Open Access Article
Open Access ArticleSensitive detection of the IS6110 sequence of Mycobacterium tuberculosis complex based on PCR-magnetic bead ELISA†
Soe Paing Kyaw ab,
Jariya Hanthamrongwit
ab,
Jariya Hanthamrongwit a,
Kulachart Jangpatarapongsa
a,
Kulachart Jangpatarapongsa c,
Prasong Khaenamd and
Chaniya Leepiyasakulchai
c,
Prasong Khaenamd and
Chaniya Leepiyasakulchai *a
*a
aDepartment of Clinical Microbiology and Applied Technology, Faculty of Medical Technology, Mahidol University, Bangkok, Thailand 73170. E-mail: chaniya.lee@mahidol.edu
bClinical Pathology Laboratory, (1000) Bedded General Hospital, Nay Pyi Taw, Myanmar 15011
cCenter for Research and Innovation, Faculty of Medical Technology, Mahidol University, Bangkok, Thailand 73170
dCenter for Standardization and Product Validation, Faculty of Medical Technology, Mahidol University, Bangkok, Thailand 73170
First published on 1st October 2018
Abstract
Tuberculosis (TB) is ranked as the top killer among infectious diseases worldwide. Early and accurate diagnosis of the disease is crucial to end the global TB epidemic. The current commercially available molecular tests are still unaffordable by most TB affected communities. Herein, we developed a novel rapid and sensitive diagnostic method to detect the IS6110 sequence of Mycobacterium tuberculosis (M. tuberculosis) complex using PCR-magnetic bead ELISA. PCR amplification ofa 123 bp repetitive sequence of the IS6110 gene was performed by using digoxigenin (DIG) and biotin-labelled primers. Streptavidin-conjugated magnetic beads were used to collect the dual-labelled amplicons and subsequently, colourimetric detection was done by using horseradish peroxidase (HRP)-conjugated anti-DIG antibody. This method is able to detect M. tuberculosis DNA down to 0.5 fg per reaction within 3 hours. The sensitivity of IS6110 PCR detection by magnetic bead ELISA is 100 times higher than that of conventional agarose gel electrophoresis. The assay specificity was determined using a panel of DNA extracted from 10 common bacteria causing lower respiratory tract infections. No cross-reactivity was detected from those bacteria by IS6110 PCR-magnetic bead ELISA. Thus, the novel highly sensitive and specific, reduced assay time and simplicity of this PCR-magnetic bead ELISA for the detection of the specific gene of M. tuberculosis complex makes it an attractive diagnostic tool for large-scale screening of tuberculosis in standard clinical laboratories.
Introduction
Tuberculosis (TB) is a major public health catastrophe with 1.6 million worldwide deaths in 2016.1 Despite the fact that TB is preventable and curable, it is still standing as the leading cause of death by communicable diseases.2 According to the World Health Organization (WHO), only 6.3 million new cases were reported in 2016 despite 10.4 million estimated incidences.1 The significant improvements in diagnostic method development of TB during recent years led to the endorsement of GeneXpert MTB/RIF and GeneXpert MTB/RIF ultra for detection of M. tuberculosis, by WHO.3 Gene Xpert MTB/RIF is a real-time PCR-based amplification technique for simultaneous detection of an M. tuberculosis-specific gene as well as gene mutations related to the rifampicin (RIF) resistance within 2 h.4 However, the implementation of GeneXpert assay for detection of M. tuberculosis in resource-limited countries is hindered especially due to its high-cost as well as the requirements of a stable electrical power supply and maintenance.5,6 Therefore, detection of TB in resource-limited high TB burden countries relies mainly upon the poor sensitive sputum smear microscopy.7 Thus, the development of rapid, sensitive and cost-effective diagnostic method is the utmost important to narrow down the substantial gap in the detection of TB.The IS6110 is an insertion sequence belongs to the IS3 family and 10–16 copies of this repetitive sequence are exclusively present in the most strains of M. tuberculosis complex, the causal organisms of TB in human.8 The polymerase chain reaction (PCR) amplification of IS6110 sequence has been successfully applied for the detection of M. tuberculosis complex from the various types of clinical specimens.9–11 However, the lower sensitivity of agarose gel electrophoresis, the commonly used method for the detection and visualization of PCR products, is the limitation of conventional PCR amplification.12 Quantitative PCR amplification technique offers the real-time quantitation of target nucleic acids during PCR reaction cycles overcoming the laborious post-PCR detection methods.13 However, real-time amplification requires the expensive PCR machine with integrated detection modules.14
Alternatively, enzyme-linked immunosorbent assay for colourimetric detection of DIG-labelled PCR amplicons (PCR-ELISA) was developed for the detection of pathogenic microorganisms in the past decades.15–17 PCR-ELISA methods allow the semi-quantitative detection of DNA on a large number of samples simultaneously.18 In standard PCR-ELISA assays, target specific biotinylated probes immobilized on the streptavidin-coated microtiter plate are used to hybridize the DIG-labelled PCR amplicons. The detection of the colourimetric signal is accomplished after incubation with the peroxidase-conjugated anti-DIG antibody and the colourimetric substrate.19 PCR-ELISA technique is a sensitive, specific and less expensive method for semi-quantitative detection of PCR products.20 However, the requirement of laborious and time-consuming hybridization steps are considered as the limitation of the conventional PCR-ELISA method.
An improved and optimized PCR-ELISA method was introduced by Kobets et al. for the detection and quantification of Leishmania parasites from tissue samples. The assay could detect 0.3 fg of Leishmania DNA per reaction which is corresponding to 0.004 parasites within 7 h after DNA preparation.21 The assay was designed to overcome the laborious hybridization step by using biotin and DIG-labelled primers in the PCR amplification.21 However, the method still required an overnight coating of a microtiter plate with streptavidin and longer incubation time to capture the biotinylated PCR amplicons to the streptavidin-coated wells of a microtiter plate.
Magnetic particles have been applied in a wide variety of biomedical applications due to their properties of the high surface area to volume ratio, stability, fast kinetic processes and high response rate to the external magnetic field.22–26 Streptavidin-conjugated magnetic beads are widely used for selective isolation of biotinylated target molecules from the complex suspension of biomaterials since the binding of streptavidin to biotin exhibits the strongest noncovalent interaction with the dissociation constant as much as 10−15 M.27,28
In the present study, taking advantages offered by the multiple copies of the repetitive IS6110 sequence, we developed a sensitive method for detection of M. tuberculosis complex using PCR and magnetic bead ELISA. Biotin and DIG-labelled IS6110 specific primers were used in PCR reaction. The amplified products were enriched by using streptavidin-conjugated magnetic beads. Subsequent colourimetric detection was directly done on the bead-bound amplicons without any pretreatments such as plate coating and blocking steps, allowing a shorter turnaround time compared to standard PCR-ELISA assay.
Method
DNA samples
M. tuberculosis DNA samples extracted from the reference strain (H37Rv, ATCC 27294) and clinical isolate were obtained from Tuberculosis Research Laboratory, National Center for Genetic Engineering and Biotechnology, and Department of Medicine, Khon Kean University, Thailand. For assay specificity testing, 10 bacteria (Klebsiella pneumoniae, Streptococcus pneumoniae, Acinetobacter baumannii, Acinetobacter lwoffii, Pseudomonas aeruginosa, Escherichia coli, Corynebacterium diphtheriae, Moraxella catarrhalis and Staphylococcus aureus and Enterobacter aerogenes) commonly recovered from the lower respiratory tract infections were tested. All the bacteria were sub-cultured and maintained in the Department. The bacteria were grown in the Luria–Bertani broth (LB) medium at 37 °C overnight. DNA extraction was performed using Quick-gDNA MiniPrep (Zymo Research, CA, USA) DNA extraction kit following the manufacturer's instruction. The DNA concentration of all Mycobacteria and non-Mycobacterium bacteria were measured with the Nanodrop 2000 (Thermo Scientific, Wilmington, USA).PCR amplification
PCR amplification was performed with IS6110 specific forward primer 5′-DIG-C6-CCTGCGAGCGTAGGCGTCGG-3′ and reverse primer 5′-biotin-C6-CTCGTCCAGCGCCGCTTCGG-3′, where C6 is the 6-carbon spacer to reduce the possible steric hindrance (Pacific Science Co. Ltd, Bang Bumru, Thailand).29 The optimal concentration of each DIG and biotin-labelled forward and reverse primer was determined by varying the concentration of primers from 80 to 200 nM per reaction. One nanogram of M. tuberculosis (H37Rv) was used as a template in PCR amplification. The optimization of primer concentration is the critical step in the PCR-magnetic bead ELISA assay since the excess biotin-labelled reverse primer in the PCR amplification can compete with the biotin-labelled PCR amplicons for the streptavidin coated on the magnetic beads. The optimal concentration of each forward and reverse primer was selected as 160 nM since the amplified products obtained from this primer set is comparable to that obtained from 200 nM (ESI Fig. S1†).A total of 25 μl reaction was set up with 1× Go TaqGreen master mix (Madison, Wisconsin, USA), 160 nM of each forward and reverse primers, and 1 μl of DNA template. PCR reaction was carried out with initial denaturation at 94 °C for 2 min, followed by 30 cycles of 15 seconds at 94 °C, 45 seconds at 64 °C and 30 seconds at 72 °C and then one-step of final extension at 72 °C for 1 min. PCR products were separated on 2% agarose gel pre-stained with GelStar nucleic acid stain (Rockland, ME, USA) at 100 V for 30 min. Then, IS6110 specific 123 bp amplicons were visualized by using Gel Doc™ EZ System (Hercules, CA, USA). Band intensities of amplicons were measured using the volume tool of Image Lab software, version 5.2.1 (Hercules, CA, USA).
Magnetic bead ELISA
DNA products obtained from PCR amplification were detected by the magnetic bead-based colourimetric enzyme substrate detection system. Briefly, Sera-Mag SpeedBead blocked streptavidin particles (1.0 μM diameter) (GE Healthcare, UK) were washed twice in 1× phosphate buffered saline (PBS) with 0.5% tween 20 with the aid of an external magnet. PCR amplicons were diluted to 5% concentration with binding buffer containing 2% bovine serum albumin (BSA) in PBS. Then, the diluted PCR amplicons were added to the washed magnetic beads and incubated for 15 min with gentle rotation to capture the biotinylated PCR amplicons on the magnetic beads. After that, the bead-bound amplicons were washed two times with the aid of an external magnetic field. Thereafter, 50 μl of HRP-conjugated mouse monoclonal anti-DIG antibody (Ab 6212, Abcam, UK) diluted with 0.5% BSA in deionized water to get a final concentration of 20 ng ml−1 was added to the amplicons bound to the magnetic beads and incubated at room temperature for 30 min. The beads were then washed for 4 times with washing buffer (0.5% tween in 1× PBS) to remove unbound excess HRP-conjugated antibodies. Fifty microliters of TMB substrate (Merck, Billerica, MA, USA) was added and incubated in the dark for 30 min to develop the colour. Finally, 50 μl of TMB stop solution (BioLegend, San Diego CA, USA) was added to stop the colour reaction. The external magnet was used to hold the magnetic beads and supernatant was collected. The optical density was measured at 450 nm using BioTek Synergy HTX multi-mode microplate reader (Winooski, VT, USA).Statistical analysis
The absorbance values were imported to GraphPad Prism version 7.00 (GraphPad Software Inc, La Jolla, CA, USA) and linear regression analysis was done to observe the relationship between the DNA concentration and absorbance values. Samples are regarded as positive for the IS6110 gene when the absorbance values are above the cut-off value derived from the average absorbance + 3 standard deviation (SD) of no template control samples.Results and discussion
Scheme design
Herein, a rapid and sensitive PCR-magnetic bead ELISA was developed for M. tuberculosis detection. The schematic illustration of the developed assay is shown in Fig. 1. PCR amplification of the IS6110 sequence was performed with target-specific forward and reverse primers modified with DIG and biotin respectively. Streptavidin-conjugated magnetic beads were used not only to capture the biotinylated PCR amplicons but also to provide the solid support for colourimetric detection of the bead-bound amplicons. The magnetic beads used in the current assay are pre-blocked with non-protein and non-surfactant blocking agent to minimize the non-specific bindings in the antigen–antibody reaction. The capture of amplicons to the beads is accomplished within 15 min as streptavidin-coated magnetic beads exhibit high affinity to the biotinylated targets. Moreover, the faster magnetic response time of magnetic beads allows the easy and quick handling of amplicons during washing steps. The turnaround time for the colourimetric detection after PCR amplification is less than 90 min.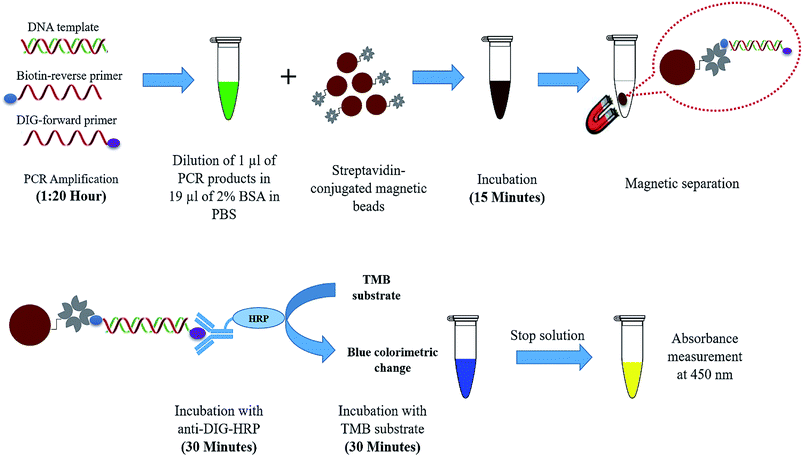 | ||
| Fig. 1 Schematic illustration of PCR-magnetic bead ELISA. The component enclosed in the dashed balloon was elaborated in the lower panel. | ||
Optimization of assay platform
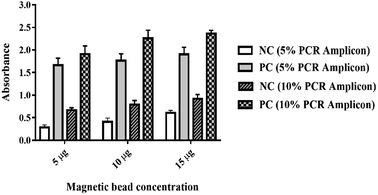
The experimental conditions including the concentration of HRP-conjugated anti-DIG, the concentration of magnetic beads and the amount of PCR amplicons were optimized to attain the maximum performance of the assay. To obtain the optimal colour intensity, proper concentration of HRP-conjugated anti-DIG antibody was ascertained as follows. HRP-conjugated anti-DIG antibody diluted to 200 ng ml−1, 100 ng ml−1, 50 ng ml−1, 25 ng ml−1 and 20 ng ml−1 in 0.5% BSA were incubated with TMB substrate for 30 min and the optical density for each concentration was measured after adding the TMB stop solution. The optimal concentration of HRP-conjugated anti-DIG antibody was selected at 20 ng ml−1 (data not shown). Then, the concentration of magnetic bead per reaction (5, 10 and 15 μg) and the amount of PCR amplicons per reaction (5%, and 10% in 2% BSA in PBS) were optimized simultaneously. The results showed that the absorbance of positive and negative control samples were increased together with the concentration of both magnetic beads and PCR amplicons while maintaining the absorbance difference between negative and positive samples (Fig. 2). Therefore, the concentration of magnetic beads and PCR amplicons that provide the lowest absorbance of negative samples were selected as the optimal concentration. The optimal concentration of magnetic beads and PCR amplicons were 5 μg and 5% (1 μl of PCR products in 19 μl of 2% BSA in 1× PBS) per reaction. The optimization of the concentration of magnetic beads and PCR products is critical for the success of the current assay since there was no difference in absorbance values from the different concentrations of undiluted PCR products (data not shown). This is plausible since highly saturated streptavidin molecules on the magnetic beads allowed the maximal binding of biotinylated PCR amplicons that provide the high signal intensities between different concentrations of PCR products. The optimal incubation time for binding HRP-conjugated anti-DIG was 30 min. Colour development was done by incubation with TMB substrate for 30 min after washing the excess antibodies with 1× PBS for 4 times.Analytical sensitivity
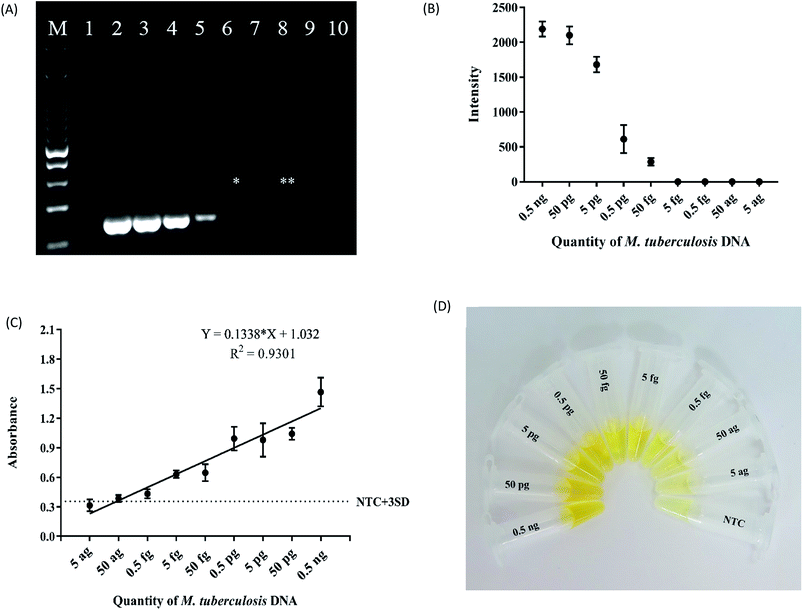
The analytical sensitivity of magnetic bead ELISA was determined using a 10-fold serial dilution of H37Rv DNA, starting from 0.5 ng to 5 ag per PCR reaction. Six and one microliter(s) of each of these products were analyzed by agarose gel electrophoresis and magnetic bead ELISA, respectively. Due to the difference in PCR efficiency, the difference in band intensities were also observed by agarose gel electrophoresis (Fig. 3A). To estimate the concentration of PCR amplicon obtained from each DNA dilution, relative DNA concentrations were calculated based on the DNA ladder concentration suggested by the manufacturer (Biotechrabbit, Berlin, Germany). However, the band intensities from 0.5 ng to 5 pg DNA input were very similar, suggesting the saturation of PCR reaction (Fig. 3B). Therefore, only 2 dilutions of DNA input, 0.5 pg, and 50 fg were used in the estimation of PCR amplicon concentration. Roughly, 4 and 2 ng μl−1 were estimated for these two dilutions, respectively (ESI Fig. S2†). Since we loaded 6 μl of PCR products onto agarose gel, the lowest quantity of PCR amplicon visible by our electrophoresis system is 12 ng.In order to determine the lowest detection limit of magnetic bead ELISA assay, the positive result is considered by comparing to the cutoff absorbance 0.355 calculated from the average absorbance + 3 SD of the negative controls (no template control). Although the variation in PCR efficiency was also detected by this assay, almost all dilutions, except 5 and 50 ag, of DNA input were considered as positive with the magnetic bead ELISA (Fig. 3C). The assay also captured slightly lower in PCR efficiency obtained from 5 fg DNA input while the band of this DNA dilution was absent by agarose gel inspection. In the other word, both 5 and 50 fg DNA input might yield approximately 2 ng amplicon but the quantity slightly less than 2 ng cannot be detected by agarose gel inspection. However, with the signal amplification of ELISA, 5 fg input DNA provides the absorbance close to that obtained from 50 fg. The mean absorbance as well as its SD of the lowest quantity of M. tuberculosis DNA above the cut-off value was 0.5 fg input. Thus, the limit of detection (LOD) of magnetic bead ELISA was 0.5 fg which is more sensitive than 1 pg detection limit of IS6110 detection using isothermal amplification method.30 Fig. 3D showed the colour intensity obtained from each concentration of H37Rv DNA samples. These results indicate that magnetic bead ELISA is 100 times more sensitive than the conventional agarose gel electrophoresis. According to this high sensitivity, our PCR ELISA-based assay shows a great promise to detect even a trace amount of PCR amplicons as long as PCR is success.
In addition, the results of our developed assay show a good linear relationship between absorbance and the quantity of M. tuberculosis DNA with the R2 = 0.9301 (Fig. 3D). This high correlation offers the good performance of the assay to distinguish between low and high DNA input albeit qualitative measurement is the expected principle of this assay. Interestingly, it has been estimated that one genome equivalent of M. tuberculosis is 5 fg of DNA.31 Therefore, it can be expected that our magnetic bead ELISA is sensitive for the detection of M. tuberculosis even if only one organism is presented in the sample.
Assay specificity
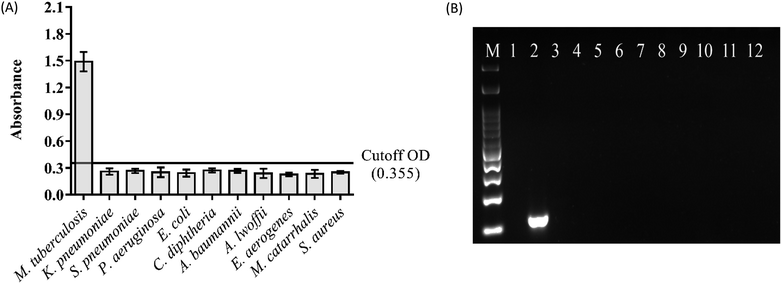
To verify the specificity, genomic DNA extracted from K. pneumoniae, S. pneumoniae, P. aerugniosa, E. coli, C. diphtheriae, A. baumannii, A. lwoffii, E. aerogenes, M. catarrhalis and S. aureus together with M. tuberculosis DNA extracted from clinical isolate were tested with PCR-magnetic bead ELISA. PCR amplification was performed with 1 ng of each bacterial DNA as the template by using IS6110 specific primers. Then, magnetic bead ELISA was performed and the results were compared with the agarose gel electrophoresis. The absorbance values of all non-Mycobacterium bacterial DNA samples were below the cut-off absorbance of 0.355, indicating that the magnetic bead ELISA is specific to IS6110 sequence only (Fig. 4A) as well as there was no non-specific background of the detecting system. Concordance with the agarose gel electrophoresis, 123 bp specific amplicon was detected only in M. tuberculosis DNA sample and exhibit the agreement of results with the magnetic bead ELISA (Fig. 4B).Conclusion
In summary, we have developed a sensitive method for the detection of the IS6110 sequence of M. tuberculosis complex. Firstly, the exponential amplification power of PCR was used to amplify the target sequence to get the biotin and DIG dual-labelled amplicons. Then, separation of dual-labelled PCR amplicons from the un-amplified products was done using streptavidin-conjugated magnetic beads and subsequent colourimetric detection was done to the amplicons bound to the beads. Utilization of specific biotin and DIG primers in the PCR reaction overcomes the requirement of laborious hybridization steps in the conventional PCR-ELISA system. As colourimetric detection was done on the magnetic beads-bound PCR amplicons in the single tube, the step of overnight coating of the ELISA plates with streptavidin is avoidable in our developed assay. The handling of magnetic beads-bound amplicons during incubation steps and washing steps was done quickly with the external magnet. The detection limit of the magnetic bead ELISA is 0.5 fg of M. tuberculosis DNA, which is 100 times more sensitive than the conventional agarose gel electrophoresis. This method also provides the semi-quantitative detection of DNA as the colour intensity is directly proportional to the amount of PCR amplicons. In addition, reagents and instruments required for magnetic bead ELISA are commercially available and affordable. Thus, our developed assay could be useful for the detection of M. tuberculosis, especially in the remote areas where the diagnosis of tuberculosis remains concerned. Finally, this assay platform could be applied to the detection of any targeted gene.Conflicts of interest
The authors have no conflicts of interest to declare.Acknowledgements
We thank senior researcher Dr Saradee Warit, Tuberculosis research unit, National Science and Technology Development Agency, Thailand and Assistant Professor Dr Kiatichai Faksri, Faculty of Medicine, Khon Kaen University for providing DNA samples from Mycobacterium tuberculosis H37Rv reference strain and from the clinical isolate. We also grateful to Assistant Professor Dr Sakda Yainoi and Assistant Professor Dr Rungrot Cherdtrakulkiat, Faculty of Medical Technology, Mahidol University for the culture isolate of non-Mycobacterium bacteria. This work is supported by Mahidol-Norway capacity building initiative for ASEAN PhD scholarship to SPK and National Science and Technology Development Agency, Thailand grant to CL. The funders had no role in study design, data analysis and decision for publication of the manuscript.References
- World Health Organization, in Global tuberculosis report 2017, 2017 Search PubMed.
- World Health Organization, Bending the curve—ending TB: annual report 2017, WHO report, 2017 Search PubMed.
- A. L. García-Basteiro, A. DiNardo and B. Saavedra, et al., Point of care diagnostics for tuberculosis, Rev. Port. Pneumol., 2018 Search PubMed.
- R. McNerney, M. Maeurer and I. Abubakar, et al., Tuberculosis diagnostics and biomarkers: needs, challenges, recent advances, and opportunities, J. Infect. Dis., 2012, 205(suppl. 2), S147–S158 CrossRef PubMed.
- C. C. Boehme, M. P. Nicol and P. Nabeta, et al., Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB/RIF test for diagnosis of tuberculosis and multidrug resistance: a multicentre implementation study, Lancet, 2011, 377(9776), 1495–1505 CrossRef.
- B. Y. Ng, E. J. Wee, N. P. West and M. Trau, Naked-Eye Colorimetric and Electrochemical Detection of Mycobacterium tuberculosis toward Rapid Screening for Active Case Finding, ACS Sens., 2015, 1(2), 173–178 CrossRef.
- A. J. Caulfield and N. L. Wengenack, Diagnosis of active tuberculosis disease: From microscopy to molecular techniques, Journal of Clinical Tuberculosis and Other Mycobacterial Diseases, 2016, 4, 33–43 CrossRef.
- B. Sekar, L. Selvaraj, A. Alexis, S. Ravi, K. Arunagiri and L. Rathinavel, The utility of IS6110 sequence based polymerase chain reaction in comparison to conventional methods in the diagnosis of extra-pulmonary tuberculosis, Indian J. Med. Microbiol., 2008, 26(4), 352 CrossRef CAS PubMed.
- A. L. Osman, N. S. Saeed and M. M. Elhassan, Polymerase Chain Reaction targeting insertion sequence IS6110 for the diagnosis of pulmonary tuberculosis among Sudanese children and young adults, Int. J. Mycobact., 2014, 3(4), 252–258 CrossRef PubMed.
- P. S. Deshpande, R. S. Kashyap and S. S. Ramteke, et al., Evaluation of the IS6110 PCR assay for the rapid diagnosis of tuberculous meningitis, Cerebrospinal Fluid Res., 2007, 4(1), 10 CrossRef PubMed.
- V. Makeshkumar, R. Madhavan and S. Narayanan, Polymerase chain reaction targeting insertion sequence for the diagnosis of extrapulmonary tuberculosis, Indian J. Med. Res., 2014, 139(1), 161 CAS.
- M. J. Sue, S. K. Yeap, A. R. Omar and S. W. Tan, Application of PCR-ELISA in molecular diagnosis, BioMed Res. Int., 2014, 2014 Search PubMed.
- E. A. Mothershed and A. M. Whitney, Nucleic acid-based methods for the detection of bacterial pathogens: present and future considerations for the clinical laboratory, Clin. Chim. Acta, 2006, 363(1–2), 206–220 CrossRef CAS PubMed.
- R. A. Babu, S. Manoharan, P. Ramadass and N. Chandran, Evaluation of RT-PCR assay for routine laboratory diagnosis of rabies in post mortem brain samples from different species of animals, Indian J. Virol., 2012, 23(3), 392–396 CrossRef PubMed.
- C. Metzger-Boddien and J. Kehle, Development and evaluation of a sensitive PCR-ELISA for detection of adenoviruses in feces, Intervirology, 2005, 48(5), 297–300 CrossRef CAS PubMed.
- G. Daeschlein, O. Assadian, F. Daxboeck and A. Kramer, Multiplex PCR-ELISA for direct detection of MRSA in nasal swabs advantageous for rapid identification of non-MRSA carriers, Eur. J. Clin. Microbiol. Infect. Dis., 2006, 25(5), 328–330 CrossRef CAS PubMed.
- A. Di Pinto, V. Terio, P. Di Pinto, V. Colao and G. Tantillo, Detection of Vibrio parahaemolyticus in shellfish using polymerase chain reaction–enzyme-linked immunosorbent assay, Lett. Appl. Microbiol., 2012, 54(6), 494–498 CrossRef CAS PubMed.
- F. Tayebeh, S. Nazarian, S. A. Mirhosseini and J. Amani, Novel PCR-ELISA Technique as a Good Substitute in Molecular Assay, J. Appl. Biotechnol. Rep., 2017, 4(2), 567–572 Search PubMed.
- C. P. Adikaram, J. Perera and S. S. Wijesundera, DNA probe based colorimetric method for detection of rifampicin resistance of Mycobacterium tuberculosis, J. Microbiol. Methods, 2014, 96, 92–98 CrossRef CAS PubMed.
- L. I. Gomes, L. H. dos Santos Marques, M. J. Enk, M. C. de Oliveira, P. M. Z. Coelho and A. Rabello, Development and evaluation of a sensitive PCR-ELISA system for detection of Schistosoma infection in feces, PLoS Neglected Trop. Dis., 2010, 4(4), e664 CrossRef PubMed.
- T. Kobets, J. Badalová, I. Grekov, H. Havelková, M. Svobodová and M. Lipoldová, Leishmania parasite detection and quantification using PCR-ELISA, Nat. Protoc., 2010, 5(6), 1074 CrossRef CAS PubMed.
- A. van Reenen, A. M. de Jong, J. M. den Toonder and M. W. Prins, Integrated lab-on-chip biosensing systems based on magnetic particle actuation–a comprehensive review, Lab Chip, 2014, 14(12), 1966–1986 RSC.
- A. Ito, M. Shinkai, H. Honda and T. Kobayashi, Medical application of functionalized magnetic nanoparticles, J. Biosci. Bioeng., 2005, 100(1), 1–11 CrossRef CAS PubMed.
- A. Akbarzadeh, M. Samiei and S. Davaran, Magnetic nanoparticles: preparation, physical properties, and applications in biomedicine, Nanoscale Res. Lett., 2012, 7(1), 144 CrossRef PubMed.
- V. Mani, B. V. Chikkaveeraiah and J. F. Rusling, Magnetic particles in ultrasensitive biomarker protein measurements for cancer detection and monitoring, Expert Opin. Med. Diagn., 2011, 5(5), 381–391 CrossRef CAS PubMed.
- N. Tran and T. J. Webster, Magnetic nanoparticles: biomedical applications and challenges, J. Mater. Chem., 2010, 20(40), 8760–8767 RSC.
- Z. Zhang, H. Zhu and Y. Tang, et al., Preparation and application of streptavidin magnetic particles, Sci. China, Ser. B: Chem., 2007, 50(1), 127–134 CrossRef CAS.
- H. Sakahara and T. Saga, Avidin–biotin system for delivery of diagnostic agents, Adv. Drug Delivery Rev., 1999, 37(1–3), 89–101 CrossRef CAS PubMed.
- K. D. Eisenach, M. Donald Cave, J. H. Bates and J. T. Crawford, Polymerase chain reaction amplification of a repetitive DNA sequence specific for Mycobacterium tuberculosis, J. Infect. Dis., 1990, 161(5), 977–981 CrossRef CAS PubMed.
- D. Liu, B. Zhao and X. Ou, et al., A novel isothermal amplification-based method to detect Mycobacterium tuberculosis complex, J. Microbiol. Methods, 2018, 145, 59–65 CrossRef CAS PubMed.
- B. Y. Ng, E. J. Wee, N. P. West and M. Trau, Rapid DNA detection of Mycobacterium tuberculosis-towards single cell sensitivity in point-of-care diagnosis, Sci. Rep., 2015, 5, 15027 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra06599c |
| This journal is © The Royal Society of Chemistry 2018 |