Open Access Article
Open Access ArticleA magnetic molecularly imprinted optical chemical sensor for specific recognition of trace quantities of virus†
Feng Zhangab,
Lianghui Luob,
Hang Gong b,
Chunyan Chen
b,
Chunyan Chen b and
Changqun Cai
b and
Changqun Cai *b
*b
aCollege of Science, Hunan Agricultural University, Changsha 410128, China
bKey Laboratory of Environmentally Friendly Chemistry and Applications of Ministry of Education, College of Chemistry, Xiangtan University, Xiangtan, Hunan 411105, China. E-mail: ccq@xtu.edu.cn; Fax: +86-731-5829-2251; Tel: +86 15273219560
First published on 18th September 2018
Abstract
A magnetic resonance light scattering (RLS) sensor based on the molecularly imprinted polymer (MIP) technique was developed for specific recognition of trace quantities of hepatitis A virus (HAV). Through a surface imprinting technique, the virus-magnetic-MIPs (virus-MMIPs) were prepared as the specific identification element, which was based on the effective synthesis of biomimetic polydopamine (PDA) inspired by mussels on the surface of Fe3O4 magnetic nanoparticles. The preparation process of the virus-magnetic-MIPs was simple and rapid under an applied magnetic field. The surface of the magnetic-MIP captured viruses through specific recognition in water, which caused changes to the particle size and shape, and subsequently resulted in changes in the intensity of the RLS. The sensor was applied to determine the amount of HAV in the linear concentration range of 0.02–1.40 nmol L−1, with a low detection limit of 6.2 pmol L−1, and it was successfully applied for the immediate detection of added HAV from a 2000-fold dilution of human serum. More importantly, the proposed strategy addressed the difficulty of virus-MIP detection in the elution process and it is rapid, easy, sensitive, and eco-friendly.
1. Introduction
Fe3O4 nano-magnetic materials are one of the most common carriers in nano-sensors because they possess the advantages of good biocompatibility, and magnetic and electrical conductivity. They are already widely used in medicine in the treatment of tumors and in molecular biology for cell separation; they are also used in other fields as catalyst carriers, microwave-absorbing materials, magnetic liquids, and magnetic recording materials.1–3 Magnetic nanoparticles modified by natural receptors are often used to identify proteins, viruses, and nucleic acids.4,5 However, the instability of these natural receptors and their high cost limits their wide application. Among the variety of biomimetic recognition systems, there has been increased interest in molecularly imprinted polymers (MIPs) because they have proven potential as synthetic receptors.6,7Molecularly imprinted polymers with magnetic nanoparticles as carriers have prominent advantages in the field of separation and identification. Because they can be easily applied to an external magnetic field to separate the target without the need of a typical filter or separation step, they possess the advantages of low cost, excellent efficiency, environmental protection, and facile separation. For example, in 2013, a simplistic strategy was presented for the selective luminescence determination of trace polycyclic aromatic hydrocarbons based on a combination of magnetic separation and the distinctive recognition of MIPs. This magnetic dissociation of the nanocomposites from the target compound prior to luminescence detection of phenanthrene affords remarkably enhanced selectivity and sensitivity, the limit of detection (LOD) was 3.64 ng mL−1.8 The difficulties encountered in imprinting virus are attributed to its large molecular size, complex structure, and conformational flexibility, which lead to highly cross-linked polymer networks and poor rebinding efficiency. Researches in this field are still achieved a great development. Altintas et al. immobilized the MIPs on a surface plasmon resonance sensor for specific recognition of waterborne viruses.9,10 Dickert et al. demonstrated MIP-based quartz crystal microbalances to detect tobacco mosaic virus by constructing mass-sensitive chemical sensors for virus.11 Shahgaldian et al. synthesized organic/inorganic imprinted polymers to recognize non-enveloped icosahedral viruses by ELISA assay.12 However, it is still urge to promote researches on molecularly imprinted polymers of viruses.
Resonance light scattering (RLS) was first reported by Pasternack in 1993,13,14 and RLS gained rapid development and was used to detect DNA,15 proteins,16 metal ions17 and so on. RLS technology has the advantages of wide detection range, rapid and simple in practical applications. According to the synchronous luminescence equation: ISL = KcbEexλexEem(λex + Δλ), it can be seen that the RLS intensity is proportional to the concentration of the scattering particles when the particle size is fixed, and vice versa, which lays a foundation for the quantitative analysis application of the RLS method.
Given the above-mentioned properties and our previous study results, a series of optical virus sensors that are based on virus-MIPs with specific recognition and sensitivity at the picomolar level has been developed by our group.18–20 However, it remains to be improved in terms of reaction kinetics, nonspecific adsorption, and elution process. Based on continued interest because of virus detection, we sought to design a biocompatible magnetic-MIP RLS virus sensor for the rapid and simple detection of virus. To the best of our knowledge, there are no reports describing a magnetic-MIP optical virus sensor that is used to detect hepatitis A virus.
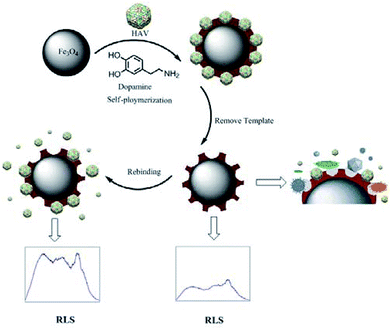
Hepatitis A virus (HAV), the causative agent of type A viral hepatitis, is an ancient human virus that was first found almost 43 years ago.21,22 Because HAV infection continues to be an important issue worldwide,23 developing a rapid and highly sensitive strategy to detect HAV is critical for the diagnosis of acute HAV infection. Herein, we describe a novel magnetic-MIP resonance light scattering (RLS) virus sensor for specific recognition of trace quantities of HAV. The core–shell virus-magnetic-MIPs (virus-MMIPs) were initially assembled by surface imprinting technique and conducted as a biomimetic designated identification molecule. By means of magnetic nanoparticles, the preparation times were shortened and the experimental apparatus simplified. Bio-inspired polydopamine (PDA) was selected as imprinting layer owing to its effective one-pot synthesis by self-polymerization capacity of dopamine (DA).24–26 HAV was treated as a model virus for specific bind with the surface of virus-MMIPs. The responded RLS signal was extremely increased with increasing concentrations of HAV. The proposed method possess the advantages of sensitivity, high selectivity, low expense, exceptional applicability, and excellent biocompatibility. More importantly, it addressed the difficulty of virus-MIP detection in the elution process and achieved the expected results.
2. Experimental
2.1 Instrumentation
The RLS spectra and intensity records on an RF-5301 fluorescence spectrofluorometer (Shimadzu, Japan) and a quartz cell (1 cm × 1 cm) was equipped. The shape and size of the nanoparticles were characterized by field emission scanning electron microscopy (FESEM, JEOL JSM-6700F, Japan). The Fourier transform infrared spectra for synthesized nanospheres were tested with an FT-IR spectrometer (IR Nicolet 6700, Shimadzu, Japan) by KBr pellets in the region 4000–400 cm−1. Absorbance spectra were measured with a UV-2450 spectrophotometer (Shimadzu, Japan). The pH values were recorded with a digital pH meter (pHs-3C, Leici, China). A 40 kHz ultrasonic cleaner water bath (KQ-25B, China) was applied for experiments. The cytotoxicity was analyzed by a MR-96A microplate reader (Mindray, China).2.2 Synthesis of virus-magnetic-MIPs
The Fe3O4 nanoparticles were prepared based on our previous work18 and a typical virus-magnetic-MIPs were prepared based on the improvement of the methods in the literature.20,25,27 The main differences were that the amounts of various particles and reagents were optimized, and the reaction time was changed based on the experimental results. In addition, magnetic separation is performed prior to elution by virtue of magnetic particle properties. In short, the resultant Fe3O4 nanoparticles (NPs) (approximately 150 mg) were dispersed in Tris buffer (pH 8.0). After adding 50 mg (2.5 mg mL−1) DA and HAV (6.85 mg mL−1), the reaction was continued via the simple and green self-polymerization of DA at 25 °C for 12 h. The resultant imprinted polymers were rapidly separated from the solution under an applied magnetic field, and then purified with deionized water to remove the excess monomers and free HAV. Then, the polymers were washed with a mixture of acetic acid (5%, v/v) and SDS (10%, w/v) to remove the bound HAV until a clear supernatant was observed. No HAV or detached PDA from the particles was detected by a UV-Vis spectrophotometer at 280 nm.25,28 The resultant NPs were thoroughly re-washed with deionized water to remove the remaining acetic acid and SDS, and then were once more subject to separation under an applied magnetic field. Finally, the products were freeze-dried for future use. Non-imprinted Fe3O4 NPs (NIPs) were prepared and treated under the same conditions but without the template virus.2.3 Resonance light scattering detection of HAV
By simultaneously scanning the excitation and emission monochromators (λex = λem) from 220.0 nm to 700.0 nm on a common spectrofluorometer, the RLS spectra of the HAV + virus-magnetic-MIPs (before virus removal), virus-magnetic-MIPs (removed virus), HAV, and bare Fe3O4 NPs were obtained. The excitation and emission slit widths were maintained at 3.0 nm and 5.0 nm, respectively, and the RLS intensities were recorded at 323 nm. The relative RLS intensity was obtained as ΔIRLS = IRLS − I0, where IRLS and I0 were the RLS intensities with and without HAV, respectively.2.4 Cytotoxicity assay
The viability of Chinese hamster fibroblast cells under different treatments was evaluated by performing the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay according to the literature.20,29,30 Briefly, 100 μL of cells (8 × 104 cells mL−1) were seeded in a 96-well plate and incubated with cell culture medium for 24 h at 37 °C in a humidified environment containing 5% CO2. The cells were further incubated with different concentrations (0, 1, 3, 10, 30, and 100 μg mL−1) of NIPs for another 24 h at 37 °C. Subsequently, 20 μL of freshly prepared MTT (5 mg mL−1) solution was added to each well, and then the plates were incubated for an additional 4 h. Formazan crystals were dissolved with DMSO, and the absorbance was measured at 490 nm with a microplate reader. Untreated cells were used as a negative control with 100% viability.3. Results and discussion
3.1 Preparation of virus-magnetic-MIPs
As shown in Scheme 1, the Fe3O4 NPs and model virus were first synthesized for the virus-magnetic-MIPs via a simple and green self-polymerization strategy using DA combined with the surface imprinting technology.24,31 Nano-sized, thin, stable and bio-inspired PDA films were formed on the surface of Fe3O4 with distinctive adhesive properties. HAV was embedded in the polymer layer by the non-covalent interaction of the amine and hydroxyl groups on the bio-inspired PDA chain. The generated multifunctional binding sites on the virus-magnetic-MIPs can identify the template virus by hydrogen and p–p bonds.18The core–shell virus-magnetic-MIPs were smoothly got by removing the model. The model virus was caught on the surface of the virus-magnetic-MIPs, which changed the intensity of the RLS. Because of the particular spatial function, the viruses, which were large or possessed different conformations, might not enter these specific recognition sites and could not be identified? Additionally, the smaller viruses maybe enter the cavities, but they might not be able to increase the size of the virus-magnetic-MIPs. This process was precise due to the great increasement of the RLS intensity when the synthetic virus-magnetic-MIPs were effectively used as the immediate quantitative detection of HAV in aqueous media by the fluorescence spectrophotometer.
3.2 Characterizations of virus-magnetic-MIPs
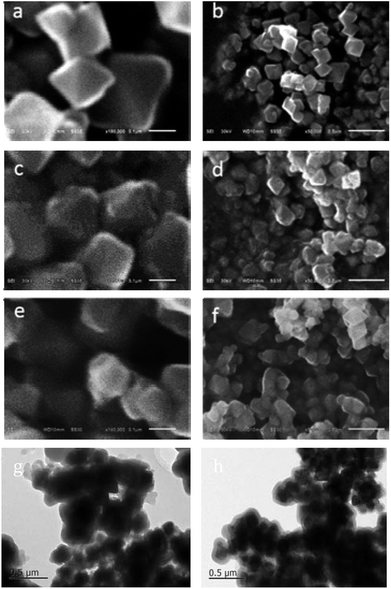
The surface appearance of the Fe3O4 NPs (Fig. 1a and b), HAV + virus-magnetic-MIPs (before the virus was removed) (Fig. 1c and d), virus-magnetic-MIPs (Fig. 1e–g),and non-imprinting (Fig. 1h) were characterized by field emission scanning electron microscopy (FESEM) and transmission electron microscope (TEM). As depicted in Fig. 1, the SEM micrographs showed that the bare Fe3O4 NPs were diamond-shaped with a relatively smooth surface and an average diameter of approximately 200 nm. The NPs without elution (HAV + virus-magnetic-MIPs) were nearly spherical. Compared with Fe3O4 NPs, the surface of the particles can be observed to be slightly larger, slightly viscous, with some spherical bulges on the surface, probably because of the PDA that was coated on the surface of the particle, and there is a granular HAV imprinted in the PDA surface. There was also a concave shape on the surface of the virus-magnetic-MIPs. As shown in the Fig. 1g and h, there were dark black cores in the middle of the particle, and they were similar in shape to Fe3O4, while the outer shell was light in color, indicating that polydopamine was successfully coated on the outside. Fig. 1g showed that there were grooves cover on outer shells and irregular structures, which indicated imprinting sites left after the eluting templates; on the contrary, the surfaces were smooth in Fig. 1h. Hence, PDA was coated onto the Fe3O4 NPs, and the virus was successfully imprinted on the surface of the NPs.To offer direct evidence for the synthetic process of the virus-magnetic-MIPs, the FT-IR spectra of the Fe3O4 NPs, NIPs, and virus-magnetic-MIPs were compared (Fig. S1†). The strong peak near 578 cm−1 was determined by the characteristic peak of the Fe–O–Fe bond. In addition, when PDA was present on the Fe3O4 NPs, peaks between 1600 and 1400 cm−1 appeared. The absorption peaks at approximately 1550 and 1457 cm−1 were attributed to aromatic C–H bending and N–H scissoring. The peak near 2927 cm−1 was caused by the expansion of the C–H stretching vibration of alkanes. These data demonstrated that the surface of the Fe3O4 NPs was successfully modified with PDA.
3.3 RLS spectral properties
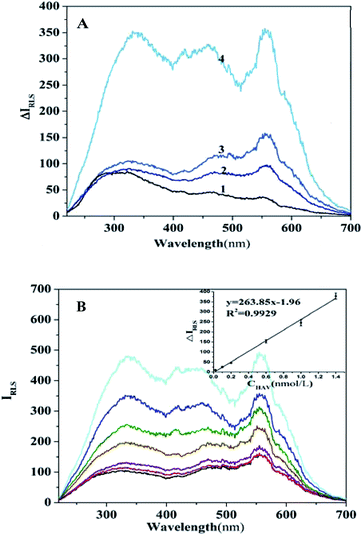
The RLS spectral curves of virus-magnetic-MIP system are shown in Fig. 2(A). Their intensities all reached the maximum (λmax) wavelength at 323 nm in the following order: HAV + virus-magnetic-MIPs > virus-magnetic-MIPs > Fe3O4 NPs > HAV. According to RLS theory, the RLS intensity is proportional to r6 (r is the radius of the granule).14 The average size of the as-prepared virus-magnetic-MIPs was approximately 200 nm, which was much greater than 1/20 of the wavelength of incident light. Consequently, the light scattering of virus-magnetic-MIPs can be regarded as Mie scattering instead of Rayleigh scattering.32 A 10 mL plastic tube was sequentially added with a certain amount of HAV-imprinted Fe3O4@PDA NPs, target virus HAV, Tris–HCl buffer. Then dilute to 5 mL with ultrapure water. The reaction system was shaken at room temperature for 150 min. Resonance light spectrum of the system was measured at 220–700 nm using a fluorescence spectrophotometer (the excitation and emission slit widths were kept at 3.0 nm and 5.0 nm,respectively, λex = λem), and the spectral intensity at a wavelength of 323 nm was recorded. As depicted in Fig. 2(B), the IRLS was proportionally improved with increasing concentrations of HAV virus, and the changes of RLS intensity depended on the adsorption affinity from the interactions between the particles and the virus.3.4 Optimization of the assay conditions
Various influencing factors such as the virus-magnetic-MIPs dosage (Fig. S2†), pH (Fig. S3†), incubation time (Fig. S4†), and temperature (25 °C) were investigated.3.5 Selectivity evaluation
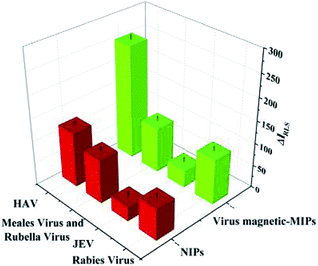
To verify the selectivity of virus-magnetic-MIPs towards the HAV template, other viruses with different molecular weights were used. These viruses included Japanese encephalitis virus, rabies virus, a mixture of measles virus and rubella virus. The selectivity of the virus-magnetic-MIPs and NIPs for these viruses with the concentration of 1.0 nmol L−1 is showed in Fig. 3. The results revealed that the virus-magnetic-MIPs exhibited outstanding selectivity for HAV with regard to NIPs. Fig. 3 shows that ΔIRLS of HAV was more superior to that of other viruses, subsequently verifying the outstanding selective recognition of HAV. There are two main reasons for the demonstration of these results. As to spatial interaction, the resultant virus-magnetic-MIPs provided a microenvironment that can precisely match the template virus by presenting the supplementary shape, functional groups, and dimension. The bio-inspired PDA chains exhibited non-covalent functionalities, such as the amine, hydroxyl groups, and p–p bonds, which were contributed to specially bind with the amino acids of proteins on the surface of the virus. Hence, HAV was specifically bound to the PDA film and able to change the RLS intensity. The results demonstrated that the MIP-based RLS assay was successfully used for selective HAV detection.3.6 Interference investigation
To evaluate the reliability of the proposed assay, the interference from substances that may be present in the real specimen was assessed with 1.0 nmol L−1 HAV under the optimized conditions. The results of multiple substances that may exist in human serum, such as Na+, CO32−, Cl−, HPO42−, K+, Mg2+, Ca2+, L-tryptophan, glucose and elastase are listed in Table 1. In this system, with the addition of various ions, the values of ΔIRLS were changed in the range from −5% to +5%. It was a result that this strategy was excellent for feasible and effective detection for target virus.| No. | Coexisting foreign substance | Concentration tolerated (×10−9 M) | Change in IRLS (%) |
|---|---|---|---|
| a Condition: nanospheres dosage: 90 ng mL−1, HAV 1.0 nmol L−1, pH 6.7, temperature: 25 °C. | |||
| 1 | Na+ | 2000 | 0.54 |
| 2 | CO32− | 2000 | −1.82 |
| 3 | L-Tryptophan | 2000 | 0.41 |
| 4 | Cl− | 2000 | 0.59 |
| 5 | HPO42− | 2000 | 0.54 |
| 6 | K+ | 1000 | 0.40 |
| 7 | Glucose | 1000 | 4.60 |
| 8 | L-Phenylalanine | 1000 | 4.77 |
| 9 | Mg2+ | 500 | 1.42 |
| 10 | Ca2+ | 1000 | −1.06 |
| 11 | Albumin | 20 | 4.49 |
| 12 | Elastase | 20 | 2.87 |
| 13 | Papain | 20 | 2.7% |
| 14 | Lysozyme | 20 | 1.53% |
3.7 Analytical performance
Calibration graphs for the relationship between the changes of RLS intensity (ΔIRLS) and increasing standard concentration of HAV (CHAV) were constructed under the optimized conditions for estimating analytical performance. As depicted in Fig. 2, a positive linear relationship between ΔIRLS and CHAV in the range from 0.02 to 1.40 nmol L−1 was showed. The linear regression equation was ΔIRLS = 263.85CHAV − 1.96 with R2 = 0.9964 (n = 7), where ΔIRLS is intensity difference in the presence and absence of HAV at 323 nm, and CHAV is the HAV concentration. The calculation formula of detection limit is LOD = 3Sb/m, where LOD, Sb, and m are detection limit, standard deviation of the blank, and slope of the calibration curve, respectively. The LOD of HAV was calculated as 6.2 pmol L−1 which was lower than our previous works,11,16,18 and the relative standard deviation, which was averaged over seven measurements with HAV concentration of 1.0 nmol L−1, was 1.2%. The obtained strategy exhibited wider linear range and relatively lower detection limit, these results indicate that the sensing strategy has an excellent sensitivity for quantitative detection of HAV. In addition, the performance of the proposed strategy have been compared with the previously reported optical sensors developed on the basis of virus-MIPs, and the corresponding results were listed in Table 2.3.8 Application to real sample analysis
To survey the feasibility of the system for realistic specimen analysis, the additional HAV in the 2000-fold diluent human serum samples and in Tris–HCl buffer solution (pH 6.2) was specifically detected, respectively. The final data and recovery rates are exhibited in Table 3. The results showed excellent recovery, thus proving that this method can be successfully used to determine HAV concentration.| Samples | The concentration of HAV (nmol L−1) | |||
|---|---|---|---|---|
| Spiked | Measured | Recovery (%) | RSD (%) | |
| a Condition: nanospheres dosage: 90 ng mL−1, pH 6.7, temperature: 25 °C. | ||||
| Human serum | 0.00 | (n = 3) | ||
| 0.10 | 0.09 ± 0.03 | 90.0 | 2.2 | |
| 0.90 | 0.89 ± 0.02 | 98.9 | 1.1 | |
| 1.10 | 1.12 ± 0.08 | 101.8 | 0.8 | |
3.9 Cytotoxicity assay
PDA materials possess wide cytocompatibility, so it can be broadly used in biological imaging and considered as biomarkers.33,34 Fe3O4 nano-magnetic materials are already widely applied to tumor treatment, cell separation, microwave-absorbing materials, magnetic recording materials, catalyst carriers, magnetic liquids, medicine and other fields.2 In this study, the MIPs and NIPs were combined with PDA and Fe3O4 NPs. The NIPs were selected for further study, and the cytotoxicity of the NIPs was tested by the common MTT assay with distinct NIP concentrations (0, 1, 3, 10, 30, and 100 mg mL−1) in Chinese hamster fibroblast cells. This strategy is ordinarily applied to assess biocompatibility of synthetic nanoparticles. It is exhibited in Fig. 4 that more than 95% of cell viability was observed after 24 h in optimal experimental conditions for co-culture. Thus, the NIP materials showed low cytotoxicity for animal cells in vitro.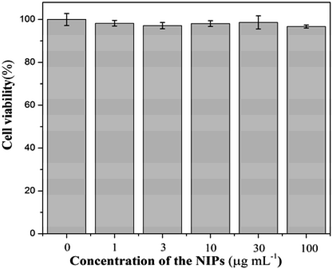 | ||
| Fig. 4 MTT assay for the viabilities of Chinese hamster fibroblast cell treated with various concentrations of NIPs (0, 1, 3, 10, 100 μg mL−1) for 24 h at 37 °C with 5% CO2. | ||
4. Conclusions
In summary, a novel virus-magnetic-MIP optical sensor was applied for the high-sensitivity and high-selectivity detection of trace quantities of HAV. The applied analytical capability of the virus-magnetic-MIPs was satisfactorily determined by evaluating the detection of HAV in human serum. The results showed that the proposed strategy was simple, rapid, and highly selective, thereby providing a novel and promising technology for clinical and bioanalytical virus quantification. But the incubation time has yet to be further shortened, which can be achieved by making the imprinting layer as thinner as possible. Based on the outstanding biocompatibility of PDA-coated and Fe3O4 molecularly imprinted NPs, this strategy is promising to further develop for cell and live imaging. Moreover, the development of correlative detection for other viruses by using the proposed strategy is presently in progress.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 21775132, 21305118), Scientific Research Foundation of Hunan Provincial Education Department (No. 16A204), and the National Natural Science Foundation of Hunan province (No. 2018JJ2388). Hunan 2011 Collaborative Innovation Center of Chemical Engineering & Technology with Environmental Benignity and Effective Resource Utilization, and “1515” academic leader team program of Hunan Agricultural University.Notes and references
- R. Chalasani and S. Vasudevan, ACS Nano, 2013, 7, 4093–4104 CrossRef PubMed.
- J. Xie, K. Chen, H.-Y. Lee, C. Xu, A. R. Hsu, S. Peng, X. Chen and S. Sun, J. Am. Chem. Soc., 2008, 130, 7542–7543 CrossRef PubMed.
- F. Pang, R. Zhang, D. Lan and J. Ge, ACS Appl. Mater. Interfaces, 2018, 10, 4929–4936 CrossRef PubMed.
- P. Hu, C. Z. Huang, Y. F. Li, J. Ling, Y. L. Liu, L. R. Fei and J. P. Xie, Anal. Chem., 2008, 80, 1819–1823 CrossRef PubMed.
- Y. Zhang, D. Li, M. Yu, W. Ma, J. Guo and C. C. Wang, ACS Appl. Mater. Interfaces, 2014, 6, 8836–8844 CrossRef PubMed.
- J. Wackerlig and R. Schirhagl, Anal. Chem., 2016, 88, 250–261 CrossRef PubMed.
- Y. Kitayama, K. Yoshikawa and T. Takeuchi, Macromolecules, 2017, 50, 7526–7534 CrossRef.
- H. Li and L. Wang, ACS Appl. Mater. Interfaces, 2013, 5, 10502–10509 CrossRef PubMed.
- Z. Altintas, J. Pocock, K.-A. Thompson and I. E. Tothill, Biosens. Bioelectron., 2015, 74, 996–1004 CrossRef PubMed.
- Z. Altintas, M. Gitttens, A. Guerreiro, K.-A. Thompson, J. Walker, S. Piletsky and I. E. Tothill, Anal. Chem., 2015, 87, 6801–6807 CrossRef PubMed.
- F. L. Dickert, O. Hayden, R. Bindeus, K. J. Mann, D. Blaas and E. Waigmann, Anal. Bioanal. Chem., 2004, 378, 1929–1934 CrossRef PubMed.
- A. Cumbo, B. Lorber, P. F.-X. Corvini, W. Meier and P. Shahgaldian, Nat. Commun., 2013, 4, 1503–1509 CrossRef PubMed.
- R. F. Pasternack and P. J. Collings, Science, 1995, 269, 935–939 CrossRef PubMed.
- R. F. Pasternack, C. Bustamante, P. J. Collings, A. Giannetto and E. J. Gibbs, J. Am. Chem. Soc., 1993, 115, 5393–5399 CrossRef.
- K. Wang, X. Qiu, C. Dong and J. Ren, ChemBioChem, 2010, 8, 1126–1129 CrossRef PubMed.
- C. Wu, X. Song, Z. Song, Z. He and J. Dai, J. Nanosci. Nanotechnol., 2018, 18, 2680–2685 CrossRef PubMed.
- Z. Chen, L. Zhu, T. Song, J. Chen and Z. Guo, Spectrochim. Acta, Part A, 2009, 72, 518–522 CrossRef PubMed.
- C. Liang, K. He, H. Wang, C. Chen, X. Chen and C. Cai, Talanta, 2016, 160, 360–366 CrossRef PubMed.
- K. He, C. Chen, C. Liang, B. Yang, C. Cai and X. Chen, Sens. Actuators, B, 2016, 233, 607–614 CrossRef.
- B. Yang, S. Lv, C. Liang, K. He, C. Chen, X. Chen and C. Cai, Biosens. Bioelectron., 2017, 87, 679–685 CrossRef PubMed.
- F. Deinhardt, Vaccine, 1992, 10, S10–S14 CrossRef PubMed.
- X. Wang, J. Ren, Q. Gao, Z. Hu, Y. Sun, X. Li, D. J. Rowlands, W. Yin, J. Wang, D. I. Stuart, Z. Rao and E. E. Fry, Nature, 2015, 517, 85–88 CrossRef PubMed.
- N. Fuster, R. M. Pintó, C. Fuentes, N. Beguiristain, A. Bosch and S. Guix, Water Res., 2016, 101, 226–232 CrossRef PubMed.
- L. Zhao, D. Chen and W. Hu, Langmuir, 2016, 32, 5285–5290 CrossRef PubMed.
- Z. Xia, Z. Lin, Y. Xiao, L. Wang, J. Zheng, H. Yang and G. Chen, Biosens. Bioelectron., 2013, 47, 120–126 CrossRef PubMed.
- M. Nurunnabi, Z. Khatun, M. Nafiujjaman, D.-G. Lee and Y.-K. Lee, ACS Appl. Mater. Interfaces, 2013, 5, 8246–8253 CrossRef PubMed.
- R. Liu, M. Sha, S. Jiang, J. Luo and X. Liu, Talanta, 2014, 120, 76–83 CrossRef PubMed.
- A. Nematollahzadeh, A. Shojaei, M. J. Abdekhodaie and B. Sellergren, J. Colloid Interface Sci., 2013, 404, 117–126 CrossRef PubMed.
- H. Li, T. Chen, L. Jin, Y. Kan and B. Yin, Anal. Chim. Acta, 2014, 852, 203–211 CrossRef PubMed.
- H. Li, C. Wang, M. She, Y. Zhu, J. Zhang, Z. Yang, P. Liu, Y. Wang and J. Li, Anal. Chim. Acta, 2015, 900, 97–102 CrossRef PubMed.
- H. Lee, S. M. Dellatore, W. M. Miller and P. B. Messersmith, Science, 2007, 318, 426–430 CrossRef PubMed.
- W. Hergert and T. Wriedt, The mie theory. Basics and applications, Springer, Berlin. 2012 Search PubMed.
- R. Mrówczyński, ACS Appl. Mater. Interfaces, 2018, 10, 7541–7561 CrossRef PubMed.
- X. Liu, J. Cao, H. Li, J. Li, Q. Jin, K. Ren and J. Ji, ACS Nano, 2013, 7, 9384–9395 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra06204h |
| This journal is © The Royal Society of Chemistry 2018 |