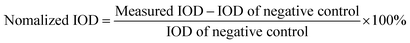Open Access Article
Open Access ArticleA disposable microcapsule array chip fabricated by ice printing combined with isothermal amplification for Salmonella DNA detection†
Enqi He‡
 bd,
Ting Cao‡a,
Liangyuan Caia,
Dan Guob,
Yinglin Zhou
bd,
Ting Cao‡a,
Liangyuan Caia,
Dan Guob,
Yinglin Zhou *a,
Xinxiang Zhang*a and
Zhihong Li*cd
*a,
Xinxiang Zhang*a and
Zhihong Li*cd
aBeijing National Laboratory for Molecular Sciences (BNLMS), Key Laboratory of Bioorganic Chemistry and Molecular Engineering of Ministry of Education, College of Chemistry, Peking University, Beijing 100871, China. E-mail: zhouyl@pku.edu.cn; zxx@pku.edu.cn; Fax: +86-10-62754112; Tel: +86-10-62754112
bState Key Laboratory of Tribology, Department of Mechanical Engineering, Tsinghua University, Beijing 100084, China
cNational Key Laboratory of Science and Technology on Micro/Nano Fabrication, Institute of Microelectronics, Peking University, Beijing 100871, China. E-mail: zhhli@pku.edu.cn; Fax: +86-10-62751789; Tel: +86-10-62766581
dCenter for Nano and Micro Mechanics, Tsinghua University, Beijing 100084, China
First published on 27th November 2018
Abstract
In this work, a novel microcapsule array chip was fabricated for the detection of Salmonella DNA by integrating an ice-printing technique with DNA isothermal amplification. Reaction solutions were previously sealed in the microcapsule array chip via ice printing. To protect the relatively fragile DNA isothermal amplification system, an extra polystyrene (PS) film was introduced to isolate the reaction solution from photopolymer precursor, which was proved to be a vital step for providing a clean and stable environment for DNA amplification reaction. Detection operation can be done by simply injecting sample DNA into the microcapsule by an easily accessible syringe, and the result can be directly obtained through color change within 90 minutes. This method shows good sensitivity, specificity and stability.
1. Introduction
Portable, disposable, and cost-effective devices that can be easily operated by inexperienced personnel with only a few steps are called for in point-of-care assays.1,2 Motivated by this need, test papers are developed and commercialized with diverse designs,3–5 among which the HCG Test Strip for early pregnancy testing6 is the most successful biochemical trial. However, most biochemical assays, especially those concerning DNA-related reactions, require strict experimental conditions that a test paper cannot provide. Firstly, a sealed environment is necessary for system storage, because fragile reagents like bio-enzymes may be deactivated in open conditions due to possible contamination or oxidation. Secondly, because most DNA-related reactions need incubation at a certain temperature, closed small vessels are required to contain reaction solutions, which should not change or volatilize during incubation. Additionally, it is advantageous if the device would endure relatively high temperature for hours. Thirdly, the device should suit signal readout methods. For example, a transparent device is preferred for colorimetric assays and non-fluorescence substrate material is preferred for fluorescence-based assays. Fourthly, if DNA amplification reaction is required, it is extremely important that the reaction device must be disposable and should not be opened when discarded to avoid the contamination of DNA amplification products. The reason is that the released product DNA may interfere with the next assays because of the high sensitivity of the amplification method.Most of the significant isothermal amplification devices are fabricated using a soft lithography method instead of conventional top-down lithography process with the advantages of low toxicity, low cost, wide substrate ranges especially non-flat surfaces, and no need for a photo-reactive surface.7 Generally, a liquid precursor of polymers is poured over the mold or the sacrificial layer and then liquid precursors are cured into the solid by incubation at some certain conditions for a period of time. For example, polydimethylsiloxane (PDMS), which has been widely used in recent isothermal amplification devices8–10 showing easy and flexible fabrication process and good biocompatibility, is cured to solid at 70 °C for almost 1 h. For most previous cases, devices are completely made before integrated with biological systems, and thus can hardly achieve pre-packaged bio-systems, for bio-reagents may suffer from deactivation under long-time incubation at high temperature, and the liquid solution cannot stay in a certain shape for the liquid precursors to be stereotyped.
Our group has developed a microcapsule chip based on 3D ice-printing technique11 that is promising address this issue. In general, a small volume of reaction solution is printed on a cooled membrane substrate and is frozen into ice. Then transparent photopolymer is poured onto the ice spot and polymerized under LED to seal the reaction solution. The fabricated microcapsules can be stored below 0 °C for long-term preservation. When in use, a small volume of sample solution (1–2 μL) is injected directly into the microcapsule for simple visual detection. This technique is potential to fit bio-reagents containing detecting devices in two ways: firstly, the low temperature of ice keeps the bio-reagents in a relatively stable physical and chemical state, making it compatible with the entire device fabrication process. Secondly, using this bottom-up one-step molding design, bio-system can be pre-packed in the device in the device fabrication process, which is significant for easy and rapid detection. Most existing isothermal DNA amplification devices cannot achieve the pre-pack function, for they have to be completely made before integrated with biological systems.
On-site point-of-care detection of pathogens has become increasingly important in the fields of food safety testing, environmental monitoring and human health care.12–16 Since the abundance of pathogens is usually very low, DNA amplification is essential in the detection process. Loop-mediated isothermal amplification (LAMP)17–20 is an isothermal method for nucleic acids amplification with high specificity and sensitivity using a set of four or six specially designed primers. Compared with the most widely applied DNA amplification method called polymerase chain reaction (PCR),21,22 which needs specific temperature cycling instrument, LAMP can be carried out in easily accessible thermal controllers such as ovens at around 65 °C. Moreover, the LAMP result can be simply determined through turbidity23,24 or color change,25,26 which makes it remarkably advantageous in point-of-care assays at home or in the field.27,28
In this work, we first integrated ice-printing technique with LAMP to fabricate a novel microcapsule array chip for Salmonella DNA detection. However, our previous ice-printing technique is not adequate for DNA isothermal amplification-based biochemical assay, mainly because raw materials of photopolymer (unpolymerized monomers and polymerization initiators) may exude during high-temperature incubation, and thus interfere with DNA isothermal amplification system. Additionally, the porous structure formed by photo polymerization might adsorb reagents during incubation process. To address this issue, we herein improved 3D ice-printing technique to fit relatively fragile DNA isothermal amplification system. A polystyrene (PS) film is fabricated encapsulating icy reaction solution before photopolymer sealing by solvent volatilization of PS solution. The PS film isolates and protects the reaction solution from sealing photopolymer, providing a clean and stable environment for bio-enzyme based reaction. Thus, this design well suits LAMP reaction and is successfully applied into Salmonella DNA detection.
2. Experimental section
2.1. Materials and reagents
DNA oligonucleotides were synthesized by Sangon Biotech. Co., Ltd. (Shanghai, China), and have the following sequences: 5′-GGCGATATTGGTGTTTATGGGG-3′, F3; 5′-AACGATAAACTGGACCACGG-3′, B3; 5′-GACGACTGGTACTGATCGATAGTTTTTC AACGTTTCCTGCGG-3′, FIP; 5′-CCGGTGAAATTATCGC CACACAAAACCCACCGCCAGG-3′, BIP; 5′-GACGAAAGAGCGTGGTAATTAAC-3′, LF; 5′-GGGCAATTCGTTATTGGCGATAG-3′, LB. Bst 2.0 WarmStart polymerase, 10× Isothermal Amplification Buffer (200 mM Tris–HCl, 500 mM KCl, 100 mM (NH4)2SO4, 20 mM MgCl2, 1% Triton X-100, pH 8.8 at 25 °C), and MgSO4 (100 mM) were obtained from New England Biolabs (Beverly, MA). Deoxynucleotide solution mixture (dNTPs) was provided by Takara Biotechnology Co. Ltd (Dalian, China). Calcein and MnCl2 were purchased from Sigma-Aldrich (St. Louis, MO, USA). Salmonella DNA, Escherichia coli DNA, and Shigella DNA were extracted right from the corresponding bacteria using bacteria genomic DNA extraction kit from Tiangen Biotech Co. Ltd (Beijing, China), and are diluted into certain concentrations. Commercial milk purchased from local supermarkets was mixed with overnight cultured Salmonella bacteria to obtain contaminated milk, which was then treated by the same bacteria genomic DNA extraction kit to get the injection sample. Other reagents were at least of analytical reagent grade. All solutions were prepared with ultrapure water purified with a Milli-Q purification system (Branstead, MA, U.S.A).2.2. Preparation of LAMP reaction solution
LAMP primers are previously dissolved into 100 μM separately in pure water, and afterwards 25× LAMP primer mix is prepared as the following concentration: c (FIP) = 40 μM, c (BIP) = 40 μM, c (F3) = 5 μM, c (B3) = 5 μM, c (LF) = 10 μM, c (LB) = 10 μM. Calcein is dissolved in 1× Isothermal Amplification Buffer into 1 mM. MnCl2 is dissolved in pure water into 10 mM. Using the above stock solutions, LAMP reaction solution is freshly prepared right before the fabrication of microcapsule array chip. The final concentrations of all reagents are listed below: Isothermal Amplification Buffer 1× (contains 2 mM MgSO4), extra MgSO4 (6 mM, 8 mM in total), LAMP primer mix 1×, c (dNTP) = 2.8 mM each, c (calcein) = 100 μM, c (MnCl2) = 1 mM, c (Bst 2.0 WarmStart polymerase) = 480 U mL−1.2.3. Chip fabrication based on ice-printing technique
The microcapsule array chip is fabricated using our homemade ice-printing instrument consisted of a set of X-Y-Z-axis platform to move the printing nozzle to a certain spot, a printing pump to pump reaction solution, a printing nozzle to drop solution and a Peltier cooler driven by condensate water to maintain cool surface (below −20 °C) in the fabrication process. The whole system is placed inside a glovebox filled with pure nitrogen to maintain dry atmosphere, avoiding frosting on the cold surface.The main frame of the chip is made by simply sticking polyethylene terephthalate (PET) film to plastic cofferdam by commercial glue. The inner side of the PET film is cleaned by ethanol and pure water before use. 30% polystyrene solution is dissolved in chloroform (different concentrations ranging from 15–40% and different solvents are prepared in optimization experiments).
The detailed fabrication procedure is illustrated as follows. Firstly, the printing nozzle is moved to a certain spot and pumps out 10 μL LAMP reaction solution onto the PET film. The solution is frozen into ice as soon as it reaches the cool surface. The radius of curvature of the ice droplet is measured to be 4.6 mm, and the contact angel is 68° (Fig. S1†). Multiple droplets can be pumped successively to form an array. Secondly, 8 μL the 30% PS solution is dropped onto the icy solution to form a contact film after solvent volatilization. It is important to make sure that the PS solution covers the ice droplet well. It takes about 20 minutes before the film gets dried throughout. Thirdly, photopolymer precursor is added slowly onto the chip to avoid bubbles, and it polymerizes under 420 nm LED for 5 minutes to seal the microcapsule. With good light transmittance property, PS film and polymer would not hinder visual colorimetric detection (Fig. S2†). In the whole process, the system is supposed to be cooled to below −20 °C to guarantee the reaction solution frozen. The chip is stored at −20 °C before use.
2.4. Direct visual detection of Salmonella DNA
The chip is taken out from refrigerator and placed in room temperature for 5 minutes to let the icy solution melt right before use. 1 μL of Salmonella DNA (or Escherichia coli DNA, and Shigella DNA) is injected into the microcapsule by a syringe, and the injecting hole is directly sealed by transparent tape. Then, the chip is incubated at 65 °C for 90 minutes. Finally, the colorimetric detection results can be simply observed by naked eyes. Photos are taken by cell phone.3. Results and discussion
3.1. Fabrication of the chip and its principle for Salmonella DNA detection
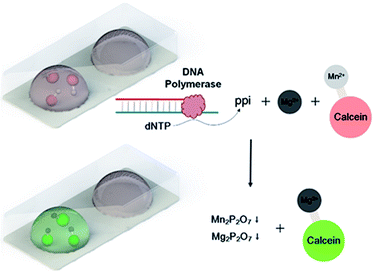
Fig. 1(A) shows the self-made instrument used in this work, consisted of X-Y-Z-axis platforms, a printing pump, a printing nozzle and a Peltier cooler. The cooler is driven by condensate water system, and can cool the surface to below −20 °C. The general procedure of fabricating ice-printing microcapsule array is explained in Fig. 1(B). First, 10 μL of reaction solution for LAMP containing Salmonella LAMP primers, DNA polymerase, colorimetric reagents and corresponding buffer is printed on the cold PET substrate and is frozen into ice (a). Then, previous dissolved PS solution is dropped on the ice droplet and a PS film is formed after solvent volatilization (b). Next, photopolymer precursor is added and polymerized under 420 nm LED to seal the microcapsule (c). Multiple microcapsules array can be fabricated at one time on a chip for multiple assays, and the whole fabrication process takes no more than 30 minutes. The obtained chips are stored at −20 °C.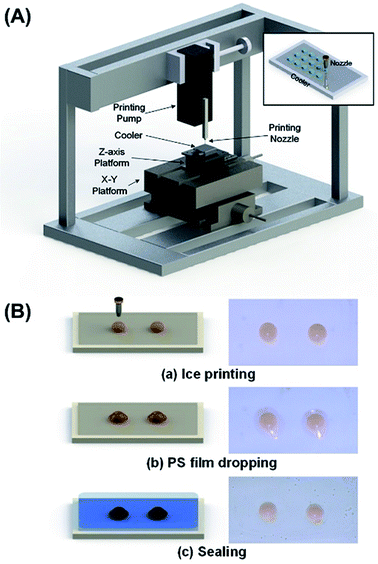 | ||
| Fig. 1 (A) Ice-printing instrument used in this work. (B) Fabrication process of microcapsule array chip. | ||
When in use, sample Salmonella DNA is injected directly into the microcapsule using a syringe after thawing the frozen chip, and the hole is simply sealed with a transparent tape (Fig. S3 and S4†). LAMP reaction is carried out in every microcapsule with 6 primers,29 consisted of two outer primers (forward primer F3 and backward primer B3), two inner primers (forward inner primer FIP and backward inner primer BIP) and two loop primers (forward loop LF and backward loop LB). The outer primers are used in initial steps to produce starting structures, while the inner primers lead the subsequent explosive cycling strand displacement reaction, generating broccoli-like final products with different lengths. Calcein (green color), a metal indicator, and MnCl2 are previously added to the reaction solution. Before amplification, the solution appears orange, which is the color for Mn2+ binding with calcein. In the intense amplification reaction process, a large amount of pyrophosphate ions are generated from dNTP, which forms insoluble salt (Mn2P2O7) with Mn2+. The released calcein binds with free Mg2+ in the solution, and appears green (Scheme 1). By simply telling the color (orange or green), the amplification conditions can be determined, which is suitable for point-of-care assays.
3.2. Function of the PS film and optimization
Notably, the PS film formation step is very important in the microcapsule fabrication process. Raw materials and polymerization residues may exude during high-temperature incubation, and thus influences the DNA isothermal amplification system, resulting in inefficient DNA amplification. Additionally, the porous structure formed by photo polymerization might adsorb reagents during incubation process, which influences the homogeneity of the reaction solution. Moreover, the metal indicator-based colorimetric system is also influenced. As Fig. 2 shows, reaction system would change color from orange to green in the absence of the target (no LAMP reaction) without fabrication of PS film, which severely interferes the colorimetric assay.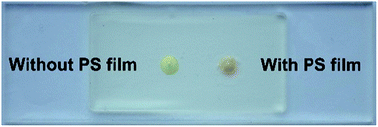 | ||
| Fig. 2 Comparison of the microcapsule solution conditions after incubation in the absence of target with and without PS film. | ||
Since the PS film is formed by solvent volatilization of the PS solution, the nature of the solvent determines the quality of the film as well as the film formation time. Four common PS solvents, namely dichloromethane (CH2Cl2), chloroform (CHCl3), tetrahydrofuran (THF), and toluene, were tried and the results are explained as follows respectively. THF may absorb water from the ice droplet due to its water-miscible property, which leads to uneven dispersion of PS, resulting in multiple small pores on the film. Because the high volatility of dichloromethane, the outer side of dissolved PS gets dried immediately, which hinders throughout volatilization of the inner side of the film. On the contrary, toluene volatilizes very slowly, and the volatilization process takes more than 1.5 h, which wastes too much time. Compared with the above three solvents, chloroform is the most appropriate PS solvent with moderate volatility, which helps to form an even and compact PS film within 20 minutes. PS concentration, and the volume of PS solution are also optimized to 30% and 8 μL to obtain best film quality.
3.3. Performance of Salmonella DNA detection
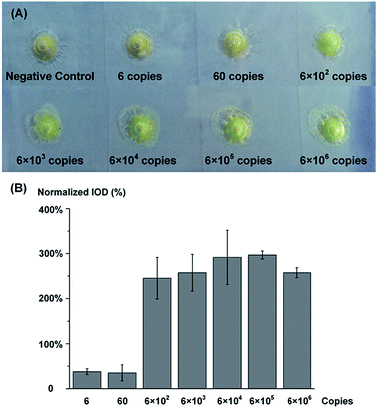
The sensitivity performance of this ice-printed microcapsule array chip for Salmonella DNA detection is shown in Fig. 3(A). 6, 60, 6 × 102, 6 × 103, 6 × 104, 6 × 105, and 6 × 106 copies of Salmonella DNA and DNA-free water (negative control) are injected into microcapsules, respectively, and incubated in an oven or any heater at hand at 65 °C for 90 minutes. Simple choices of “orange or green” can be immediately and clearly made by naked eyes just on the spot owing to the colorimetric principle of LAMP and the transparent property of the whole chip. Notably, down to 600 copies of Salmonella DNA can be detected, which is comparable to the limit of detection of other visual detection methods based on LAMP and better than conventional PCR29–31 (Table S1†). To better illustrate this test, we measure the Integrated optical densities (IOD) of the green color (parameter: hue 48-70) using software Image J. The difference of green color IOD between experimental group and negative control is calculated and normalized by dividing by the IOD of negative control, i.e.,The processed normalized IODs are showed in Fig. 3(B), which indicates no gradual color change among different concentrations, suggesting that our device a “yes or no” sensor like most LAMP based colorimetric sensors25,26 rather than a quantitative sensor.
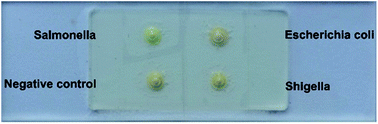
Fig. 4 shows the specificity test of Salmonella DNA assay against DNA-free water, Escherichia coli DNA, and Shigella DNA. After LAMP reaction, the microcapsule with Salmonella DNA injection turns green, however, other microcapsules stay orange. This visual result indicates good DNA discrimination ability of our detection. The stability performance, which is tested after storing the microcapsule chip in freeze for 0–15 days, is shown in Fig. S5.† Distinguished color change of the Salmonella DNA assay is observed compared with the negative control, which demonstrates good stability. Furthermore, the stability of the materials used is also tested (Fig. S6†).
To further verify its ability for Salmonella detection in real samples, we treat commercial milk using our device. Salmonella bacteria is cultured overnight and mixed with commercial milk. The mixture is treated following DNA extraction process to obtain the injection sample as usual. In Fig. 5, the left microcapsule injected the contaminated milk sample gives a positive green color.
 | ||
| Fig. 5 Visual detection of Salmonella contaminated milk. DNA-free water is used as negative control. | ||
4. Conclusions
We proposed a microcapsule array chip for visual Salmonella DNA detection based on improved ice-printing technique, which meets the needs of point-of-care systems with fragile bio-agents on all aspects including storage, incubation needed reaction and sealed discard without contamination. The chip fabrication process is simple and fast without traditional complicated lithography techniques. Only 10 μL reaction solution is contained in each microcapsule, which gives rise to reagents saving. Notably, with the isothermal amplification method, no annealing and temperature cycling steps are needed in the whole DNA amplification process, which stays away from instrumental constrains and can be easily carried out in ordinary heating conditions like water baths or ovens. LAMP also enables colorimetric detection based on orange to green color change, leading to easy, rapid, and on-spot result determination. With excellent sensitivity, specificity and stability, this system is promising in real point of-care detection of Salmonella DNA as well as other pathogen DNA.Hopefully, automated production can be realized to save labor and further improve efficiency owing to the simple and programmable chip fabrication process. Moreover, by fabricating multiple microcapsules and using LAMP solutions targeting different analytes on one chip, high-throughput and multiple-target assays can be further performed. This novel device also provides an alternative method on other bio-assays with fragile reagents, for instance, nucleic acids and proteins. Our future work will focus on integrating DNA extraction into our device by designing microfluidics systems.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key Research and Development Program of China (No. 2016YFA0200802) and the National Natural Science Foundation of China (No. 21675004 & No. 21575005).Notes and references
- E. Petryayeva and W. R. Algar, RSC Adv., 2015, 5, 22256–22282 RSC.
- Z. Fang, W. Wu, X. Lu and L. Zeng, Biosens. Bioelectron., 2016, 56, 192–197 CrossRef PubMed.
- N. K. Thom, G. G. Lewis, K. Yeung and S. T. Phillips, RSC Adv., 2014, 4, 1334–1340 RSC.
- Q. Ju, M. O. Noor and U. J. Krull, Analyst, 2016, 141, 2838–2860 RSC.
- J. R. Choi, J. Hu, R. Tang, Y. Gong, S. Feng, H. Ren, T. Wen, X. Li, W. A. B. W. Abas, B. Pingguan-Murphy and F. Xu, Lab Chip, 2016, 16, 611–621 RSC.
- C. Gnoth and S. Johnson, Geburtshilfe Frauenheilkd., 2014, 74, 661–669 CrossRef CAS PubMed.
- Y. Xia and G. M. Whitesides, Annu. Rev. Mater. Sci., 1998, 28, 153–184 CrossRef CAS.
- K. Hsieh, A. S. Patterson, B. S. Ferguson, K. W. Plaxco and H. T. Soh, Angew. Chem., Int. Ed., 2012, 124, 4980–4984 CrossRef.
- Y. Liu, Y. Zhao, Y. Qin, X. Du, Q. Wang and J. Lyu, RSC Adv., 2016, 6, 13399–13406 RSC.
- X. Fang, Y. Liu, J. Kong and X. Jiang, Anal. Chem., 2010, 82, 3002–3006 CrossRef CAS PubMed.
- H. Z. Zhang, F. T. Zhang, X. H. Zhang, D. Huang, Y. L. Zhou, Z. H. Li and X. X. Zhang, Anal. Chem., 2015, 87, 6397–6402 CrossRef CAS PubMed.
- L. Schwenkbier, S. Pollok, A. Rudloff, S. Sailer, D. Cialla-May, K. Weber and J. Poppa, Analyst, 2015, 140, 6610–6618 RSC.
- S. J. Oh, B. H. Park, G. Choi, J. H. Seo, J. H. Jung, J. S. Choi, D. H. Kim and T. S. Seo, Lab Chip, 2016, 16, 1917–1926 RSC.
- Z. Fang, W. Wu, X. Lu and L. Zeng, Biosens. Bioelectron., 2014, 56, 192–197 CrossRef CAS PubMed.
- Y. Xia, Z. H. Liu, S. Q. Yan, F. Yin, X. J. Feng and B. F. Liu, Sens. Actuators, B, 2016, 228, 491–499 CrossRef CAS.
- M. Pacheco, B. Jurado-Sanchez and A. Escarpa, Anal. Chem., 2016, 90(4), 2912–2917 CrossRef PubMed.
- T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino and T. Hase, Nucleic Acids Res., 2000, 28, e63 CrossRef CAS PubMed.
- C. D. Guevara, V. V. Swaminathan, B. Reddy, J. Huang, Y. Liu and R. Bashir, RSC Adv., 2016, 6, 103872–103887 RSC.
- M. S. Draz and X. Lu, Theranostics, 2016, 6, 522–523 CrossRef CAS PubMed.
- L. Zhu, Y. Xu, N. Cheng, P. Xie, X. Shao, K. Huang, Y. Luo and W. Xu, Sens. Actuators, B, 2017, 242, 880–888 CrossRef CAS.
- R. Saiki, D. Gelfand, S. Stoffel, S. Scharf, R. Higuchi, G. Horn, K. Mullis and H. Erlich, Science, 1988, 239, 487–491 CrossRef CAS PubMed.
- H. A. Erlich, D. Gelfand and J. J. Sninsky, Science, 1991, 252, 1643–1651 CrossRef CAS PubMed.
- Y. Mori, K. Nagamine, N. Tomita and T. Notomi, Biochem. Biophys. Res. Commun., 2001, 289, 150–154 CrossRef CAS PubMed.
- Y. Mori, M. Kitao, N. Tomita and T. J. Notomi, J. Biochem. Biophys. Methods, 2004, 59, 145–157 CrossRef CAS PubMed.
- N. Tomita, Y. Mori, H. Kanda and T. Notomi, Nat. Protoc., 2008, 3, 877–882 CrossRef CAS PubMed.
- H. Nikbakht, P. Gill, A. Tabarraei and A. Niazi, RSC Adv., 2014, 4, 13575–13580 RSC.
- M. Safavieh, M. U. Ahmed, E. Sokullu, A. Ng, L. Braescu and M. Zourob, Analyst, 2014, 139, 482–487 RSC.
- X. Fang, H. Chen, S. Yu, X. Jiang and J. Kong, Anal. Chem., 2010, 83, 690–695 CrossRef PubMed.
- C. Techathuvanan, F. A. Draughon and D. H. D'Souza, J. Food Sci., 2010, 75, M165–M172 CrossRef CAS PubMed.
- J. L. Yang, G. P. Ma, R. Yang, S. Q. Yang, L. Z. Fu, A. C. Cheng, M. S. Wang, S. H. Zhang, K. F. Shen and R. Y. Jia, J. Appl. Microbiol., 2010, 109, 1715–1723 CAS.
- J. Li, L. Zhai, X. Bie, Z. Lu, X. Kong, Q. Yu, F. Lu, C. Zhang and H. Zhao, Food Control, 2016, 60, 230–236 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra07045h |
| ‡ These two authors contribute equal to this work. |
| This journal is © The Royal Society of Chemistry 2018 |