Open Access Article
Open Access ArticleHomoduplex to i-motif structural switch exhibited by a cytosine rich strand of the MYH7 heavy chain β gene promoter at physiological pH†
Anju Singh and
Shrikant Kukreti
and
Shrikant Kukreti *
*
Nucleic Acids Research Lab, Department of Chemistry, University of Delhi, Delhi-110007, India. E-mail: skukreti@chemistry.du.ac.in
First published on 4th October 2018
Abstract
Genomic locations such as promoter, exon, intron, telomeric and non-telomeric regions are rich in GC-rich sequences with the potential to form G- and C-tetraplexes on both strands independently. Herein, we employed biophysical and biochemical methods to study a 34-mer C-rich DNA sequence of the myosin heavy chain β gene (MYH7β) promoter, namely HM34C for humans and the rabbit counterpart, RM34C, which differs from HM34C at three positions (three bases). Circular dichroism (CD), UV-thermal denaturation and native gel electrophoresis studies demonstrated that both the C-rich promoter segments form C-tetraplex (i-motif) structures. The CD studies revealed that HM34C forms the i-motif structure at acidic pH (5.2) in the presence of 0.1 M NaCl but remains unstructured at physiological pH. Interestingly, RM34C can form the stable i-motif structure in acidic as well as physiological pH. A shift in the positive peak from 280 nm to 275 nm with the increase in temperature from 4 °C to 30 °C was observed in temperature-dependent CD studies. UV-melting studies showed a biphasic transition for RM34C, indicating the existence of two structural species at neutral pH. In view of these findings we suggest that at physiological pH, the RM34C sequence exists in equilibrium between two structural motifs, i.e. the i-motif and homoduplex structure. This study may add to the understanding of the i-motif/homoduplex in equilibrium in physiological environments.
Introduction
Nucleic acids can adopt a variety of secondary structures depending on their sequence and their environment (solvent, ionic conditions, pH etc.). Apart from the twisted ladder-like double helical structure, DNA can adopt various non-B-DNA structures such as G-quadruplex, i-motif, H-DNA and cruciforms. Under physiological conditions, these non-B-DNA structures can prevail over DNA duplexes to regulate DNA processing or gene expression.1 It is well documented that consecutive runs of guanine, by virtue of self-association properties, have the tendency to fold into the G-quadruplex structure. With the formation of a G-quadruplex, the complementary C-rich strand becomes single-stranded, which should facilitate the folding into the i-motif form. For several G-rich sequences, including c-MYC, BCL-2, Rb, VEGF-A, RET, KRAS and c-Kit, the complementary cytosine-rich strand has been shown to form the i-motif structure. Initially, it was proposed that polycytidylic acid would form parallel duplexes at low pH.2 The first detailed structure of the cytosine-rich sequence, d(TCCCCC), forming a tetrameric DNA structure with hemiprotonated C–C+ base pairs, called the i-motif was reported in 1993.3,4 The two parallel stranded C·C+ (one cytosine must be hemiprotonated at the N3 position) base-paired duplexes intercalate into each other in an anti-parallel orientation. i-motif structures are the only known nucleic acid structures that involve base intercalation. It is interesting to note that C-rich sequences with the potential to form i-motif structures are frequently found in eukaryotic genomes.5 The existence of the i-motif demonstrates most dramatically the wide range of structures that DNA can form. The three known topologies of i-motif structures are usually generated involving one, two or four strands. The most interesting differences are in the end effects observed in the loops of the folded structures and at the ends of the C-tetrameric structure.6A short DNA oligomer d(TCC) having just two cytosines form the tetrameric i-motif. It is stabilized by a network of C–H⋯O hydrogen bonding between neighboring deoxyriboses. Other stabilizing factors in i-motif that have been proposed in addition to hydrogen bonding between sugar residues are interactions between stacked C+·C base pairs, stereoelectronic effects involving the sugar moiety, hydration, and hemiprotonated C+·C pairs. However, very few reports are known about the formation of i-motif structures at neutral pH.7–10 Mergny and co-workers have shown that a stretch of cytidines can form the i-motif at slightly acidic or even neutral pH.7 It is reported that the DNA fragments of fragile X chromosomes can fold into i-motif structures at neutral pH.8 Interestingly, Waller et al. have reported a silver cation induced folding of a C-rich sequence into a stable i-motif structure.11 Zhou et al. have demonstrated that the i-motif structure can also be formed at neutral and slightly alkaline pH at 4 °C, using CD (circular dichroism), UV absorption spectroscopy and PAGE (polyacrylamide gel electrophoresis).12
The present study is an attempt to see the possibility of the formation of i-motif structures by a genomic sequence at physiological pH and salt conditions. Here, we probed a C-rich sequence located in the promoter region of the Myosin Heavy Chain β gene (MYH7β), for its i-motif-forming ability at neutral pH. These sequences always co-exist with the G-rich counterpart in a gene. The biophysical studies were performed in varied solution conditions. Their structure, molecularity and stability were determined by non-denaturating gel electrophoresis (PAGE), UV-thermal denaturation and circular dichroism (CD) studies. This study may add to the understanding of the i-motif/homoduplex in equilibrium under physiological solution environments.
Materials and methods
All the DNA sequences synthesized at 1 μM scale were procured from Helix Biosciences (Delhi, India) in PAGE purified lyophilized form. Oligonucleotides were stored at −20 °C and were used in the study without further purification. The concentrations of the DNA oligonucleotides were determined spectrophotometrically by using extinction coefficient (ε) calculated by nearest neighbour approximation and measuring the absorbance at 260 nm at elevated temperature (95 °C), following the method described in literature.13–15 The stock solutions of all the DNA oligonucleotides including control size markers were prepared by directly dissolving the lyophilized powder in MilliQ water. All other fine chemicals were of analytical grade. The DNA sequences used in this study are summarized in Table 1.| S. no. | Name | Oligonucleotide sequences | Size | ε (M−1 cm−1) |
|---|---|---|---|---|
| 1 | HM34C | 5′-AGGGCACCCCCACCCCCAACGGCACCCAGTCCCT-3′ | 34 | 302, 100 |
| 2 | RM34C | 5′-AGGGCGCCCCCACCCCCACCTGCACCCAGTCCCT-3′ | 34 | 291, 800 |
| S. no. | Name | Control size marker | Size | ε (M−1 cm−1) |
|---|---|---|---|---|
| 1 | PAL12 | 5′-CTTGAGCTCAAG-3′ | 12 | 113, 700 |
| 2 | PAL24 | 5′-CTTGAGCTTGAGCTCAAGCTCAAG-3′ | 24 | 226, 100 |
| 3 | M35 | 5′-GACTGACTTAAGCGCATAGCTAGCTAGCTCGATAGCTGA-3′ | 342, 600 | |
| 4 | M35C | 5′-TCAGCTATCGAGCTAGCTAGCTATGCGCTTAAGTCAGTC-3′ | 35 | 333, 100 |
Non-denaturating gel electrophoresis
The commercially procured oligonucleotides were subjected to denaturating gel electrophoresis to check the purity, by running them on 20% PAGE using 7 M urea. After being assured about the purity, non-denaturating gel electrophoresis was carried out on 15% polyacrylamide gels (19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 acrylamide/bisacrylamide) to study their structural status. DNA samples of 15 μM concentration were prepared in 20 mM sodium cacodylate buffer (pH 5.7 & 7.4), 100 mM NaCl and 0.1 mM EDTA. The final volumes of the samples in the buffer (20 μL) were heat treated at 95 °C for 5 min and then slowly cooled to room temperature and were incubated overnight at 4 °C. We used DNA oligonucleotides PAL12 (5′-CTTGAGCTCAAG-3′), PAL24 (′5-CTTGAGCTTGAGCTCAAGCTCAAG-3′), M35 (5′-GACTGACTTAAGCGCATAGCTAGCTAGCTCGATAGCTGA-3′) and M35C (5′-TCAGCTATCGAGCTAGCTAGCTATGCGCTTAAGTCAGTC-3′) as control size markers. These deoxyoligonucleotides (control size markers) have frequently been used in the author's laboratory.16–19 PAL12 and PAL24 are palindromic DNA sequences that form 12-bp and 24-bp perfect duplexes in native conditions, whereas M35 and M35C are random markers complementary to each other. Samples were mixed with an aliquot of loading dye orange-G before loading on the gel. The gel was pre-equilibrated for one hour before loading the samples. After electrophoresis, the gel was stained with Stains-All solution and finally visualized under white light and images were taken by an Alphaimager™ 2200 (Alpha Infotech Corp.).
1 acrylamide/bisacrylamide) to study their structural status. DNA samples of 15 μM concentration were prepared in 20 mM sodium cacodylate buffer (pH 5.7 & 7.4), 100 mM NaCl and 0.1 mM EDTA. The final volumes of the samples in the buffer (20 μL) were heat treated at 95 °C for 5 min and then slowly cooled to room temperature and were incubated overnight at 4 °C. We used DNA oligonucleotides PAL12 (5′-CTTGAGCTCAAG-3′), PAL24 (′5-CTTGAGCTTGAGCTCAAGCTCAAG-3′), M35 (5′-GACTGACTTAAGCGCATAGCTAGCTAGCTCGATAGCTGA-3′) and M35C (5′-TCAGCTATCGAGCTAGCTAGCTATGCGCTTAAGTCAGTC-3′) as control size markers. These deoxyoligonucleotides (control size markers) have frequently been used in the author's laboratory.16–19 PAL12 and PAL24 are palindromic DNA sequences that form 12-bp and 24-bp perfect duplexes in native conditions, whereas M35 and M35C are random markers complementary to each other. Samples were mixed with an aliquot of loading dye orange-G before loading on the gel. The gel was pre-equilibrated for one hour before loading the samples. After electrophoresis, the gel was stained with Stains-All solution and finally visualized under white light and images were taken by an Alphaimager™ 2200 (Alpha Infotech Corp.).
Circular dichroism spectroscopy
The CD spectra of oligonucleotides were recorded on a CD spectropolarimeter, JASCO J-815, at room temperature (24 °C) using a quartz cuvette of 10 mm path length. Temperature-dependent CD experiments were performed at various temperatures. The cell holder was thermostated by a JASCO PTC-348 temperature controller and to avoid water condensation in the cuvette chamber, it was flushed with a constant purging of N2 gas. The CD spectra were collected from 330 to 200 nm at wavelength steps of 1 nm, and the final reported spectra correspond to the average of three scans. The scans of buffer alone were subtracted from the average scans for each sample for baseline correction. Data were collected in units of millidegrees and were normalized to total DNA strand concentrations. Samples were prepared in 20 mM sodium cacodylate buffer (pH 5.7 & 7.4) containing 100 mM NaCl and 0.1 mM EDTA. All samples used for CD were heat treated to 95 °C and slowly cooled at room temperature followed by overnight incubation at 4 °C to ensure that the C-rich sequences folded to form i-motif structures or other stable higher secondary structures. It is a well-documented fact that G-quadruplex and C-tetraplex (i-motif) formation occur with a slow kinetics. We generally heat the samples up to 95 °C and then allow slow cooling at room temperature. As a result, all hydrogen bonds break during the nucleation process and then with slow cooling the proper renaturation/reannealing takes place, facilitating the formation of a thermodynamically stable conformation/structure. This step is required specifically for G/C rich oligos before every experiment (PAGE, Tm, CD etc.) so that unwanted/irregular secondary structure formation due to random hydrogen bonding can be avoided, along with facilitation of the formation of the stable secondary structure.UV thermal denaturation
To elucidate the thermal stabilities of the secondary structures adopted by C-rich sequences (HM34C and RM34C), UV thermal melting was carried out in 20 mM sodium cacodylate buffer (pH 5.7 & 7.4) containing 100 mM NaCl. Melting profiles were measured at 265 and 295 nm by using Shimadzu UV spectrophotometer 1800 equipped with a temperature controller. The stoppered quartz cuvette of 1 cm optical path length with 110 μL was used for the melting experiments. The temperature was scanned at a heating rate of 0.5 °C min−1. The analysis of the melting curves for the recorded melting profile was carried out using the first derivative.Results and discussion
This study reports on the biophysical and biochemical investigations of C-rich DNA oligonucleotide sequences of promoter locations of the Myosin Heavy Chain β gene (MYH7) extracted from human (HM34C) and rabbit (RM34C) by using NCBI BLAST (https://blast.ncbi.nlm.nih.gov). The HM34C (AGGGCAC5AC5AACGGCACCCAGTCCCT) sequence differs from RM34C (AGGGCGC5AC5ACCTGCACCCAGTCCCT) at three positions, viz. 6, 19, 21. These C-rich sequences were characterized under varied pH and temperature conditions, generating distinct sequence-dependent topologies including the i-motif and homoduplex structure at neutral pH (7.4).Polyacrylamide gel electrophoresis (PAGE)
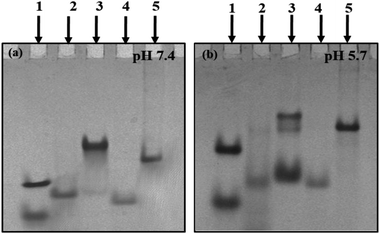
Structural status of C-rich sequences at neutral pH (7.4). Non-denaturating gel electrophoresis is a sensitive and informative technique used to infer the molecularity/strandedness and folding status of oligonucleotides. This technique has been efficiently used to determine and differentiate various multistranded structures. After checking the purity of C-rich sequences, further native gel electrophoresis was performed to establish their structural status at varied solution conditions. Next, 15% non-denaturating PAGE was carried out in 20 mM sodium cacodylate buffer (pH 7.4 & 5.7) containing 0.1 M NaCl and 0.1 mM EDTA. Fig. 1(a) shows the electrophoretogram of HM34C and RM34C at neutral pH (7.4). PAL12 (12-mer duplex-Fig. 1(a)); lower band, lane (1), PAL24 (24-mer duplex-Fig. 1(a)); upper band, lane (1) are self-complementary palindromic duplexes, running as 24-base and 48-base oligomers in the native PAGE. Also, a random 35-mer single strand, size marker M35 (Fig. 1(a), lane (4) and a duplex of M35 + M35C (70 nt-Fig. 1(a)-lane (5)) were used as control oligomer marker. HM34C showed a single band (lane 2) migrating equivalent to M35, which indicates the unstructured status of C-rich strand. In contrast, RM34C (lane 3) exhibited two distinct bands, where the upper band was intense in comparison to the lower band. When compared to the gel markers, the mobility of the upper band of RM34C was found to be retarded in comparison to PAL24 (24-mer duplex) and the duplex of M35 + M35C (lane 5). Thus, the upper band of RM34C can be assigned to a bimolecular structure. The lower faint band migration corresponding to M35 can be assigned as unstructured RM34C. It is worth mentioning again here that RM34C and HM34C differ by three nucleotides. It was intriguing to observe that out of the two C-rich sequences of identical length (34-mer), the RM34C sequence displayed retarded gel mobility at neutral pH 7.4 (Fig. 1(a). This is only possible if the sequence experiences C-protonation at pH 7.4 and is involved in higher order structuration. We demonstrate here that RM34C adopts a bimolecular structure at neutral pH.
Structural status of C-rich sequences at acidic pH (5.7). As evident from the literature, cytosine protonation is a prerequisite for the formation of the i-motif, prone to form at low or acidic pH. On the basis of the knowledge from the existing literature, gel studies were carried out at acidic pH 5.7 to facilitate cytosine protonation. Fig. 1(b) illustrates the mobility pattern of HM34C and RM34C in 20 mM sodium cacodylate buffer (pH 5.7) containing 0.1 M NaCl and 0.1 mM EDTA. Lanes 1, 4 and 5 were comprised of control size markers PAL12, PAL24 (lane 1), M35 (lane 4) and M35 + M35C (lane 5), respectively. Lane 3 showed two bands for HM34C, reflecting the presence of two structural species in solution. Based on their mobility pattern with respect to the size markers, the upper faint band (HM34C) migrates with the duplex of M35, indicating the bimolecular nature of HM34C. On considering the strand stoichiometry, it was thought to be the dimeric C-tetraplex (i-motif). The fast migrating band of HM34C can be of a folded unimolecular structure, as it was seen to migrate faster than M35. A lower intense band indicates a higher population of unimolecular species. In contrast, being of same length and sequence (except differing in three bases), RM34C (lane 3) exhibited three bands reflecting three distinct structural species in solution. The lower band (lane 3) migrated slower than M35 but faster than PAL24, and could be assigned to either unimolecular species or an unstructured single strand, while the middle band (lane 3) moved equivalent to the M35 + M35C duplex (lane 6) and could be the folded bimolecular structure involving two strands of RM34C, generating two interdigitated parallel duplexes (involving C:CH+). The upper band, being intense, indicates that the bimolecular species is the dominant structure at low pH. The uppermost band, when compared with the control size marker M35 + M35C, is shown to migrate slightly slower than the M35 duplex. Here, the possibility of formation of either the homoduplex or i-motif cannot be ruled out, as the same was found as the predominant species at physiological pH (Fig. 1(a), lane 3). It is, therefore, speculated that the uppermost intense band of bimolecular species (Fig. 1(b), lane 3), is structurally different from the middle band.
Further, to check the stability of the duplex and i-motif formed, the native gel was also run at 37 °C at varied pH (pH 5.7 and 7.4) (Fig. S2(a & b)†). The gel image showed the identical mobility pattern at 37 °C, as was obtained at 4 °C (Fig. 1(a and b)). The only difference observed was in the status of both the sequences (HM34C and RM34C) at pH 5.7. Here, at 37 °C, only a single intense band was observed, while the lower bands disappeared. However, HM34C (supplementary Fig. 2(a); lane 2) displayed a very intense band at 37 °C, indicating that only one structural species is stable at this temperature, in contrast to the 4 °C gel (Fig. 1(b); lane 2) where two bands were observed. The mobility patterns of HM34C and RM34C oligomers at physiological pH (7.4) were found to be almost similar at both the temperature values (4 °C and 37 °C).
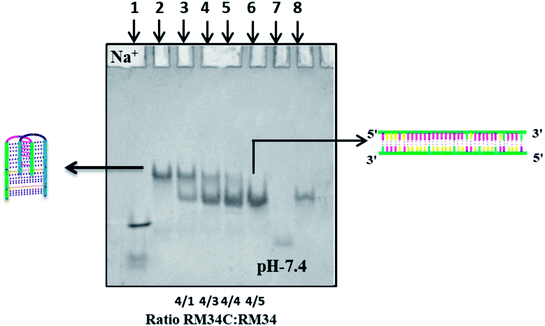
Structural status of RM34C in the presence of its G-counterpart RM34 at neutral pH (7.4). After examining the electrophoretic pattern of HM34C and RM34C at acidic as well as physiological pH, to further establish the structural status of RM34C, we carried out a control gel experiment. To see whether RM34C exists as a self-associated structured species, which might further hinder its duplex forming ability with the complementary strand, it was titrated with increasing concentrations (50 picomoles to 250 picomoles) of G-rich RM34 [5′-TC3GCG5TG5TGGACGTGGGTCAG3A-3′] in (Fig. 2).
Fig. 2 displays the gel image of RM34C alone and its mixture with G-rich RM34 at various ratios in 20 mM sodium cacodylate buffer (pH 7.4), 0.1 M Na+ and 0.1 mM EDTA. On analysis, RM34C was found to exhibit its identical structural status as discussed earlier in gel, Fig. 1(a) (lane 3), where it migrated as a retarded band to the duplex (M35 + M35C size marker), clearly indicating self-structuration of RM34C at neutral pH. Lane 3 → 5 displays two bands reflecting the presence of two structural species in solution. The lower band migration, equivalent to the M35 + M35C duplex, corresponds to the duplex species of RM34C + RM34, and the upper retarded band, migrating with the RM34C band, is interpreted as a bimolecular complex (Fig. 1(a)). It was observed that the intensity of the lower band increased, whereas the upper band disappeared with the addition of RM34 in increasing concentration. This clearly reflects the decrease in the population of self-associated bimolecular species of RM34C. Lane 6 showed a single intense band, migrating with the duplex of M35, consequently confirming the formation of a perfect Watson–Crick duplex between RM34C and RM34. The gel assay clearly demonstrates that due to the binding of the complementary RM34 to RM34C in varied ratios, the self-associated structure of RM34C is slowly destabilized resulting, in the formation of a perfect duplex with RM34. This control experiment further established the formation of a self-associated complex by RM34C, which is most likely a bimolecular i-motif species, at neutral pH.7 It is important to mention here that although both the structures formed by RM34C (Fig. 2, lane-2) and the Watson–Crick duplex of RM34C + RM34 (Fig. 2, lane-6) are biomolecular (involving two strands), the difference in their gel mobility comes from the fact that while RM34C + RM34 is a compact Watson–Crick duplex, the self-associated biomolecular complex RM34C could be a partially folded interdigitated structure of two parallel duplexes involving C·C+ pairs. Such a self-associated structure, known as the i-motif, may have a retarded mobility compared to a compact duplex.
Circular dichroism studies
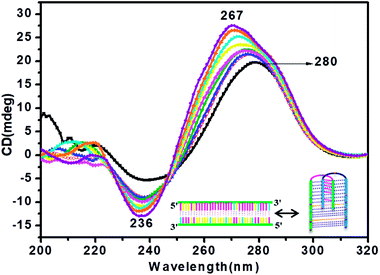
CD spectroscopy was carried out for secondary structure analysis and information on the conformational characteristics of the C-rich strands (HM34C and RM34C). The CD spectra of stacked C·C+ base pairs in an i-motif are characterized by a positive band at 285–290 nm and a negative band centred at 260–268 nm.20,21 The CD spectra of HM34C and RM34C in 20 mM sodium cacodylate (pH 7.4 & 5.7) containing 0.1 M NaCl and 0.1 mM EDTA are shown in Fig. 3(a and b). It is well documented that i-DNA is characterized by a typical positive CD signal at 286 nm followed by a negative peak at 265 nm, whereas B-DNA-like structures exhibit a positive peak at 275 nm with a negative peak centred around 254 nm.22 The presence of a large positive peak at acidic pH 5.7 towards longer wavelength near 286 nm, followed by the appearance of a negative band around ∼260 nm, is consistent with the well-documented features of stacked C·C+ base pairs in an i-motif structure for all said oligonucleotides. i-Motif formation is sensitive to pH conditions and it is clearly shown in Fig. 3 depicting the CD status of all C-rich sequences at two different pH values 7.4 and 5.7. The amplitude of the positive band at 286 nm drastically increased when the pH of the sample solution was decreased from 7.4 to 5.7. The highest CD amplitude at pH 5.7 corresponds to the formation of a maximum number of i-motif species (involving C·C+ pairing) compared to their population at physiological pH (7.4). In the case of HM34C at pH 7.4, the positive maxima shifted towards 275 nm with a concomitant decrease in amplitude, indicating the lack of secondary structure, possibly due to deprotonation, resulting in unstructured C-rich single strands, whereas a sharp peak at 275 nm with significant amplitude was observed for RM34C (Fig. 3(b)), reflecting the presence of some self-associated structural species.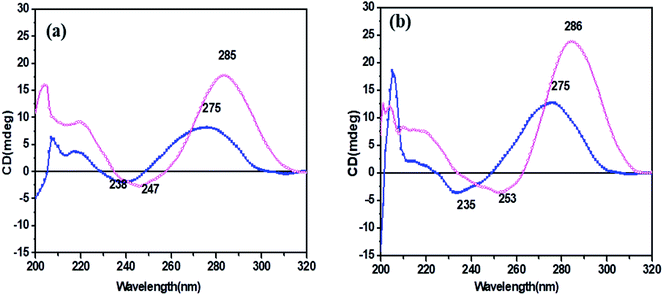 | ||
Fig. 3 CD spectra of (a) HM34C and (b) RM34C in 20 mM sodium cacodylate buffer, pH 7.4 (- -) and pH 5.7 (- -) and pH 5.7 (- -), containing 0.1 M NaCl and 0.1 mM EDTA. -), containing 0.1 M NaCl and 0.1 mM EDTA. | ||
We presumed a fair possibility of parallel-stranded homoduplex formation here by RM34C involving C·C+ base pairing. This possibility was further investigated by temperature dependent CD measurements to clarify the structural status of RM34C.
Temperature-dependent CD studies
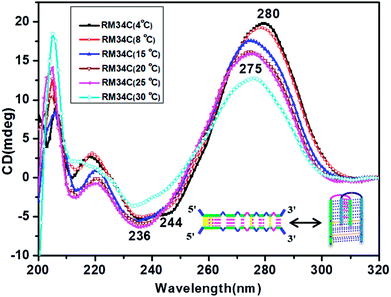
Fig. 4 depicts the temperature-dependent CD spectra of RM34C at neutral pH of 7.4. A positive peak at 280 nm with a negative band centred at 244 nm at 4 °C indicate the cytosine–cytosine+ base pairing. Thus, the CD demonstrates that RM34C can form i-motif structures under neutral conditions at 4 °C; however, the shift in the positive peak from 286 nm to 280 nm could be due to the decreased cytosine protonation at neutral pH in comparison to acidic pH. With a gradual increase in temperature from 4 °C to 30 °C, the 280 nm positive band is blue shifted to 275 nm, following a decrease in the ellipticity.At 30 °C, the CD spectrum displayed positive and negative bands at 275 nm and 240 nm, respectively. The amplitude of the CD spectrum does not fully correspond to any random coil/unstructured species but indicates the possibility of formation of the homoduplex of RM34C. The size of this most probable homoduplex structure was established by native gel (Fig. 1(a), lane 3). Further, the occurrence of two isoelliptic points at 244 nm and 265 nm entail the transition between two discrete conformations (i-motif ↔ homoduplex) and is a two-state process. Since we see a clear temperature dependence of CD signals, this undoubtedly reflects the transition of the i-motif to a homoduplex state at 30 °C. The stabilities of these proposed structures adopted by RM34C at neutral pH were further checked by thermal denaturation studies.
The structure of RM34C at physiological temperature, in the presence of its G-rich counterpart
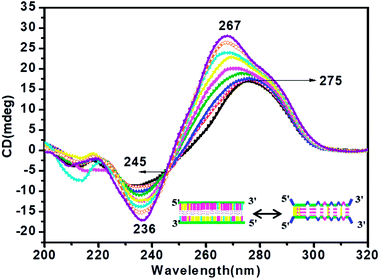
CD titration studies were carried out to gain more insight into the structural switching shown by RM34C at physiological pH, 7.4. Experimental solution conditions were kept similar to those used in the earlier section at 37 °C. This study involved CD measurements of gradual additions of G-rich RM34 to RM34C. The CD spectra manifested significant ellipticity, with a positive peak at 275 nm along with a negative peak centred at 236 nm, indicating the formation of a structure adopted by RM34C at physiological pH and temperature (Fig. 5). CD spectra displayed in Fig. 5, clearly show that on successive increments (0.25 μM to 3.5 μM) of RM34 to RM34C, positive CD peaks showed a further blue shift from 275 nm to 267 nm. This transition is due to the formation of a new structural species, possibly a duplex formed between fully complementary sequences of RM34 and RM34C. The positive peak at 267 nm can be clearly assigned to the Watson–Crick duplex species of RM34C.Interestingly, it is worth noting here that even when RM34 was added in excess to the RM34C, a minor population of structural species adopted by RM34C was also present (∼275 nm), along with the perfect duplex of RM34 + RM34C (∼267 nm) (Fig. 5). The spectra depicted two positive peaks at 267 nm and 275 nm with an isodichroic point centred at 245 nm. The isodichroic point indicates the existence of two structural species in equilibrium. The CD titration results are in good agreement with the gel results, where RM34C is demonstrated as a bimolecular structured species and migrates slower than the compact perfect duplex of M35 + M35C. On the basis of the CD titration results, we propose that RM34C exists as two structural species at physiological pH and temperature, i.e., a bimolecular species (with bulges), and as a perfect duplex in the presence of its G-counterpart.
The structure of RM34C at 4 °C in the presence of its G-rich counterpart
For information on the structure of RM34C at physiological pH (pH 7.4), CD titration studies were also carried out at 4 °C. CD spectra of RM34C in the presence of its G-rich counterpart RM34 are depicted in Fig. 6. CD spectra at 4 °C were apparent with a positive peak at 280 nm and a negative peak centred at 244 nm; as suggested earlier, this can be assigned to i-motif species. On the successive addition of complementary RM34 to RM34C, a prominent CD peak at 267 nm with increasing ellipticity and a blue shift was observed. There was also a minor positive peak at 280 nm. The presence of the isodichroic point at 245 nm again reflects the existence of two structural species in equilibrium. Based on CD titration and gel studies at 4 °C, it was established that RM34C might adopt the i-motif structure at low temperature, which turns into a duplex structure in the presence of its counterpart. This experiment confirms the structural transition of RM34C from i-motif to duplex species.UV-thermal denaturation studies
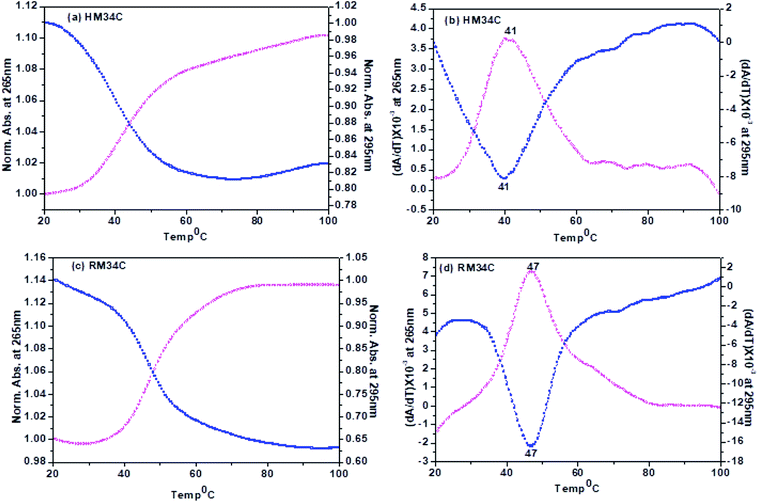
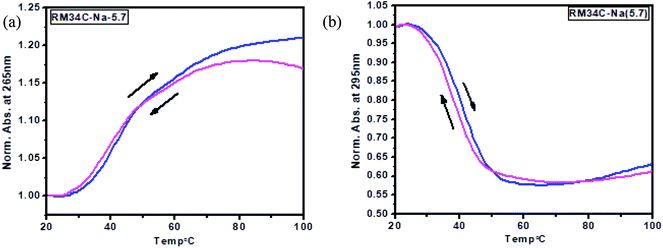
A nice cross-over was observed at 265 and 295 nm for all the Tm curves. Broad monophasic melting curves monitored at 265 and 295 nm were obtained for HM34C (Fig. 7(a)). The melting temperature calculated by the first derivative plot of both curves was found to be 41 °C (Fig. 7(b)). Thus, it can be hypothesized that various structural species exist in solution conditions, which have approximately the same stability in solution and they unfold as a single transition in melting studies. The Tm profiles were also obtained for RM34C at acidic pH. Fig. 7(c) displays the biphasic transition at both wavelengths for RM34C. Tm calculated by the first derivative plot shows a prominent peak at 47 °C with a slight hump at 65 °C (Fig. 7(d)). It is clear from the thermal melting studies that all the C-rich sequences used in the present study (HM34C and RM34C) had an inverted melting transition, indicating the unfolding of i-motif structures with a fair possibility of the co-existence of said structures having varied molecularity/strandedness. The data on the thermal denaturation and renaturation of RM34C was recorded at pH 5.7 at 265 nm as well as 295 nm and given in Fig. 8. Near-reversible superimposition of heating and cooling traces indicates that the transitions were kinetically reversible.
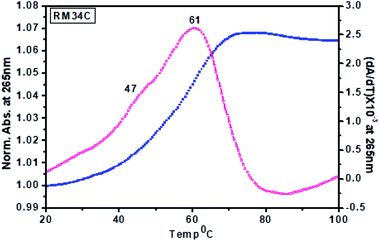 | ||
| Fig. 9 Thermal denaturation profiles of RM34C in 20 mM sodium cacodylate buffer (pH 7.4) containing 0.1 M NaCl and 0.1 mM EDTA, monitored at 265 nm. | ||
Tm calculated for the melting profile of RM34C was found to be 61 °C for the higher temperature transition, and 47 °C for the lower temperature transition (small hump on the curve). There is a fair possibility that uncoiling of the homoduplex species occurred at 61 °C, whereas disordering of the i-motif structure occurred at 47 °C. The small hump observed on the melting curve (Tm = 47 °C) reflects the minor population of the i-motif in comparison to the homoduplex species. The results of thermal melting studies are in good agreement with temperature dependence CD studies. An overlay of the thermal melting profile of RM34C at both pH values (5.7 and 7.4) is also provided in the ESI as Fig. S3.†
Proposed models for the co-existence of the biomolecular homoduplex and i-motif structures
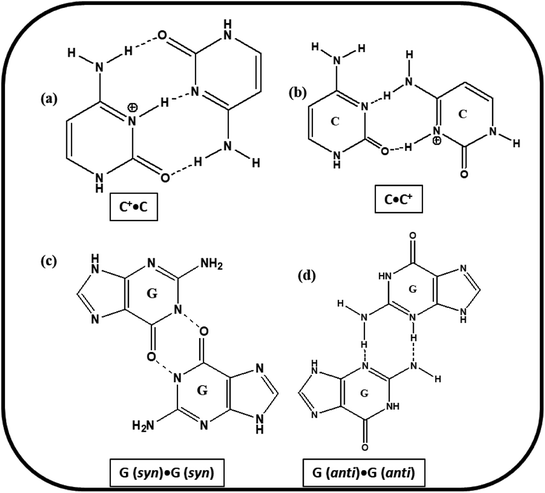
Based on the experimental results of PAGE, Tm and CD studies, the most possible structural models for the studied C-rich sequences are proposed. The cartoons of the proposed structures formed by HM34C and RM34C at acidic, as well as neutral pH are presented in Fig. 11. Two gel bands obtained for HM34C migrated as bimolecular and unimolecular species. The possibility of non-Watson–Crick hydrogen bonding providing stability to proposed structures cannot be ruled out. It is a well-documented fact that G·G mispairing contributes to greater thermal stability.24,25 The possible base pairing involved in i-motif structures adopted by HM34C and RM34C are schematically presented in Fig. 10.The formation of parallel bimolecular species (homoduplex) can be justified, as it can involve ten C·C+ and two G·G hydrogen bonds, whereas the parallel dimeric i-motif may contain five C·C+ pairs, which can further be stabilized by the possibility of the mismatch of two G·G, providing extra stability to the structure. It is important to remember that the CD spectra exhibited by C-rich sequences have also been observed by others for a number of oligonucleotides containing varied numbers of cytosine residues. These reported structures, stabilised by C·C+ pairs, include parallel26,27 and antiparallel duplexes,26 as well as parallel duplexes with looped out bases.28 Analogous models including hairpin structures have also been put forward based on experimental evidence.29 While the triple hydrogen-bonded C·C+ trans pair, concomitant with anti N-glycosidic conformations for all the cytosines, leads to the parallel alignment of the sugar-phosphate backbone chains, a different C·C+ pair with only two hydrogen bonds (Fig. 10) could provide an antiparallel duplex26 as well as a hairpin structure. However, NMR studies on a number of cytosine-rich oligonucleotides, d(C12), d(TC3), d(TC3T), d(C4TC4), d(T2C8T2), d(TC4), d(TC5), and d(C4) have revealed a novel and unusual four-stranded structure arising from the antiparallel intercalation of two C·C+ paired (triple hydrogen bond) parallel stranded duplexes, called i-DNA.3,30
Moreover, evidence has been provided that sequences containing four blocks of CCC of the human telomeric repeat adopt an intramolecular i-motif based structure.29,30 Here, the intramolecular i-motif is formed by C3 blocks of HM34C with two narrow loops harbouring CCA, AGT bases, and with a 9-bases-wide loop CCAACGGCA (Fig. 11(a)). Interestingly, RM34C adopts the i-motif structure in acidic as well as neutral or physiological pH (7.4). On the basis of the molecularity/strandedness of the structure through PAGE, the i-motif structure of RM34C was detected to be bimolecular. However, at neutral pH, the possibility of homoduplex formation by two RM34C strands cannot be ruled out. Accordingly, the bimolecular parallel homoduplex proposed may contain four G·G hydrogen bonds at the 5′-end and two G·G mismatch at the 3′-side. These G·G mismatches could provide stability to these structures. The proposed model for RM34C at acidic pH and at neutral pH is represented in Fig. 11(b).
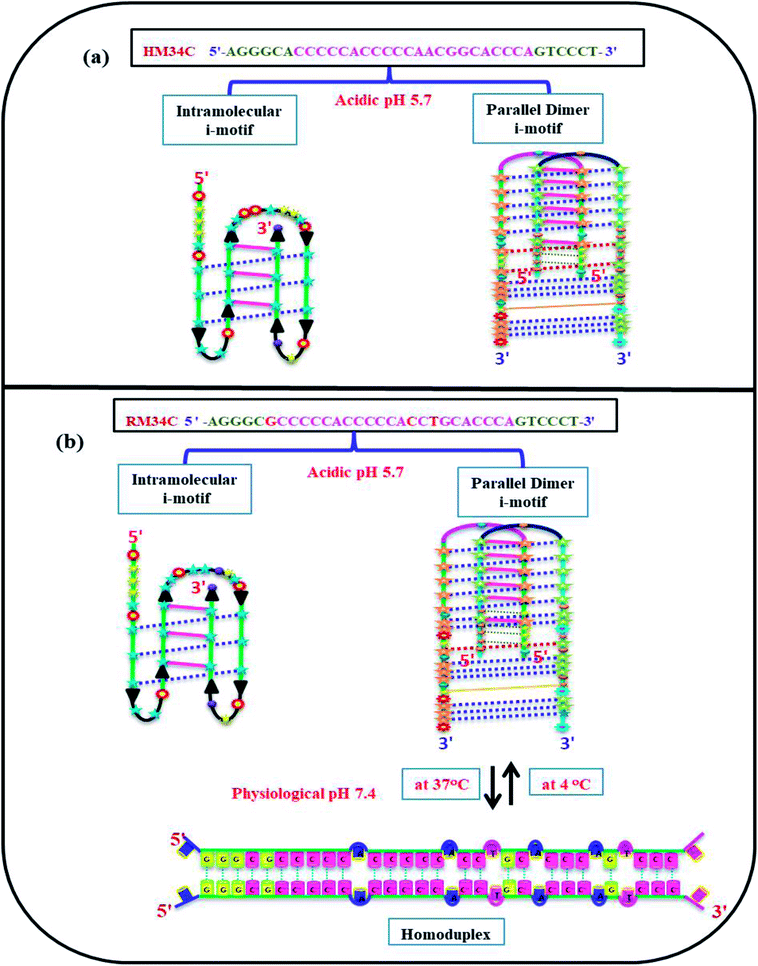 | ||
| Fig. 11 Proposed model adopted by HM34C at acidic pH (5.7) (a) and RM34C at acidic pH (5.7) as well as physiological pH (7.4) (b). | ||
In theory, cytosine deprotonation should result in the instability of the i-motif, or even transition into a random coil at higher pH than 7. However, previous reports have demonstrated that C·C+ base pairs can be formed in hairpin and parallel duplexes at neutral and slightly alkaline pH.31,32 Therefore, there is a fair possibility that C·C+ base pairs are formed under normal physiological cellular conditions. Indeed, the stability of C·C+ base pairs would be weak under such conditions. We observed the formation of the i-motif at low temperature in the case of RM34C, while at 37 °C, the i-motif structures were converted to homoduplexes. Thus, besides the well-known effect of pH, the temperature is also an important factor for the formation of i-motif structures.
Biological relevance
Non-canonical DNA topologies, such as DNA triplex, G-quadruplex, i-motifs, variants of unusual DNA structures, have attracted considerable attention from the scientific community for their potential role in gene regulation. The enrichment of guanine-rich sequences at promoters, telomeres, as well as various hot spot regions proved the substantial role of the G-quadruplex in the regulation of gene expression.33 Complementary counterparts of these G-rich sequences also tend to adopt C-tetraplex or i-motif structures, containing stacks of hemiprotonated C–C+ pairing. Under physiological conditions, G and C rich strands can adopt normal Watson–Crick duplex structures. Although both G and C-rich sequences experience the same cations and microenvironment, G-quadruplexes are unaffected by pH, whereas cytosines get protonated at acidic pH and adopt intercalative DNA (i-motif) structures. It is well documented that while G-quadruplexes inhibit gene expression in some cases, i-motifs have the tendency to enhance transcriptional activity as well as their use as pH-responsive switches in macromolecular assemblies.34–36 It has been reported that in the influence of negative supercoiling, the i-motif formation takes place under physiological conditions.37 Moreover, since the i-motif has been shown to induce a pH-dependent conformational switch, these transient structures have potential applications in nanotechnology as nanomolecular devices.38–40 Recently, i-motif DNA nanomachines, functional between pH 5.5 and 6.8, were constructed for application as pH sensors inside living cells.41 The architecture of C-tetraplexes was found to be more complicated than G-tetraplexes due to the existence of C-rich sequences in i-motifs, hairpin and homoduplex structures in equilibrium.35,36 Uribe et al. have reported that i-motif structures formed within the C-rich sequence of the HIF-1α promoter can exist under physiological conditions and that hnRNP K can bind to this C-rich sequence.42 Yunxi et al. have reported that small molecules like IMC-48, IMC-76 and hnRNP LL can interact with the i-motif and modulate the transcription of the BCL-2 gene.43Conclusion
Based on the existing knowledge in literature, PAGE, circular dichroism (CD), and thermal denaturation profiles, it was concluded that all the C-rich sequences from the MYH7 gene promoter sequence studied here at acidic pH can adopt i-motif structures of varied molecularity/strandedness. The HM34C and RM34C sequences depict the presence of two C5 and two C3 stretches separated by thymine (T). It is evident here that RM34C forms the i-motif structure at acidic, as well as at neutral pH. A very recent study by the Waller group demonstrated that i-motif formation and its stability can be achieved without molecular crowding agents at physiological pH, provided that there is a minimum tract length of C5. It is also established that more than half of the genomic oligonucleotides were shown to adopt the i-motif at neutral pH. These C-rich sequences are not randomly distributed in the genome but they are predominantly found in gene promoters.44 This study strengthens our results that the MYH7 gene promoter sequence RM34C having C5 blocks can adopt the i-motif as well as homoduplex at physiological pH, whereas HM34C exists as an unstructured single strand at physiological pH. These sequence motifs differ from each other by three bases. It is well documented that a single base difference in sequence is crucial for the regulation of its polymorphic nature and its pivotal role played in biological processes.45 The CD spectra cannot predict the molecularity/strandedness of the i-motif topology (intramolecular, dimer or tetramer status), as all the three structures are formed by the interdigitation of two parallel duplexes involving C·C+ pairs, which give identical CD bands irrespective of their molecular architecture. However, CD depicted all the signatures of the structures reported here, i.e., the i-motif, homoduplex and Watson–Crick duplex. Thus, it is clear from the temperature dependent CD studies that the structural status of the dimeric i-motif changes at 37 °C. The Tm value of 61 °C may be assigned to the structure that is stable at 37 °C and is characterized by the +275/−240 nm CD signal. Most probably, this could be a homoduplex structure stabilized by G·G and C·C+ base pairs.Although we are far from the clear picture of in vivo biological processes and with our studies we can only give an approximate idea about the structural polymorphism of said cytosine-rich sequences, we can still suggest that i-motifs can be formed not only under acidic conditions, but also at physiological pH and so may affect their possible biological roles. Nevertheless, more studies are required for the detailed description of i-motif formation and its topology in in vivo conditions for a better understanding of their recognition by proteins or any other important biological application.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Authors would like to acknowledge University of Delhi for the research grant R&D (RC/2015/9677-882) and DU-DST/PURSE Grant (CD/2015/1534).References
- C. Yunxi, K. D. Kong, G. Chiran, X. Cuixia and M. Hanbin, Mutually Exclusive Formation of G-Quadruplex and i-Motif is a General Phenomenon Governed by Steric Hindrance in Duplex DNA, Biochemistry, 2016, 55(15), 2291–2299 CrossRef PubMed.
- R. Langridge and A. Rich, Molecular structure of helical polycytidylic acid, Nature, 1963, 198, 725–728 CrossRef CAS PubMed.
- K. Gehring, J. L. Leroy and M. A. Gueron, Tetrameric DNA structure with protonated cytosine-cytosine base pairs, Nature, 1993, 363, 561–565 CrossRef CAS PubMed.
- J. Fegion, A new DNA quadruplex, Curr. Biol., 1993, 3, 611–613 CrossRef.
- P. Catasti, X. Chen, S. V. Mariappan, E. M. Bradbury and G. Gupta, DNA repeats in the human genome, Genetica, 1999, 106, 15–36 CrossRef CAS PubMed.
- D. E. Gilbert and J. Fegion, Multistranded DNA structures, Curr. Opin. Struct. Biol., 1999, 9, 305–314 CrossRef CAS PubMed.
- J. L. Mergny, L. Lacroix, X. Han, J. L. Leroy and C. Helene, Intramolecular folding of pyrimidine oligodeoxynucleotides into anti-DNA motif, J. Am. Chem. Soc., 1995, 117, 8887–8898 CrossRef CAS.
- P. Fojtik and M. Vorlickova, The fragile X chromosome (GCC) repeat folds into a DNA tetraplex at neutral pH, Nucleic Acids Res., 2001, 29, 4684–4690 CrossRef CAS PubMed.
- X. Li, Y. Peng, J. Ren and X. Qu, Carboxyl-modified single-walled carbon nanotubes selectively induce human telomeric i-motif formation, Proc. Natl. Acad. Sci. U.S.A., 2006, 103, 19658–19663 CrossRef CAS PubMed.
- C. Zhao, J. Ren and X. Qu, Single-walled carbon nanotubes binding to human telomeric i-motif DNA under molecular-crowding conditions: more water molecules released, Chem.–Eur. J., 2008, 14, 5435–5439 CrossRef CAS PubMed.
- H. A. Day, C. Huguin and Z. A. E. Waller, Silver cations fold i-motif at neutral pH, Chem. Commun., 2013, 49, 7696–7698 RSC.
- J. Zhou, C. Wei, G. Jia, X. Wang, Z. Feng and C. Li, Formation of i-motif structure at neutral and slightly alkaline pH, Mol. BioSyst., 2010, 6, 580–586 RSC.
- J. D. Puglisi and I. Tinoco Jr, Absorbance melting curves of RNA, Methods Enzymol., 1989, 180, 304–325 CAS.
- N. Sugimoto, S. Nakano, S. Kotah, A. Matsumura, H. Nakamura, T. Ohmichi, M. Yoneyama and M. Sasaki, Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes, Biochemistry, 1995, 34, 11211–11216 CrossRef CAS PubMed.
- S. Nakano, T. Kanzaki and N. Sugimoto, Influences of ribonucleotide on a duplex conformation and its thermal stability: study with the chimeric RNA–DNA strands, J. Am. Chem. Soc., 2004, 126, 1088 CrossRef CAS PubMed.
- M. Kaushik and S. Kukreti, Structural polymorphism exhibited by a quasipalindrome present in the locus control region (LCR) of the human b-globin gene cluster, Nucleic Acids Res., 2006, 34(12), 3511–3522 CrossRef CAS PubMed.
- M. Kaushik, A. Bansal, S. Saxena and S. Kukreti, Possibility of an Antiparallel (Tetramer) Quadruplex Exhibited by the Double Repeat of the Human Telomere, Biochemistry, 2007, 46, 7119–7131 CrossRef CAS PubMed.
- M. Kaushik, M. Prasad, S. Kaushik, A. Singh and S. Kukreti, Structural transition from dimeric to tetrameric i-motif, caused by the presence of TAA at the 3′-end of human telomeric C-rich sequence, Biopolymers, 2010, 93(2), 150–160 CrossRef CAS PubMed.
- S. Kaushik, M. Kaushik, F. Svinarchuk, C. Malvy, S. Fermandjian and S. Kukreti, Presence of divalent cation is not mandatory for the formation of intramolecular purine-motif triplex containing human c-junprotooncogene target, Biochemistry, 2011, 50(19), 4132–4142 CrossRef CAS PubMed.
- G. Manzini, N. Yathindra and L. E. Xodo, Evidence for intramolecularly folded i-DNA structures in biologically relevant CC-repeat sequences, Nucleic Acids Res., 1994, 22, 4634–4640 CrossRef CAS PubMed.
- H. Kanehara, M. Mizuguchi, K. Tajima, K. Kanaori and K. Makino, Spectroscopic evidence for the formation of four-stranded solution structure of oligodeoxycytidinephosphorothioate, Biochemistry, 1997, 36, 1790–1797 CrossRef CAS PubMed.
- J. Kypr, I. Kejnovská, D. Renčiuk and M. Vorlíčková, Circular dichroism and conformational polymorphism of DNA, Nucleic Acids Res., 2009, 37(6), 1713–1725 CrossRef CAS PubMed.
- J.-L. Leroy, J.-L. Mergny, M. Gueron and C. Helene, Intramolecular folding of a fragment of the cytosine-rich strand of telomeric DNA into an i-motif, Nucleic Acids Res., 1994, 22, 1600–1606 CrossRef CAS PubMed.
- S. H. Ke and R. M. Wartell, Influence of nearest neighbor sequence on the stability of base pair mismatches in long DNA: determination by temperature-gradient gel electrophoresis, Nucleic Acids Res., 1993, 21, 5137–5143 CrossRef PubMed.
- E. M. Evertsz, K. Rippe and T. M. Jovin, Parallel-stranded duplex DNA containing blocks of trans purine–purine and purine–pyrimidine base pairs, Nucleic Acids Res., 1994, 22, 3293–3303 CrossRef CAS PubMed.
- D. M. Gray, T. Cui and R. L. Ratliff, Circular dichroism measurements show that C.C+ base pairs can coexist with A.T base pairs between antiparallel strands of an oligodeoxynucleotide double-helix, Nucleic Acids Res., 1984, 12, 7565–7580 CrossRef CAS PubMed.
- E. L. Edwards, M. H. Patrick, R. L. Ratliff and D. M. Gray, A.T and C.C+ base pairs can form simultaneously in a novel multistranded DNA complex, Biochemistry, 1990, 29, 828–836 CrossRef CAS PubMed.
- D. M. Gray and M. Vaughan, Circular dichroism spectra show that repeating dinucleotide DNAs may form helices in which every other base is looped out, Nucleic Acids Res., 1980, 8(16), 3695–3707 CrossRef PubMed.
- S. Ahmed and E. Henderson, Formation of novel hairpin structures by telomeric C-strand oligonucleotides, Nucleic Acids Res., 1992, 20(3), 507–511 CrossRef CAS PubMed.
- J.-L. Leroy, K. Gehring, A. Kettani and M. Gueron, Acid multimers of oligodeoxycytidine strands: stoichiometry, base-pair characterization, and proton exchange properties, Biochemistry, 1993, 32, 6019–6031 CrossRef CAS PubMed.
- A. Yu, M. D. Barron, R. M. Romero, M. Christy, B. Gold, J. Dai, D. M. Gray, I. S. Haworth and M. Mitas, At physiological pH, d(CCG)15 forms a hairpin containing protonated cytosines and a distorted helix, Biochemistry, 1997, 36, 3687–3699 CrossRef CAS PubMed.
- M. Vorlickova, I. Kejnovska, M. Tumova and J. Kypr, Conformational properties of DNA fragments containing GAC trinucleotide repeats associated with skeletal displasias, Eur. Biophys. J., 2001, 30, 179–185 CrossRef CAS PubMed.
- V. S. Chambers, G. Marsico, J. M. Boutell, M. D. Antonio, G. P. Smith and S. Balasubramanian, High-throughput sequencing of DNA G-quadruple structures in the human genome, Nat. Biotechnol., 2015, 33, 877–881 CrossRef PubMed.
- S. Kendrick and L. H. Hurley, The role of G-quadruplex/i-motif secondary structures as cis-acting regulatory elements, Pure Appl. Chem., 2010, 82, 1609–1621 CAS.
- S. Kendrick, H.-J. Kang, M. P. Alam, M. M. Madathil, P. Agrawal, V. Gokhale, D. Yang, S. M. Hecht and L. H. Hurley, The Dynamic Character of the BCL2 Promoter i-Motif Provides a Mechanism for Modulation of Gene Expression by Compounds That Bind Selectively to the Alternative DNA Hairpin Structure, J. Am. Chem. Soc., 2014, 136, 4161–4171 CrossRef CAS PubMed.
- H.-J. Kang, S. Kendrick, S. M. Hecht and L. H. Hurley, The Transcriptional Complex between the BCL2 i-Motif and hnRNP LL Is a Molecular Switch to Control Gene Expression, J. Am. Chem. Soc., 2014, 136, 4172–4185 CrossRef CAS PubMed.
- D. Sun and L. H. Hurley, The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: implications for drug targeting and control of gene expression, J. Med. Chem., 2009, 52, 2863–2874 CrossRef CAS PubMed.
- T. Liedl and F. C. Simmel, Switching the conformation of a DNA molecule with a chemical oscillator, Nano Lett., 2005, 5(10), 1894–1898 CrossRef CAS PubMed.
- E. Guittet, D. Renciuk and J.-L. Leroy, Junctions between i-motif tetramers in supramolecular structures, Nucleic Acids Res., 2012, 40, 5162–5170 CrossRef CAS PubMed.
- Y. C. Dong, Z. Q. Yang and D. S. Liu, DNA nanotechnology based on i-motif structures, Acc. Chem. Res., 2014, 47, 1853–1860 CrossRef CAS PubMed.
- S. Modi, M. G. Swetha, D. Goswami, G. D. Gupta, S. Mayor and Y. Krishnan, A DNA nanomachine that maps spatial and temporal pH changes inside living cells, Nat. Nanotechnol., 2009, 4, 325–330 CrossRef CAS PubMed.
- D. J. Uribe, Y.-J. Shin, E. Lau, S. W. Ebbinghaus and D. Sun, Heterogeneous Nuclear Ribonucleoprotein K Binds to the Cytosine-Rich Sequence of the Hypoxia Inducible Factor 1 Alpha Proximal Promoter that Forms a Stable i-motif at Neutral pH, J. Phys. Chem. Biophys., 2013 DOI:10.4172/2161-0398.S5-001.
- Y. Cui, D. Koirala, H. Kang, S. Dhakal, P. Yangyuoru, L. H. Hurley and H. Mao, Molecular population dynamics of DNA structures in a bcl-2 promoter sequence is regulated by small molecules and the transcription factor hnRNP LL, Nucleic Acids Res., 2014, 42, 5755–5764 CrossRef CAS PubMed.
- E. P. Wright, J. L. Huppert and Z. A. E. Waller, Identification of multiple genomic DNA sequences which form i-motif structures at neutral pH, Nucleic Acids Res., 2017, 45(6), 2951–2959 CrossRef CAS PubMed.
- D. Miyoshi, H. Karimata and N. Sugimoto, Drastic Effect of a Single Base Difference between Human and Tetrahymena Telomere Sequences on Their Structures under Molecular Crowding Conditions, Angew. Chem., Int. Ed., 2005, 44, 3740–3744 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra05179h |
| This journal is © The Royal Society of Chemistry 2018 |