary to the PNA units of the probes, which rendered it useful for selective detection of target siRNAs. Moreover, the use of Py-AA-TO as an affinity-labeling agent for the siRNAs enabled the selective visualization of siRNAs encapsulated in the carriers in the living cells.7 One of the key issues is to enhance the binding ability of the probe for target siRNAs with a view toward the goal of sensitive siRNA analytical assays suitable for practical use.
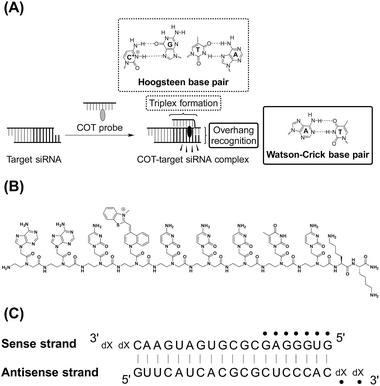
In this work, we developed a new fluorescent PNA probe with the improved binding affinity and selectivity for siRNAs, where the probe can simultaneously recognize 3′-overhang and dsRNA sequences near the overhang of target siRNAs (Fig. 1A). In case of Py-AA-TO, TO unit linked with PNA dinucleotide was designed to intercalate into the dsRNA region of target siRNAs, which results in little selectivity to dsRNA sequence. Instead, we explored triplex-forming PNAs (TFPs) as the dsRNA-binding units so as to achieve sequence-selective recognition of dsRNA region.9 TFPs are fundamentally composed of homopyrimidine PNA oligomers and can strongly and selectively form triplex structures with the homopurine sequences of target dsRNA tracts at acidic pH, by forming T (U)·A–U and C+·G–C base triples through Hoogsteen base pairing (Fig. 1A). Besides the sequence-selective recognition property, TFPs feature the strong binding to target dsRNAs, where even 6-mer oligomers show the dissociation constant (Kd) less than 100 nM.9 This affinity is superior to against ds-nucleic acids (Kd = 1.0–10 μM).10 Therefore, we expect that the integration of TFP as a dsRNA-binding unit into siRNA-targeting probe enables to achieve stronger affinity compared to Py-AA-TO as well as the sequence-selective binding to dsRNA region of target siRNAs. Specifically, a TFP possessing TO as a base surrogate was utilized in the probe design. This class of TFPs named as triplex-forming forced intercalation (tFIT) probes exhibit the significant light-up response of the TO unit upon triplex formation with dsRNA tracts, as demonstrated in our previous work.11 tFIT probes were shown to be useful for analyzing dsRNA sequences at single-base pair resolution. Here, such a tFIT probe was directly attached to C-terminus of PNA dinucleotide capable of recognition of 3′-overhang sequence of target siRNA. The resulting probe which we call COT (combination of overhang recognition and triplex formation) probe was discussed based on the examination of the binding and fluorescence sensing of target siRNAs.
COT probe (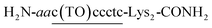 , Fig. 1B) was designed for targeting siRNA sequence that can knockdown red fluorescent protein (RFP) gene (Fig. 1C).12 Overhang recognition unit (italic base) would recognize 3′-overhanging dTdT in the antisense strand while tFIT unit (underlined base) would recognize the purine-rich sequence of the sense strand. TO base surrogate was placed so as to face the uracil nucleotide in dsRNA region because it can function as a universal base that non-discriminatorily binds to all four kinds of base pairs in the triplex.11 Furthermore, two lysine residues were introduced to the C-terminus of the probe in order to increase the solubility as well as the binding affinity to target siRNA through electrostatic interaction.13 We also designed the control probe that lacks the overhang recognition unit (
, Fig. 1B) was designed for targeting siRNA sequence that can knockdown red fluorescent protein (RFP) gene (Fig. 1C).12 Overhang recognition unit (italic base) would recognize 3′-overhanging dTdT in the antisense strand while tFIT unit (underlined base) would recognize the purine-rich sequence of the sense strand. TO base surrogate was placed so as to face the uracil nucleotide in dsRNA region because it can function as a universal base that non-discriminatorily binds to all four kinds of base pairs in the triplex.11 Furthermore, two lysine residues were introduced to the C-terminus of the probe in order to increase the solubility as well as the binding affinity to target siRNA through electrostatic interaction.13 We also designed the control probe that lacks the overhang recognition unit ( ; Fig. S1†). These probes were manually synthesized by solid-phase synthesis, purified by reverse phase HPLC, and characterized by MALDI-TOF-MS (ESI†). Besides target siRNA, we examined a control siRNA with noncognate dsRNA sequence and 3′-overhanging dTdT (noncognate siRNA;6 5′-CGU ACG CGG AAU ACU UCG AdTdT-3′/3′–dTdTG CAU GCG CCU UAU GAA GCU -5′). In addition, we used another control siRNAs having fully-matched dsRNA sequence with no overhang (no overhang siRNA) or 3′-overhanging dAdA that was mismatched with the overhang recognition unit of COT probe (mismatch siRNA), as shown in Fig. 1C.
; Fig. S1†). These probes were manually synthesized by solid-phase synthesis, purified by reverse phase HPLC, and characterized by MALDI-TOF-MS (ESI†). Besides target siRNA, we examined a control siRNA with noncognate dsRNA sequence and 3′-overhanging dTdT (noncognate siRNA;6 5′-CGU ACG CGG AAU ACU UCG AdTdT-3′/3′–dTdTG CAU GCG CCU UAU GAA GCU -5′). In addition, we used another control siRNAs having fully-matched dsRNA sequence with no overhang (no overhang siRNA) or 3′-overhanging dAdA that was mismatched with the overhang recognition unit of COT probe (mismatch siRNA), as shown in Fig. 1C.
First, UV melting experiments were performed in order to evaluate the binding of COT probe to target siRNA, in 10 mM sodium acetate (pH 5.5) buffer solutions containing 100 mM NaCl and 1.0 mM EDTA (Fig. S4†). We monitored absorbance change at 300 nm, where the triplex-duplex transition of tFIT unit can be selectively detected.11a,14 As the temperature increased from 10 °C to 80 °C, the absorbance remarkably decreased. This is most likely due to deprotonation of N3 protonated PNA cytosines (C+) involved in Hoogsteen base pairs with G–C (C+·G–C triplets) in the triplex structures. The melting temperature (Tm) was obtained as 66 ± 0.5 °C, which indicates the tFIT unit can form a thermally stable triplex with dsRNA region of target siRNA. Triplex formation is also supported by the results of circular dichroism (CD) spectral change of target siRNAs upon addition of the probe (Fig. S5†). We found the Tm value was significantly reduced for noncognate siRNA containing unrelated dsRNA sequence (ΔTm > 30 °C) whereas the mismatches (mismatch siRNA) or deletion of 3′-overhang sequence (no overhang siRNA) led to little influence on triplex stability (Table S2†). This result indicates that tFIT unit of COT probe retains the sequence selectivity for dsRNA region of target siRNA.
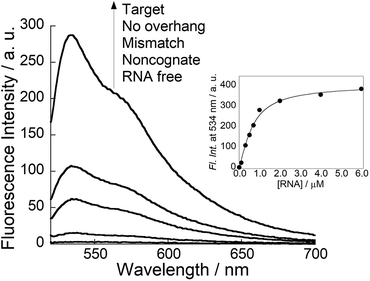
Next, we examined the fluorescence response of COT probe for target siRNA in pH 5.5 buffer at 25 °C (Fig. 2). In the absence of target siRNA, the probe (100 nM) showed negligible fluorescence (Φfree < 0.01), because of non-radiative energy loss by free rotation of the benzothiazole and quinoline rings in the TO base surrogate.8 The addition of the equimolar target siRNA caused the remarkable light-up response of the TO unit, in which the light-up factor (I/I0, where I and I0 denote the fluorescence intensities in the presence and absence of a target, respectively) was more than 104-fold. The fluorescence quantum yield of COT probe bound to target siRNA reaches 0.29. The light-up property is almost comparable to those of tFIT probes for dsRNA tracts.11b In contrast, almost no response was found at pH 7.0 where cytosines are hardly protonated (Fig. S6†). This is consistent with that the COT probe binding involves triplex formation of tFIT unit by Hoogsteen base pairing of C+ with G. Fluorescence titration experiments were carried out to estimate the binding affinity of COT probe for target siRNA (inset of Fig. 2). The resulting titration curve was well fitted by a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding isotherm, which gave the dissociation constant (Kd) of 340 ± 43 nM (n = 3). Significantly, this affinity is one order of magnitude larger than that of Py-AA-TO (Kd = 3.5 ± 0.40 μM).7a It suggests that the use of tFIT unit as dsRNA-binding unit leads to the improved binding affinity for target siRNAs.
1 binding isotherm, which gave the dissociation constant (Kd) of 340 ± 43 nM (n = 3). Significantly, this affinity is one order of magnitude larger than that of Py-AA-TO (Kd = 3.5 ± 0.40 μM).7a It suggests that the use of tFIT unit as dsRNA-binding unit leads to the improved binding affinity for target siRNAs.
Fluorescence response of COT probe was found to be very sensitive to the overhang sequence as well as dsRNA sequence of target siRNAs. As shown in Fig. 2, the light-up response is much pronounced for target siRNAs over three kinds of control siRNAs under the identical conditions. As for noncognate siRNA, very small response can be attributed to little formation of triplex structure at 25 °C (Table S2†). On the other hand, it was shown that both mismatch siRNA and no overhang siRNA were able to form a thermally stable triplex with the probe (Table S2†). Thus, we reasoned small response for these control siRNAs resulted from high flexibility of the TO unit in the resulting complexes.11a Considering that TO unit is located close to the terminal of triplex-forming region, the intramolecular rotation of TO unit would be less restricted in the complex between the probe with these siRNAs. Accordingly, the fluorescence response of COT probe allows to discriminate not only the overhanging sequence but also dsRNA sequence near the overhang of target siRNAs.
The observed abilities of COT probe for target siRNA were compared with those of control probe having no overhang recognition units. While the control probe could form the triplex structure (Table S2†), its light-up response upon binding to siRNAs was very low (Fig. S7†). This can be attributed to weaker binding affinity (Kd = 2.3 ± 0.36 μM) as well as the increased flexibility of the TO unit in the resulting triplex discussed above, due to the loss of overhang recognition. Also, we found relatively large response for mismatch and no overhang siRNAs whereas the response for noncognate siRNA was very weak. This resulted in the reduced selectivity for target siRNA over control siRNAs compared to COT probe. These results indicate that the overhang recognition in COT probe is crucial for high affinity and selectivity to overhang and dsRNA sequences for target siRNAs.
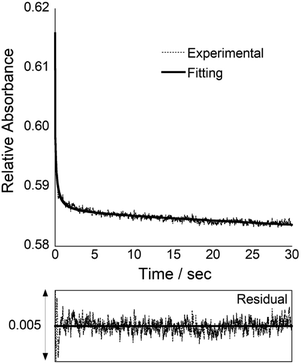
In order to understand more details about COT probe binding to target siRNAs, we characterized the binding kinetics by stopped-flow experiments (Fig. 3). The absorbance change at 260 nm was monitored upon mixing COT probe with target siRNA, which enables to estimate the association rate constant (kon) by a nonlinear least-squares regression analysis.11a,15 We then calculated the dissociation rate constant (koff) by the equation (koff = Kd × kon), where we assume two-state binding for the biomolecular complexes. The obtained kinetic parameters were summarized in Table 1. kon value was determined as 3.0 ± 0.31 × 106 M−1 s−1, which indicates COT probe binding rapidly proceeds. It is noteworthy that this value is 5.7-fold larger than that of control probe without overhang recognition units (kon = 5.2 ± 0.45 × 105 M−1 s−1; Fig. S8†). Meanwhile, koff values were comparable (koff/s−1; COT probe, 1.0; control probe, 1.2). Therefore, the overhang recognition is highly likely to be important for the rapid association for the binding to target siRNA, which would be responsible for strong binding affinity of COT probe over control probe.
Further kinetic analysis of COT probe provided the insights into the binding pathway for target siRNA. We estimated the dissociation constants and kinetic parameters for mismatched and no overhang siRNAs (Fig. S9 and S10†). It was clearly seen that kon values were reduced for these controls siRNAs relative to target siRNA whereas koff values were comparable. This again points out that the binding preference for target siRNA over control siRNAs was characterized by more rapid association due to the overhang recognition. In addition, these results showed that the dissociation of COT/siRNA complexes were insensitive to overhang recognition (koff = 1.0–1.2 s−1), as observed for that of control probe without overhang recognition units (cf. Fig. S8†). Therefore, it is highly likely that the dissociation of triplex structure is rate-limiting in the binding event of COT probe.
According to the principle of microscopic reversibility, the reaction necessarily follows the same pathway in the forward and reverse directions. Since both reactions pass through a common intermediate, the rate-limiting step is the same.16 Thus, the rate-limiting step of the association reaction would be triplex formation in this system. Taken together with the obtained binding kinetics, we could propose the association mechanism for COT probe with target siRNA. Here, it should be recalled that kon values are highly correlated with the 3′-overhang sequence of target siRNA (Table 1). If the first associative step involves rate-limiting triplex formation with dsRNA region near the overhang of target siRNAs, we should observe the kon values that correlated only with the sequence of dsRNA region, not overhang sequence. Therefore, we reasoned that overhang recognition first occurred and subsequently, tFIT unit formed triplex with dsRNA region for COT probe binding to target siRNA. This is quite characteristic compared to triplex tethered oligonucleotide probes (TOPs)17 which simultaneously bind to one single-stranded RNA and one dsRNA region of target RNA structures by Watson–Crick and Hoogsteen base-pairing, respectively, similar to the present probe design. In case of a triplex TOPs having 8-mer ssRNA-binding unit and 12-mer dsRNA-binding unit, it was shown that duplex formation was rate-limiting and preceded triplex formation.18 The observed difference in the kinetic nature would partly result from the length of single-stranded RNA region that can be recognized by Watson–Crick base-pairing between our probe and triplex TOPs.
In summary, we described useful binding and fluorescence sensing abilities of COT probe that was developed based on the combination of overhang recognition and triplex formation for target siRNA. To the best of our knowledge, this is the first report on fluorescent probes capable of simultaneous discrimination of the overhanging sequence as well as dsRNA sequence near the overhang of target siRNAs. COT probe displayed the improved binding affinity and selectively to target siRNAs compared to Py-AA-TO that was previously developed in our group (Table S3†). Thus, this class of COT probes can find various analytical applications, such as in vitro siRNA detection and quantification,4 with a view toward the implementation of siRNA therapeutics. In this context, as has been demonstrated for TOPs,18 the introduction of the spacer linking the overhang recognition unit and tFIT unit is a possible approach for further enhancement of the binding abilities for target siRNAs, where the spacer should be carefully designed (Fig. S11†). Meanwhile, we observed the light-up response of COT probe for single-stranded RNAs in addition to target siRNA due to possible duplex formation under the present condition (Fig. S12†). Thus, the selectivity to target siRNA over single-stranded RNAs should be improved for the practical use, for which the connection of TO base surrogate through the propyl spacer would be useful, as shown in our previous study.11b It should be noted that the present COT probe can work only at acidic pH because the protonation of cytosine is required for effective triplex formation. Therefore, the incorporation of cytosine nucleotide analogues such as thio-pseudoisocytosine19 in place of cytosine would be useful for the probe design which achieved the binding ability at neutrality toward in vivo applications. We are now undertaking further studies in these directions.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Grant-in-Aid for Scientific Research (B) (No. 16H04159) and for Young Scientists (A) (No. 17H04881), from Japan Society for the Promotion of Science (JSPS).Notes and references
- M. D. Shortridge and G. Varani, Curr. Opin. Struct. Biol., 2015, 30, 79–88 CrossRef CAS PubMed.
- K. D. Warner, C. E. Hajdin and K. M. Weeks, Nat. Rev. Drug Discovery, 2018, 17, 547–558 CrossRef CAS PubMed.
- (a) Y. Sato, T. Ichihashi, S. Nishizawa and N. Teramae, Angew. Chem., Int. Ed., 2012, 51, 6369–6372 CrossRef CAS PubMed; (b) Y. Sato, Y. Toriyabe, S. Nishizawa and N. Teramae, Chem. Commun., 2013, 49, 9983–9985 RSC; (c) Y. Sato, H. Saito, D. Aoki, N. Teramae and S. Nishizawa, Chem. Commun., 2016, 52, 14446–14449 RSC.
- I. M. van der Wiel, J. Cheng, R. Koukiekolo, R. K. Lyn, N. Stevens, N. O'Connor, N. J. Turro and J. P. Pezacki, J. Am. Chem. Soc., 2009, 131, 9872–9873 CrossRef CAS PubMed.
- K. Zhou, P. Kos, Y. Yan, H. Xiong, Y.-L. Min, K. A. Kinghorn, J. T. Minnig, J. B. Miller and D. J. Siegwart, Chem. Commun., 2016, 52, 12155–12158 RSC.
- S. M. Elbashir, J. Harborth, W. Lendeckel, A. Yalcin, K. Weber and T. Tuschl, Nature, 2001, 411, 494–498 CrossRef CAS PubMed.
- (a) T. Sato, Y. Sato, K. Iwai, S. Kuge, S. Nishizawa and N. Teramae, Chem. Commun., 2015, 51, 1421–1424 RSC; (b) T. Sato, Y. Sato, K. Iwai, S. Kuge, N. Teramae and S. Nishizawa, Anal. Sci., 2015, 31, 315–320 CrossRef CAS PubMed; (c) Y. Sato, M. Kaneko, T. Sato, S. Nakata, Y. Takahashi and S. Nishizawa, ChemBioChem, DOI:10.1002/cbic.201800560.
- J. Nygren, N. Svanvik and M. Kubista, Biopolymers, 1998, 46, 39–51 CrossRef CAS PubMed.
- M. Li, T. Zengeya and E. Rozners, J. Am. Chem. Soc., 2010, 132, 8676–8681 CrossRef CAS PubMed.
- I. Lubitz, D. Zikich and A. Kotlyar, Biochemistry, 2010, 49, 3567–3574 CrossRef CAS PubMed.
- (a) T. Sato, Y. Sato and S. Nishizawa, J. Am. Chem. Soc., 2016, 138, 9397–9400 CrossRef CAS PubMed; (b) T. Sato, Y. Sato and S. Nishizawa, Chem. - Eur. J., 2017, 23, 4079–4088 CrossRef CAS PubMed; (c) T. Chiba, T. Sato, Y. Sato and S. Nishizawa, Org. Biomol. Chem., 2017, 15, 7765–7769 RSC.
- Y.-L. Chiu and T. M. Rana, Mol. Cell, 2002, 10, 549–561 CrossRef CAS PubMed.
- O. Muse, T. Zengeya, J. Mwaura, D. Hnedzko, D. W. McGee, C. T. Grewer and E. Rozners, ACS Chem. Biol., 2013, 8, 1683–1686 CrossRef CAS PubMed.
- B.-W. Sun, B. R. Babu, M. D. Sørensen, K. Zakrzewska, J. Wengel and J.-S. Sun, Biochemistry, 2004, 43, 4160–4169 CrossRef CAS PubMed.
- T. Sato, N. Sakamoto and S. Nishizawa, Org. Biomol. Chem., 2018, 16, 1178–1187 RSC.
- R. L. Burwell and R. G. Pearson, J. Phys. Chem., 1966, 70, 300–302 CrossRef CAS.
- A. C. Moses and A. Schepartz, J. Am. Chem. Soc., 1996, 118, 10896–10897 CrossRef CAS.
- A. C. Moses and A. Schepartz, J. Am. Chem. Soc., 1997, 119, 11591–11597 CrossRef CAS.
- G. Devi, Z. Yuan, Y. Lu, Y. Zhao and G. Chen, Nucleic Acids Res., 2014, 42, 4008–4018 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedure, probe synthesis, UV melting curves, CD spectra, fluorescence response of probes, stopped-flow analysis and examination of spacer introduction. See DOI: 10.1039/c8ra08759h |
| This journal is © The Royal Society of Chemistry 2018 |