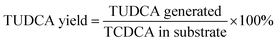Open Access Article
Open Access ArticleCo-immobilised 7α- and 7β-HSDH as recyclable biocatalyst: high-performance production of TUDCA from waste chicken bile†
Qingzhi Ji *a,
Bochu Wang
*a,
Bochu Wang b,
Chou Lic,
Jinglan Haoa and
Wenjing Fenga
b,
Chou Lic,
Jinglan Haoa and
Wenjing Fenga
aSchool of Pharmacy, Yancheng Teachers' University, No. 2, Hope Avenue, Yancheng, Jiangsu 224051, P. R. China. E-mail: jiqingzhi202@126.com; Fax: +86-0515-88258773; Tel: +86-0515-88258773
bKey Laboratory of Biorheological Science and Technology (Chongqing University), Ministry of Education, College of Bioengineering, Chongqing University, Chongqing 400030, P. R. China
cCollege of Marine and Bio-engineering, Yancheng Teachers' University, Yancheng, Jiangsu 224051, P. R. China
First published on 4th October 2018
Abstract
Chicken gallbladder has long been considered to be worthless and discarded as waste. The main composition of chicken bile is taurochenodeoxycholic acid (TCDCA), which is the isomeride of tauroursodeoxycholic acid (TUDCA). TUDCA has been effectively used for treatment of many diseases. In this paper, 7α- and 7β-hydroxysteroid dehydrogenases (HSDH) were co-immobilised on modified chitosan microspheres, and used as recyclable biocatalyst for the catalysis of chicken bile. The catalytic reaction reached equilibrium within 4 h compared with 1 h using TCDCA as substrate. After four continuous batch reactions, the conversion of TCDCA was lower than 40% and TUDCA yield was about 15% for the catalysis of chicken bile. TUDCA yield was approximately 62% after equilibrium and the content of TUDCA in reaction product was as high as 33.16%. Furthermore, the experiments showed that activity of enzymes were significantly inhibited by bilirubin, Cu2+ and Ca2+ in complex substrate. The research described not only widens the utilization of chicken bile, but also provides a clean way for the preparation of TUDCA.
1. Introduction
Tauroursodeoxycholic acid (TUDCA) and its hydrolysate ursodeoxycholic acid (UDCA) are secondary bile acids, which are synthesized from primary bile acid chenodeoxycholic acid (CDCA) in enterohepatic circulation.1 These are candidate medicines commonly used for the treatment of hepatobiliary disease.2–4 Furthermore, TUDCA can prevent hearing and vision loss, and slow down retinal degeneration.5,6 Recent papers have reported that TUDCA and UDCA are neuroprotective in Parkinson's, Huntington's and Alzheimer's diseases, and also in humans with amyotrophic lateral sclerosis.7–9 There are many additional ancillary features of TUDCA, such as promoting blood vessel repair,10 inhibiting obesity11 and enhancing islet function.12 Even TUDCA and UDCA can be prepared through a series of chemical processes; it is therefore very meaningful to obtain TUDCA in a clean and high-performance way.There are few reports about the acquisition of TUDCA through the transformation of bile acids in vitro, especially using a complex as substrate. The structure of some bile acids are similar, such as TUDCA and taurochenodeoxycholic acid (TCDCA), UDCA and CDCA, both of them are epimers with the difference of –OH at 7 site as Fig. 1. Chicken is a kind of delicious food eaten around the world. However, chicken gallbladder is often abandoned, which is rich in TCDCA. If the chicken bile can be used reasonably, it will become an abundant bioresource. The value of chicken bile will be greatly increased once TCDCA is transformed into TUDCA. Further exploitation of such a bioresource will be promoted, as well. The biotransformation of chicken bile not only provides a new way to produce TUDCA, but also provides benefit for environment protection. Torres et al. had reported one similar example. Soybean peroxidase was immobilized onto activated carbon obtained from agricultural waste with a total utilization of biomass and minimal waste generation.13
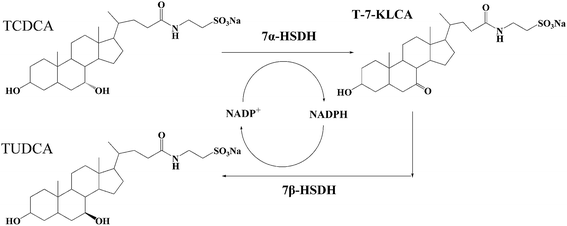 | ||
| Fig. 1 The reaction process of TCDCA to TUDCA catalysed by 7α- and 7β-HSDH. TCDCA and TUDCA are the isomeride at 7 site. 7α- and 7β-HSDH can isomerize the –OH of 7 site for TCDCA or TUDCA. | ||
The epimerization of TCDCA to TUDCA can be catalysed by 7α- and 7β-hydroxysteroid dehydrogenases (7α- and 7β-HSDH).14,15 7α- and 7β-HSDH belong to short chain dehydrogenase/reductase (SDR) family and exist in some gut microbes, such as Clostridium absonum,15 Collinsella aerofaciens16 and Bacteroides fragilis.17 The transformation of CDCA to UDCA, TCDCA to TUDCA have been proved to be feasible.18,19 The reactions catalyzed by both free enzymes and immobilised enzymes have been studied. However, in all the former related studies, CDCA or TCDCA was used as the catalytic substrate. We recognize that there has not been any reports on the biotransformation of TCDCA in complex substrate to get high-value TUDCA. In nature, most of the substance exist in the form of mixtures. It is more meaningful to obtain TUDCA using chicken bile as substrate.
This research was focus on the transformation of TCDCA in chicken bile to obtain high valuable TUDCA with the catalysis of co-immobilised 7α-, 7β-HSDH. The co-immobilised 7α-, 7β-HSDH was prepared using modified chitosan microspheres as carrier. Both the stability of enzymes and the effects of complex substrate were carefully studied. The reaction time and the concentration of coenzyme were also optimized. The possible compositions that affected enzyme activity in complicated substrate were confirmed. Molecular docking was used to explain the possible mechanism of bilirubin or complex substrate on enzyme activity.
2. Experimental section
2.1 Materials
Fresh poultry gallbladder was obtained from supermarket (Chongqing, China). The chitosan (particle size, 40–100 mesh) from Sangon Biotech (Shanghai, China) was used to prepare the carrier of enzymes. BCA Protein Assay Reagent from Beyotime (Shanghai, China) was used to detect the protein concentration. TCDCA and TUDCA standards were purchased from the National Institutes for Food and Drug Control (Beijing, China). NADP-Na2 and NADPH-Na4 (purity ≥ 97%) produced by Roche (Switzerland). Total Cholesterol Assay Reagent Kit (F0021-1), and Bilirubin Assay Reagent Kit (C018) were purchased from Jiancheng Bioengineering Institute (Nanjing, China). All the other chemicals used in this study were of analytical grade and used without further purification. Double-distilled water was used in all experiments.2.2 Preparation and component analysis of poultry bile
Fresh poultry gallbladder was cut and excluded membrane after sterilized by 75% ethanol. The mixture solution was filtrated by a filter with 100 μm pore size. The filtrate was freezed at −80 °C for 12 h and freeze drying for 48 h. After the sample was completely dry, it was stored in desiccator for the following experiments.The content determination of bile acid in poultry bile used high performance liquid chromatography coupled with evaporative light scattering detector (HPLC-ELSD). The content of cholesterol and bilirubin was detected using appropriate kits according to instructions.
2.3 Enzyme immobilisation and activity analysis
The process of immobilisation between chitosan and enzymes was shown in Fig. 2. The chitosan microspheres were prepared according to ref. 14 and 20, with slight modifications: a chitosan solution was prepared by dissolving 0.5 g of chitosan flakes into 50 mL of 2% (v/v) acetic acid solution. The resulting solution was dropped into 3% (w/v) sodium tripolyphosphate (STPP) and incubated for 3 h. Then the chitosan microspheres were activated with 0.125% glutaraldehyde (GA) solution at 25 °C and 130 rpm for 3 h. The carrier was separated by filtration and washed three times with water to remove redundant GA.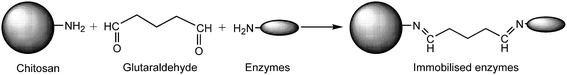 | ||
| Fig. 2 The process of enzyme immobilisation between chitosan and enzymes. Chitosan microspheres were firstly activated by glutaraldehyde. | ||
The activated microspheres (approximately 3 g) were incubated with 6 mL (1 mg mL−1) of 7α-HSDH or 7β-HSDH solution in phosphate buffer (50 mM, pH 7.2–7.4) for 4 h at 16 °C. Finally, the chitosan microspheres with immobilised enzymes were filtered and washed with phosphate buffered saline solution (pH, 7.2). The amount of immobilised protein was determined by testing the concentration of initial protein solution (mg mL−1) and the protein remaining in the supernatant (mg mL−1) with BCA Protein Assay Kit. Bovine serum albumin was used as the standard.
To determine the activity of immobilised 7α-HSDH (or 7β-HSDH), the enzyme assay mixture in a total volume of 3 mL contained 0.5 mM TCDCA (or TUDCA), 0.2 mM NADP+ and 1.5 g immobilised 7α-HSDH (or 7β-HSDH). When free enzymes was used as catalyst, the final concentration was 1 mg mL−1. The amount of NADPH was proportional to the ultraviolet absorption at 340 nm. So change of ultraviolet absorbance at 340 nm was recorded to measure enzyme activity. One unit of enzyme activity was defined as the amount of enzyme needed to convert 1 μM substrate (TCDCA/TUDCA) within 1 minute at 25 °C in a buffer containing 50 mM Tris–HCl (pH 8.5).
2.4 Enzymatic reaction
The reaction was carried out on the condition of 25 °C, pH 8.5 Tris–HCl (50 mM), using TCDCA (8 mM) and chicken bile (equal TCDCA amount) as substrate, respectively. The reaction was continued for 5 hours with the catalysis of co-immobilised 7α-, 7β-HSDH, six consecutive batch reactions were conducted. TCDCA conversion and TUDCA yield were analyzed. The catalytic efficiency were also studied with different concentration of TCDCA (2–24 mM) and NADP+ (0.1–4 mM). The other reaction conditions were consistent.2.5 The effect of complex substrate on the activity of enzymes
The effects of chicken bile on the activity of free 7α-HSDH and immobilised 7α-HSDH were conducted using TCDCA and chicken bile (containing equal TCDCA). The chicken bile powder was dissolved by 50 mM Tris–HCl (pH 8.5) and sonicated for 30 min. The solution was centrifuged for 10 min at 8000 rpm. The supernatant was used as substrate. The detection of enzyme activity was according to 2.3.The effect of bilirubin on the activity of 7α-HSDH and 7β-HSDH was studied according to the following methods. Bilirubin was added to the solution of TCDCA or TUDCA (8 mM), and insure that the final concentration of bilirubin was 0.457% (mass ratio) equal to the amount of chicken bile. Then the substrate was catalyzed by free enzymes or immobilised enzymes. The activity of enzymes was detected according to 2.3.
The metal ions usually affected the activity of enzymes. According to the content of chicken bile, each metal ions were added to TCDCA solution, respectively, and the effect on enzyme activity was detected according to 2.3.
2.6 High performance liquid chromatography (HPLC)
TCDCA conversion and TUDCA yield were detected using HPLC-ELSD. The HPLC analysis was performed with a Waters Acquity SQD (Santa Clara, CA, USA) equipped with a 150 × 2.1 mm, 3 μm Welch Ultimate XB-C18 column. The injection volume was 5 μL. Two solvents (A: 100% methanol; B: 50 mM aqueous ammonium acetate solution) were used at a constant flow rate of 0.8 mL min−1. Elution was performed using a linear gradient from 60 to 90% A for 15 min, from 90 to 60% A immediately, and then held constant for 10 min. The ELSD detection temperature was 80 °C. The detection was carried out in nitrogen environment and the speed of nitrogen was 3.0 L min−1.212.7 Docking of bilirubin and enzymes
The molecular docking of bilirubin to 7α-HSDH or 7β-HSDH was performed using the 3D crystal structure of these proteins (PDB codes 5EPO and 5GT9) obtained from the Protein Data Bank.22,23 The ionizable residues were set to their pH 7.4 protonation states; the His, Arg and Lys were protonated while those of Asp and Glu were deprotonated. The Autodock 4.2 plugin was used for all dockings in this study. The docking parameters for AutoDock 4.2 were kept to their default values. The docking results were ranked by the binding free energy. The binding modes with the lowest binding free energy and the most cluster members were chosen for the optimum docking conformation. The binding results were illustrated using the PyMOL Molecular Graphics System Version 1.5.3. Results and discussion
3.1 The activity of immobilised enzymes
The results of immobilisation and activity for the 7α-HSDH and 7β-HSDH were shown in Table 1. After 4 h immobilisation at 16 °C, the protein loading yields of 7α-HSDH and 7β-HSDH. were about 93.50% and 84.50%, respectively. The activity of immobilised 7α-HSDH was about 9.87 U g−1-support, which was higher than immobilised 7β-HSDH (about 8.75 U g−1-support). However, the specific activity yields of 7α-HSDH and 7β-HSDH were only 42.26% and 49.12%, which demonstrated that more than half activity of two enzymes lost in the process of immobilisation. This phenomenon is so common in enzyme immobilisation.| Activity (U mg−1) | Protein loading (mg-protein per g-support) | Protein loading yield (%) | Immobilized enzyme activity (Ug−1) | Activity yield (%) | |
|---|---|---|---|---|---|
| a Conditions for immobilization: GA-modified chitosan microspheres (3 g) were reacted with 7α- or 7β-HSDH (6 mL, 1.0 mg mL−1) in phosphate buffer (50 mM, pH 7.2) at 16 °C for 4 h. Conditions for enzyme detection: TCDCA or T-7-KLCA (0.5 mM) catalysed by immobilized 7α- or 7β-HSDH (3 g) with NADP(H) (0.5 mM) in Tris–HCl (50 mM, pH 8.5) at 25 °C for 1 min. | |||||
| 7α-HSDH | 12.49 ± 4.62 | 1.87 ± 0.08 | 93.50 ± 4.00 | 9.87 ± 1.89 | 42.26 ± 8.09 |
| 7β-HSDH | 10.54 ± 2.74 | 1.69 ± 0.14 | 84.50 ± 7.00 | 8.75 ± 3.41 | 49.12 ± 19.14 |
3.2 The effect of time and NADP+ concentration on transformation
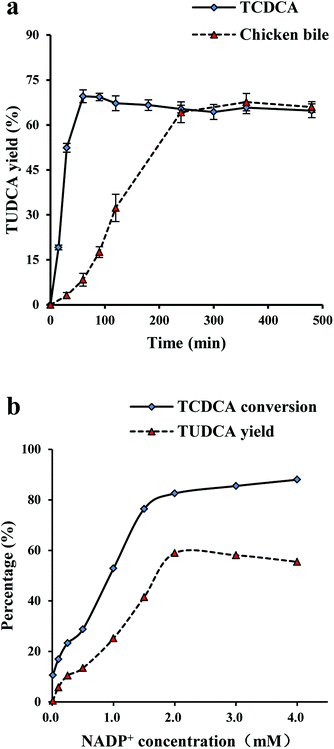
At the equilibrium, TUDCA yields were about 70% both TCDCA and chicken bile as substrates with the catalysis of co-immobilised 7α-, 7β-HSDH as Fig. 3a. However, it took longer time to reach equilibrium using chicken bile solution as substrate compared with TCDCA. It was found that the catalytic reaction of TCDCA reached equilibrium within 60 min, while the catalysis of chicken bile was as long as 240 min. The longer reaction time may due to the low activity of 7α- and 7β-HSDH to TCDCA in chicken bile. 7α- or 7β-HSDH may be inhibited by some components in complex substrate.As shown in Fig. 3b, the optimum concentration of NADP+ was 2 mM for the catalysis of TCDCA. When the concentration of NADP+ was less than 2 mM, TCDCA conversion and TUDCA yield gradually increased with NADP+. The highest TCDCA conversion was about 82%, and the highest TUDCA yield was about 58%. When the concentration of NADP+ was more than 2 mM, the changes were minimal for both TCDCA conversion and TUDCA yield. The effect of NADP+ concentration on TCDCA conversion and TUDCA yield may due to the reaction is reversible. The catalytic equilibrium can be regulated by the concentration of NADP+ to some extent.
3.3 Operation stability of catalysis
In order to examine the catalytic stability of co-immobilised 7α- and 7β-HSDH, both TCDCA and chicken bile were used as substrates for experiments. In six continuous batch reactions, the changes of TCDCA conversion and TUDCA yield were not significant using TCDCA as substrate. TCDCA conversion was more than 85% and TUDCA yield was about 50–60%. However, the catalysis of chicken bile solution was not as stable as TCDCA. After four continuous reactions, TCDCA conversion was lower than 40% and TUDCA yield was about 15%. In the fifth and sixth reactions, the TUDCA generated could not detected by HPLC-ELSD as described in Fig. 4b.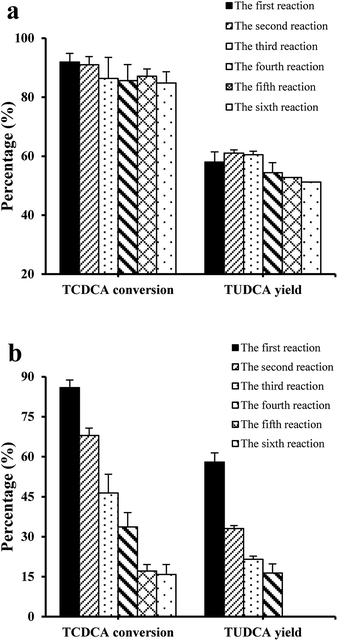 | ||
| Fig. 4 The effect of reaction times on the transformation of TCDCA in six continuous reactions, (a) TCDCA (8 mM) as substrate; (b) chicken bile with equal TCDCA (8 mM) as substrate. | ||
The operation stability of immobilised enzymes is dependent on many factors such as carrier, immobilisation methods and reaction medium, in addition to the structure of free enzymes. The projects to improve the stability of immobilised enzymes had been reviewed by Bommarius et al.24 and Stepankova et al.25 The number of recycling times of co-immobilised 7α- and 7β-HSDH for the catalysis of chicken bile solution was less than catalysis of TCDCA. It may be in relation to the complexity of the substrate chicken bile. Using continuous stirred tank membrane reactors, the stability of β-glucosidase immobilised on chitosan pellets was studied by Gallifuoco et al.26 Fructose did not decrease biocatalyst stability, while alcohol affected enzyme half-life from 2586 h at 3% (w/v) ethanol to 1378 h at 12% (w/v). The addition of terpenes to solution containing 10% (w/v) alcohol reduced the half-life by a further 10%. So further exploration was conducted in order to explain which composition affected the activity of enzymes.
3.4 The effect of complex substrate on activity of enzymes
The different activity of 7α-HSDH for the catalysis of TCDCA and chicken bile revealed that some compositions in complex substrate have negative effect on enzymes activity. Except bile acid, the other four main components of chicken bile are protein, amino acid, cholesterol and bilirubin. Due to the species diversity of protein and amino acid, we focused on the effect of cholesterol and bilirubin. From Fig. 5, we found that the activity of enzymes were significantly affected by bilirubin (p < 0.1). Inhibition rate of bilirubin was about 21% to free 7α-HSDH, and about 15% to immobilised 7α-HSDH. To free 7β-HSDH, the inhibition rate of bilirubin was about 25%, and about 19% to immobilised 7β-HSDH. The effect of cholesterol was not as significant as bilirubin, the inhibition rate was about 5–10%. Most of metal ions in chicken bile had no effect on the activity of 7α-HSDH, however, Cu2+ and Ca2+ had significant effect on the activity of 7α-HSDH, especially, Cu2+ to free 7α-HSDH. The inhibition rate of Cu2+ to free 7α-HSDH was about 29%, and about 20% to immobilised 7α-HSDH as Fig. 6a. The inhibition to 7β-HSDH even more. The inhibition rate of Cu2+ to free 7β-HSDH was about 30%, and about 24% to immobilised 7β-HSDH (Fig. 6b). The different inhibition of metal ions to free enzymes and immobilised enzymes may attribute to the partly adsorption of modified chitosan carrier.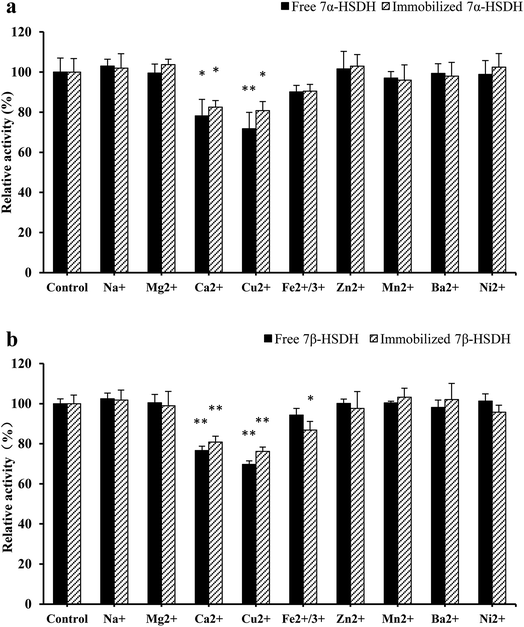 | ||
| Fig. 6 The effect of metal ions on the activity of immobilised 7α- and 7β-HSDH. The concentration of each metal ions was determined according to the content of chicken bile when the concentration of TCDCA defined as 8 mM. The study of Li et al. was referred.27 TCDCA and TUDCA were used as the substrates for the activity detection of 7α- and 7β-HSDH as (a) and (b), respectively. *p < 0.05, **p < 0.01. | ||
The effect of bilirubin on 7α-HSDH activity may attribute to the binding of bilirubin with protein. Bilirubin can be combined with a variety of proteins according to references. Orlov et al. reported that bilirubin can bind to different binding sites of bovine serum albumins and human serum albumins.28 Furthermore, bilirubin can also bind to hepatic Y-Protein and Z-Protein.29 So it was no wonder that 7α- and 7β-HSDH were partly inhibited by bilirubin.
7α- and 7β-HSDH are both belong to short-chain dehydrogenase/reductase (SDR). Some metal ions have an impact on the activity of SDR enzymes, such as, Cu2+, Ca2+ and Ni2+. Chen et al. reported a novel 3α-HSDH from Pseudomonas aeruginosa, which inhibited by Cu2+, Ca2+ and Co2+. The strongest inhibiting ion Cu2+ decreased the enzyme activity to only 67% of the control. 3α-HSDH can be activated by Zn2+, K+, Mn2+, Fe3+ and Na+, among which Zn2+ exhibited the strongest activating effect by increasing the enzyme activity 1.75-fold. Some other metal ions Mg2+, Ni2+ and Al3+ have no effect on 3α-HSDH.30 ChKRED20 one SDR, was cloned from Chryseobacterium sp. CA49 by Tang.31 The experiments demonstrated that Ag+, Cu2+, or Fe3+ negatively impacted the activity of ChKRED20. Some of metal ions showed clear stimulative effect, such as Ni2+, Mg2+, Zn2+, K+, Co2+ and Rb+.
3.5 Exploration of inhibition mechanism for 7α- and 7β-HSDH
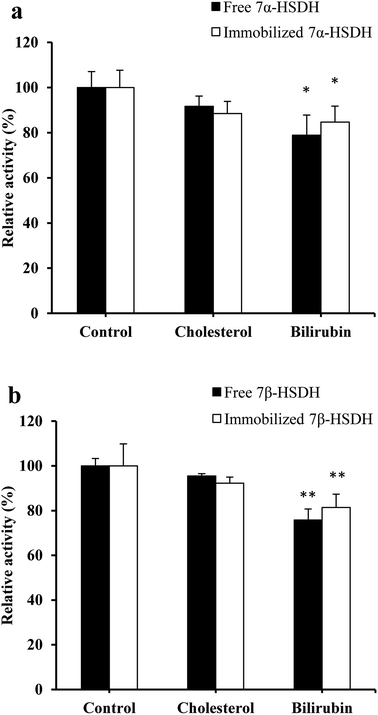
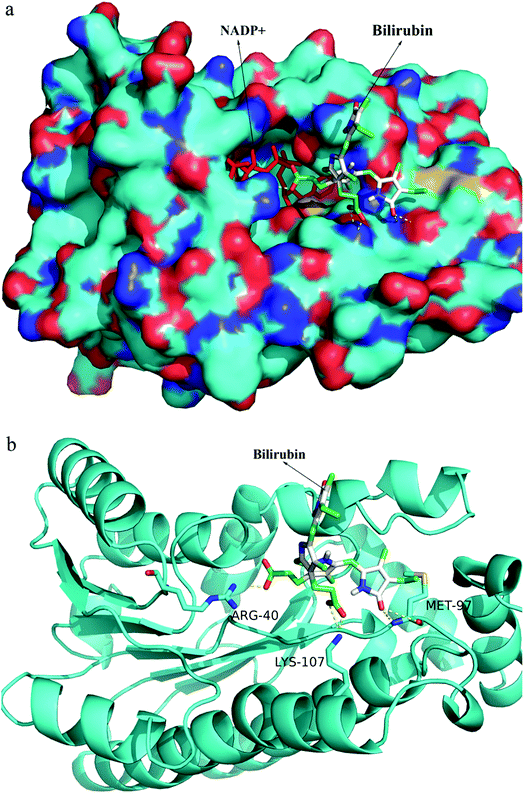
After random docking of bilirubin to 7α- or 7β-HSDH, the combination with high probability was analyzed as Fig. 7. Bilirubin was most probably combined with Arg (16), Arg (20) and Arg (194) of 7α-HSDH as Fig. 6b. Arg (16) and Arg (194) are the important residues of catalytic process which had been demonstrated by Lou et al.22 It was no wonder that the activity of 7α-HSDH was effected by the combination of bilirubin. For 7β-HSDH, the binding site of bilirubin to 7β-HSDH were Arg (40), Met (97) and Lys (107) which were located at the exit of substrate and product (Fig. 8). The activity decrease of 7β-HSDH may attributed to the access blocking of substrates and products.In this paper, inhibition mechanism for 7α- and 7β-HSDH was simply studied by docking. The docking results were ranked by binding free energy. Further detailed studies are under way. It is well known that both ligands and receptors are flexible, molecular dynamics simulations is a good way for conformational changes and virtual screening studies. In order to better investigate the mechanism of human butyryl cholinesterase inhibited by cyclosarin and sarin, docking and mixed quantum and molecular mechanics combined with principal components analysis were performed to evaluate the docking results.32 Theoretical strategy combining docking (MM), chemometric analysis and QM calculations was employed by de Lima et al. to check out the association and kinetic reactivation coefficients associated to oximes. The docking results were selected by means of the principal components analysis and submitted to QM calculations.33
3.6 The analysis of product composition
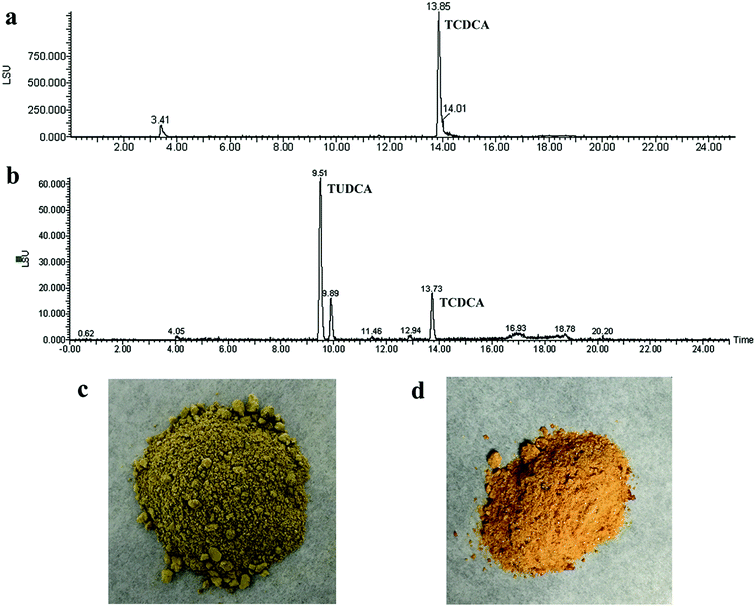
The composition of reaction product was analysed by HPLC-ELSD. The peak positions of TCDCA (about 13.8 min) and TUDCA (about 9.5 min) were presented in Fig. 9a and b. Most of TCDCA in chicken bile was transformed and high-value TUDCA generated successfully. The analysis of bile acid in reaction product showed that the proportion of TUDCA was as high as 33.16% and attained to 57.21% to the total bile acid in product (as Table 2). The nature chicken bile is dark green as Fig. 9c, after transformation by co-immobilised 7α- and 7β-HSDH, the color of sample changed into reddish-brown as Fig. 9d. This result may be attributed to the generation of TUDCA and partly adsorption of bilirubin.| Content (%) | Chicken bile | Reaction product |
|---|---|---|
| a TUDCA: tauroursodeoxycholic acid. TCDCA: taurochenodeoxycholic acid. The content of bile acid in chicken bile and reaction product were detected by HPLC-ELSD. Protein, amino acids, cholesterol and bilirubin content are quantified by test kits. “—”stands for this composition detected but not quantified. | ||
| TUDCA | — | 33.61 ± 0.17 |
| TCDCA | 42.57 ± 0.93 | 12.40 ± 0.10 |
| Total bile acid | 42.57 ± 0.93 | 57.21 ± 0.44 |
| Protein | 7.76 ± 0.14 | 3.24 ± 0.16 |
| Amino acids | 11.00 ± 0.57 | 19.20 ± 3.33 |
| Cholesterol | 0.50 ± 0.07 | 0.19 ± 0.01 |
| Bilirubin | 0.46 ± 0.02 | 0.25 ± 0.05 |
According to the literature, 7α- and 7β-HSDH from Xanthomonas maltophilia has been purified and immobilised on resin by Pedrini et al. and used for UDCA preparation.18 It took 7 days to obtain 75% UDCA through epimerization based on immobilised 7α- and 7β-HSDH. Zheng et al. adopted a two-step reaction strategy for the synthesis of UDCA from CDCA,19 where 7α-HSDH from Clostridium absonum, 7β-HSDH from Ruminococcus torques. 7α-HSDH, lactate dehydrogenase (LDH), 7β-HSDH and glucose dehydrogenase (GDH) were co-immobilised onto an epoxy-functionalized resin by Zheng et al.34 Continuous production of UDCA was achieved with the catalysis of two cascade reactors. Lasted for at least 12 h in the packed bed reactors, the yield of UDCA reached nearly 100%. In this study, the TUDCA was prepared within 4 h with the catalysis of co-immobilised 7α- and 7β-HSDH using chicken bile as substrate. The TUDCA yield was approximately 62% and the content of TUDCA in reaction product was about 33.16%. This research provided a novel approach for the exploitation of waste chicken bile, which complies with green chemistry principles.
4. Conclusions
In conclusion, 7α- and 7β-HSDH from Clostridium absonum were co-immobilised on glutaraldehyde modified chitosan microspheres. We are sure high-value TUDCA was the first time prepared using waste chicken bile as substrate. The catalytic reaction reached equilibrium within 4 h. TUDCA yield was approximately 62% after equilibrium and the content of TUDCA in reaction product was about 33.16%. TUDCA was prepared high-performance with the catalysis of recyclable biocatalyst. The effect of complex substrate on the activity of enzymes demonstrated that 7α-HSDH is significantly inhibited by bilirubin, Cu2+ and Ca2+. The inhibition mechanism of bilirubin was explored by freedom docking of moleculars. Further research on chicken bile treatment and bioreactor design will be gradual promoted.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by Supported by Visiting Scholar Foundation of Key Laboratory of Biorheological Science and Technology (Chongqing University), Ministry of Education (CQKLBST-2018-018), National Science and Technology Major Projects for “Major New Drugs Innovation and Development” (2014ZX09301306-007).References
- J. M. Ridlon, D.-J. Kang and P. B. Hylemon, J. Lipid Res., 2006, 47, 241–259 CrossRef CAS PubMed.
- G. Alpini, L. Baiocchi, S. Glaser, Y. Ueno, M. Marzioni, H. Francis, J. L. Phinizy, M. Angelico and G. LeSage, Hepatology, 2002, 35, 1041–1052 CrossRef CAS PubMed.
- L. Baiocchi, G. Tisone, M. A. Russo, C. Longhi, G. Palmieri, A. Volpe, C. Almerighi, C. Telesca, M. Carbone, L. Toti, F. De Leonardis and M. Angelico, Transplant Int., 2008, 21, 792–800 CrossRef CAS PubMed.
- W. Wang, J. Zhao, W. Gui, D. Sun, H. Dai, L. Xiao, H. Chu, F. Du, Q. Zhu and B. Schnabl, Br. J. Pharmacol., 2018, 175, 469–484 CrossRef CAS PubMed.
- J. Hu, M. Xu, J. Yuan, B. Li, S. Entenman, H. Yu and Q. Y. Zheng, Neuroscience, 2016, 316, 311–320 CrossRef CAS PubMed.
- L. Fernandez-Sanchez, P. Lax, A. Noailles, A. Angulo, V. Maneu and N. Cuenca, Molecules, 2015, 20, 13875–13893 CrossRef CAS PubMed.
- A. E. Elia, S. Lalli, M. R. Monsurro, A. Sagnelli, A. C. Taiello, B. Reggiori, V. La Bella, G. Tedeschi and A. Albanese, Eur. J. Neurol., 2016, 23, 45–52 CrossRef CAS PubMed.
- L. M. Cortez, J. Campeau, G. Norman, M. Kalayil, J. Van der Merwe, D. McKenzie and V. L. Sim, J. Virol., 2015, 89, 7660–7672 CrossRef CAS PubMed.
- A. I. Rosa, S. Duarte-Silva, A. Silva-Fernandes, M. J. Nunes, A. N. Carvalho, E. Rodrigues, M. J. Gama, R. Cmp, P. Maciel and M. Castro-Caldas, Mol. Neurobiol., 2018, 1–17 Search PubMed.
- J. G. Cho, J. H. Lee, S. H. Hong, H. N. Lee, C. M. Kim, S. Y. Kim, K. J. Yoon, B. J. Oh, J. H. Kim, S. Y. Jung, T. Asahara, S. M. Kwon and S. G. Park, Stem Cells, 2015, 33, 792–805 CrossRef CAS PubMed.
- Q. Y. Guo, Q. D. Shi, H. X. Li, J. L. Liu, S. F. Wu, H. Z. Sun and B. Zhou, Int. J. Endocrinol., 2015, 687938, DOI:10.1155/2015/687938.
- Y. Y. Lee, S. H. Hong, Y. J. Lee, S. S. Chung, H. S. Jung, S. G. Park and K. S. Park, Biochem. Biophys. Res. Commun., 2010, 397, 735–739 CrossRef CAS PubMed.
- J. A. Torres, F. G. E. Nogueira, M. C. Silva, J. H. Lopes, T. S. Tavares, T. C. Ramalho and A. D. Corrêa, RSC Adv., 2017, 7, 16460–16466 RSC.
- Q. Z. Ji, J. Tan, L. C. Zhu, D. S. Lou and B. C. Wang, Biochem. Eng. J., 2016, 105, 1–9 CrossRef CAS.
- E. E. Ferrandi, G. M. Bertolesi, F. Polentini, A. Negri, S. Riva and D. Monti, Appl. Microbiol. Biotechnol., 2012, 95, 1221–1233 CrossRef CAS PubMed.
- T. Akao, T. Akao and K. Kobashi, J. Biochem., 1987, 102, 613–619 CrossRef CAS PubMed.
- M. J. Bennett, S. L. McKnight and J. P. Coleman, Curr. Microbiol., 2003, 47, 475–484 CrossRef CAS PubMed.
- P. Pedrini, E. Andreotti, A. Guerrini, M. Dean, G. Fantin and P. P. Giovannini, Steroids, 2006, 71, 189–198 CrossRef CAS PubMed.
- M. M. Zheng, R. F. Wang, C. X. Li and J. H. Xu, Process Biochem., 2015, 50, 598–604 CrossRef CAS.
- S. A. Cetinus and H. N. Oztop, Enzyme Microb. Technol., 2003, 32, 889–894 CrossRef.
- Y. Zhao, L. X. Zan and W. J. Sun, Chin. J. Pharm. Anal., 2006, 26, 127–129 CAS.
- D. S. Lou, B. C. Wang, J. Tan, L. C. Zhu, X. X. Cen, Q. Z. Ji and Y. Wang, Sci. Rep., 2016, 6, 22885 CrossRef CAS PubMed.
- R. Wang, J. Wu, D. K. Jin, Y. Chen, Z. Lv, Q. Chen, Q. Miao, X. Huo and F. Wang, Acta Crystallogr., 2017, 73, 246–252 CAS.
- A. S. Bommarius and M. F. Paye, Chem. Soc. Rev., 2013, 42, 6534–6565 RSC.
- V. Stepankova, S. Bidmanova, T. Koudelakova, Z. Prokop, R. Chaloupkova and J. Damborsky, ACS Catal., 2013, 3, 2823–2836 CrossRef CAS.
- A. Gallifuoco, F. Alfani, M. Cantarella, G. Spagna and P. G. Pifferi, Process Biochem., 1999, 35, 179–185 CrossRef CAS.
- F. Li, B. C. Wang and L. C. Zhu, Chin. Tradit. Pat. Med., 2015, 37, 2555–2558 CAS.
- S. Orlov, I. Goncharova and M. Urbanova, Spectrochim. Acta, Part A, 2014, 126, 68–75 CrossRef CAS PubMed.
- D. R. Davis and R. A. Yeary, Proc. Soc. Exp. Biol. Med., 1975, 148, 9–13 CrossRef CAS PubMed.
- J. M. Chen, X. F. Gao, L. Hong, L. T. Ma and Y. S. Li, Protein Expression Purif., 2015, 115, 102–108 CrossRef CAS PubMed.
- T. X. Tang, Y. Liu and Z. L. Wu, J. Mol. Catal. B: Enzym., 2014, 105, 82–88 CrossRef CAS.
- T. C. Ramalho, E. F. F. D. Cunha, A. A. D. Castro, A. F. Pereira and W. E. A. D. Lima, Lett. Drug Des. Discovery, 2016, 13, 360–371 CrossRef.
- W. E. A. D. Lima, A. Francisco, E. F. F. D. Cunha, Z. Radic, P. Taylor, T. C. C. França and T. C. Ramalho, J. Biomol. Struct. Dyn., 2017, 35, 1272–1282 CrossRef PubMed.
- M. M. Zheng, F. F. Chen, H. Li, C. X. Li and J. H. Xu, ChemBioChem, 2017, 19, 347–353 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra06798h |
| This journal is © The Royal Society of Chemistry 2018 |