Open Access Article
Open Access ArticleScreening of potential IL-tolerant cellulases and their efficient saccharification of IL-pretreated lignocelluloses†
Yi-Xin Sun a,
Bing-Bing Shena,
Hui-Ying Hana,
Yuan Lua,
Bi-Xian Zhangb,
Yun-Fei Gaob,
Bao-Zhong Hu*c and
Xiao-Mei Hu*a
a,
Bing-Bing Shena,
Hui-Ying Hana,
Yuan Lua,
Bi-Xian Zhangb,
Yun-Fei Gaob,
Bao-Zhong Hu*c and
Xiao-Mei Hu*a
aCollege of Life Science, Northeast Agricultural University, Harbin, 150030, China. E-mail: huxiaomei1982@163.com
bHeilongjiang Academy of Agricultural Sciences, Harbin, 150086, China
cHarbin University, Harbin, 150086, China. E-mail: bzhu@neau.edu.cn
First published on 3rd September 2018
Abstract
Lignocellulosic biomass as one of the most abundant and renewable resources has great potential for biofuel production. The complete conversion of biomass to biofuel is achieved through the effective pretreatment process and the following enzyme saccharification. Ionic liquids (ILs) are considered as a green solvent for lignocellulose pretreatment. However, ILs exhibit an inhibitory effect on cellulase activity, leading to a subsequent decrease in the efficiency of saccharification. The screening of new potential IL-tolerant cellulases is important. In the current study, a fungal strain with a relatively high cellulase production was isolated and identified as Penicillium oxalicum HC6. The culture conditions were optimized using corn stover and peptone as the carbon source and nitrogen source at pH 4.0 and 30 °C with an inoculation size of 2% (v/v) for 8 days. It was found that P. oxalicum HC6 exhibited potential salt tolerance with the increase of the enzyme production at a salt concentration of 5.0% (w/v). In addition, high enzyme activities were obtained at pH 4.0–6.0 and 50–65 °C. The crude enzyme from P. oxalicum HC6 with good thermal stability was also stable in the presence of salt and ILs. Good yields of reducing sugar were obtained by the crude enzyme from P. oxalicum HC6 after the saccharification of corn stover that was pretreated by ILs. P. oxalicum HC6 with potentially salt-tolerant and IL-tolerant enzymes has great potential application in the enzymatic saccharification of lignocellulose.
1. Introduction
Lignocellulosic biomass can be considered as the most abundant polymer found on Earth. A substantial amount of lignocellulosic materials such as agricultural residues, agricultural by-products, and woody biomass are produced worldwide and their annual production rates are around 200 billion.1 Among a variety of lignocellulosic biomasses, agricultural stover is essentially potential raw material for the production of high value products such as chemical reagents, biosurfactants and biofuel.2–4 Lignocellulosic biomass is mainly composed of three main biopolymers including cellulose, hemicellulose and lignin, which form a complex and recalcitrant structure.5Generally, cellulose and hemicellulose, which comprise nearly two-thirds of lignocellulosic biomass, are preferred as the potential feed-stock for biofuel production.6 A complete cellulase system is required for the synergic action to further convert cellulose into monomeric sugars for the effective production of biofuels. These cellulases comprise three major groups of enzymes: (1) endoglucanases (EC 3.2.1.4), which attack regions of low crystallinity in cellulose fibers, creating free chain-ends, (2) cellobiohydrolases (EC 3.2.1.91), which further degrade the molecule by cleaving cellobiose from the free-chain ends, and (3) β-glucosidases (EC 3.2.1.21) which hydrolyze cellobiose to produce glucose.7
Numbers of microorganisms are known as efficient cellulase producers and fungi are considered as potential sources for cellulase production, because of the large embedded accumulation of mycelium which is easy to separate and hence it helps to reduce the cost of separation process from the fermentation medium.8 Aspergillus sp. and Trichoderma sp. are known as the model fungi among a variety of fungal organisms.9 Trichoderma reesei and its mutants, including certain hyper-producing strains for example, Rut C-30, possess low β-glucosidase activity relative to the total cellulase activity, which impairs performance during the hydrolysis of lignocellulosic materials.10 Considering the lack of a complete cellulase system offered by these fungi, researchers are focusing on the isolation and screening of novel fungi with improved cellulase systems which are capable of acting synergically on the cellulosic biomass.8
Nevertheless, the recalcitrant nature of lignocelluloses is the obstacle in the production of biofuels. The amount of lignin present in the biomass and the de-polymerization (DP) of cellulose limit the transformation efficiency.8 The pretreatment process such as steam explosion, acid or alkaline hydrolysis and ionic liquids-pretreatment are always employed.
Ionic liquids (ILs) have been recognized as an alternative green solvent for lignocellulose pretreatment, which possess several advantages including high thermal stability, negligible vapor pressure, no flammability, and widely accessible temperature range.11,12 For instance, 1-ethyl-3-methylimidazolium acetate ([EMIM]CH3COOH) was reported widely to perform the effective decomposition of lignocelluloses and to provide the high cellulose digestibility.13–16
Although ILs are effective for breaking down lignocelluloses, they can also inhibit the enzymes used in the subsequent saccharification, resulting the inefficient saccharification of cellulose. Therefore, a washing process is required to remove any residual ILs after ILs-pretreatment. Large scale washing requires extra energy, increasing more processing cost.17,18 However, there are very limited reports concerning on the ILs-tolerant cellulase.
In this study, a cellulose-degradable strain was isolated and identified. Factors governing the enzyme production were investigated. Studies on evaluation of the capability of salt-tolerant microbes were carried out. The capability of the potential microbes for producing salt-tolerant and ILs-tolerant cellulase was investigated. We also compared the enzymatic hydrolysis of lignocellulosic substrates by the crude enzyme and commercial cellulase.
2. Materials and methods
2.1 Samples collection
The soil was collected from wheat and corn rotation farm of Shanxian county in Shandong province in China. The surface soil (1–5 cm) was collected and stored at 4 °C. Corn stover and rice straw were provided by Xiangfang farm in Harbin city in China and they were dried, ground and passed through 40 mesh sieves. Wheat bran was provided by Xiangfang Flour Factories of Harbin in China. All chemicals were purchased from Aladdin company in China and were used as received.2.2 Screening procedure
The colonies of various fungi were isolated from the soil using PDA medium. The isolated single colony was incubated on the CMC medium (K2HPO4 1.0 g, NaCl 0.5 g, (NH4)2SO4 2.0 g, MgSO4 0.5 g, CMC-Na 10 g, agar 15 g) at 30 °C for 5 days. Following that, all plates were stained with Congo-red solution (0.1%, w/v) for 15 min and were discolored with 1 mol L−1 NaCl.19 The strain with a large hydrolysis zone was selected and cultured on PDA medium for 4 days. After that, 1 cm of agar piece was taken and transferred into 30 mL of PD medium. The solutions were then cultured at 30 °C for 3 days with shaking at 200 rpm. 1 mL of the suspension (4–6 mg dry mycelia wt per mL) was added into 100 mL of Hutchinson inorganic salt medium (KH2PO4 1.0 g, MgSO4 0.3 g, NaCl 0.1 g, CaCl2 0.1 g, FeCl3 0.01 g) with cover stover of 1% (w/v) and peptone of 0.2% (w/v), which was incubated at 30 °C for 5 days with shaking at 150 rpm. The resulting solution was then centrifuged at 8000 rpm for 5 min at 4 °C to give the crude enzyme.2.3 Enzyme assays
Enzyme assays, including filter paper cellulase (FPase), endoglucanase (CMCase) and β-glucosidase, were determined as International Union of Pure and Applied Chemistry (IUPAC) standard procedure.20 FPase activity was assayed by incubating the 0.5 mL of suitably diluted enzyme with Whatman no. 1 filter paper (1.0 × 6.0 cm) containing 1.0 mL of sodium citrate buffer (pH 4.8) for 60 min at 50 °C. The CMCase activity was determined using (1%, w/v) sodium carboxymethyl cellulose (CMC-Na) for 30 min at 50 °C. β-glucosidase activity was measured using salicin solution (0.5%, w/v) for 30 min at 50 °C. The reducing sugar was determined by 3,5-dinitrosalicylic acid (DNS) method.21 One unit (U) of enzyme activity was defined as the amount of the enzyme which released 1 μmol of reducing sugar/min under the conditions indicated.2.4 Molecular identification
The DNA of the enriched fungus was extracted using a DNA kit according to the manufacturer's instructions (OMEGA, USA). Universal primers of ITS1: (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4: (5′-TCCTCGCCTTATTGATATGC-3′) were used for fungal analysis. The ITS sequence of the gene was amplified by polymerase chain reaction (PCR). The reaction mixture of 50 μL contained 2 μL template DNA, 2 μL forward primer, 2 μL reverse primer, 25 μL 2 × Taq PCR MasterMix (TIANGEN, China) and 19 μL ddH2O. The samples were amplified with the following conditions: initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 35 s, and elongation at 72 °C for 2 min, and final elongation at 72 °C for 10 min. The PCR products were sequenced in Huada gene company (Beijing, China). The sequence was compared to the sequences in the GenBank database. A homology search was performed using bioinformatics tools available online: BLAST (www.ncbi.nlm.nih.gov/BLAST). MEGA program (version 5.0) with the neighbor-joining (NJ) algorithm was used to build phylogenetic trees based on the ITS sequence.22 The ITS sequence data for fungal sample were deposited into the GenBank database.2.5 Optimization of the conditions for cellulase production
Various factors were investigated in order to optimize the environmental conditions of cellulase production. The effects of carbon sources (corn stover, rice straw, wheat bran, CMC-Na, avicel) at a concentration of 1% (w/w) and nitrogen sources (peptone, beef extract, yeast extract, NH4NO3, urea) at a concentration of 0.2% (w/w) on the enzyme activity were determined on the basis of Hutchinson inorganic salt medium. Other factors including pH value, inoculum size, incubation temperature and time were evaluated.2.6 Determination of salt-tolerance of the fungi
The capability of the fungi on cellulase production under different salt concentrations were determined. According to Wang et al. and Taechapoempol et al.,23,24 a modified method was used in our work. Based on the optimal conditions for cellulase production, different NaCl concentrations including 0%, 2.5%, 5.0%, 7.5% and 10.0% (w/v) was added and investigated. FPase, CMCase and β-glucosidase activity were measured as the method described in 2.3.2.7 Optimization of the conditions for enzyme activities for enzymatic hydrolysis
The crude enzyme solution was obtained under the optimal conditions for cellulase production. The effect of pH value in the range of pH 3.0–11.0 was estimated, which was prepared by citrate buffer (pH 3.0–6.0), sodium phosphate buffer (pH 7.0–8.0) and glycine–NaOH buffer (pH 9.0–11.0). The incubation temperature (30–80 °C) with 5 °C intervals was studied. The mixture was cultured for 1 h. FPase, CMCase and β-glucosidase activities were measured as the method described in 2.3.2.8 Determination of enzyme activities in the presence of salt and ionic liquid
The 1-ethyl-3-methylimidazolium acetate ([EMIM]CH3COOH) was prepared using the published methods in the literature.25 According to Gunny et al.,26 a modified method was used in our work. To evaluate the stability of cellulase at different salt and ILs concentrations, the crude enzyme solution was mixed with citrate buffer at the pH 5.0 with the addition of NaCl or [EMIM]CH3COOH in the range of 0–15.0% (w/v) and was cultured for 1 h at 60 °C. FPase, CMCase and β-glucosidase activities were measured as the method described in 2.3.2.9 Pretreatment of corn stover
The pretreatment of corn stover (1 g) was performed in [EMIM]CH3COOH (20 g) at 120 °C for 3 h in an 100 mL autoclave. After that, the mixture was cooled to room temperature quickly and 80 mL of acetone/deionized water (1/1, v/v) was added into the mixture. The resulting solid was filtered and washed with 80 mL of acetone/deionized water (1/1, v/v), followed by 300 mL of deionized water in order to remove any residual ionic liquid. The solid was then dried in a vacuum at 60 °C to give the cellulose-rich materials. The yield of cellulose-rich materials was determined as the ratio of the mass of cellulose-rich materials in the mass of the corn stover that was subjected to the pretreatment process according to eqn (1), and the yield was 60.23%.
 | (1) |
2.10 Analysis of corn stover after ILs-pretreatment
According to National Renewable Energy Lab (NREL) method, 300 mg of corn stover that was pretreated by [EMIM]CH3COOH or corn stover was mixed with 3 mL 72% H2SO4 in a water bath at 30 °C for 60 min. The mixture was stirred strongly every 10 min. Following that, 84 mL deionized water was added into the mixture, which is diluted into 4% H2SO4 aqueous solution. The mixture was put in an autoclave at 121 °C for 60 min. After the mixture was cooled to room temperature, the mixture was centrifuged. The filtrate was then determined for glucose and xylose using HPLC analysis. Measure the absorbance of the sample at 320 nm with ε=(30 L g−1 cm−1) on a UV-visible spectrophotometer. The filtered solids were put into a muffle furnace at 575 °C to give the acid-insoluble lignin and ash. The content of cellulose and hemicellulose was calculated from glucose and xylose by conversion factors of 0.90 and 0.88, respectively.11,27 There are cellulose (30.55%), hemicellulose (14.40%), lignin (22.47%) and ash (1.3%) in original corn stover. There are cellulose (33.20%), hemicellulose (20.54%), lignin (21.98%) and ash (1.0%) in regenerated cellulose-rich materials.2.11 Enzymatic hydrolysis
50 mg of corn stover that was pretreated by [EMIM]CH3COOH was mixed with an appropriate amount of the crude enzyme (0.02 U FPase/mg of substrate) that was filtered through 0.22 μm filter membrane in a 50 mL Erlenmeyer flask. The total volume of the reaction mixture was replenished to 20 mL using citrate buffer (0.05 mol L−1) at pH 5.0. The reaction mixture was incubated under a rotary shaker at 60 °C for 72 h with 150 rpm. The commercial cellulase (from Trichoderma reesei ATCC26921) was used as a positive control. A commercial cellulase reaction system (enzyme dosage equal to crude enzyme) in 20 mL of citrate buffer (0.05 mol L−1, pH 4.8) served as a substitute of the culture supernatants from T. reesei ATCC26921. The enzymatic hydrolysis was carried out for 72 h at 50 °C with 150 rpm. The control experiments were carried out under the same conditions using the denatured commercial enzymes or the denatured crude enzyme that was boiled for 30 min. The reducing sugars were measured by 3,5-dinitrosalicylic acid (DNS) method as described by Miller.21 The yields of glucose, xylose and cellobiose were measured by HPLC. The sugar saccharification rate of 1U FPase was calculated according to eqn (2).
 | (2) |
3. Results and discussion
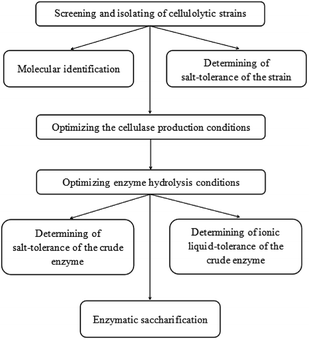
The detailed schematic representation has been provided as Fig. 1.3.1 Isolation and identification of the microbe
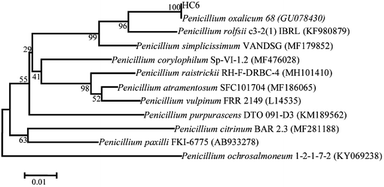
Five microbial strains with discoloration zones of Congo-red were isolated as the positive microbes, indicating their ability to secrete cellulase. The microbe with a large discoloration zone was selected and determined for its enzyme activities, which were 0.11 U mL−1 (FPase), 0.21 U mL−1 (CMCase) and 0.43 U mL−1 (β-glucosidase), respectively.This fungal strain was determined based on its ITS sequence of DNA. The obtained nucleotide sequence has been submitted to NCBI GenBank with under accession no. MH558553. Phylogenetic tree was shown in Fig. 1. The ITS sequence of HC6 showed 100% sequence similarity with that of Penicillium oxalicum strain 68 (accession no. GU078430). Thus, this fungal strain was identified as Penicillium oxalicum HC6.
Many strains of Penicillium sp. such as Penicillium janthinellum EMS-UV-8 and Penicillium decumbens JU-A10 were increasingly being reported to produce cellulase at a commercial level.28,29 13 species of Penicillium sp. exhibiting avicelase activity among 31 fungal strain were isolated from subtropical and tropical forests of China.30 The newly isolated P. oxalicum HC6 has a potential to possess the cellulase production (Fig. 2).
3.2 Optimization of the conditions for the cellulase production
Recently, lignocellulosic biomasses have been studied as potential substrates for the enzyme production using microorganisms, especially filamentous fungi. Similar results were reported that high amounts were obtained from Penicillium oxalicum GZ-2 and Penicillium sp. GDX01 from untreated lignocellulosic materials compared with commercial polymeric materials.31,32 The chemical composition of lignocellulosic biomass and the accessibility of various components of corn stover and rice straw, especially hemicelluloses, improved the enzyme hydrolysis by P. oxalicum HC6. The use of economical agricultural wastes instead of expensive cellulose is of the great interest for industrial application.
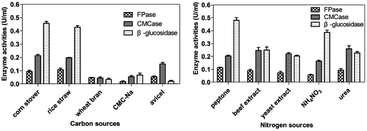
Nitrogen source is also one of the crucial factors that affect microbial metabolism.33 In this work, three nitrogen organic sources (peptone, beef extract, yeast extract) and two inorganic nitrogen sources (NH4NO3, urea) were selected for the optimization of nitrogen sources at 30 °C and pH 7.2 with corn stover and incubation size of 1% (v/v) under 150 rpm for 5 days. As shown in Fig. 3b, urea was the most effective nitrogen source for CMCase production of P. oxalicum HC6. High production of FPase and β-glucosidase were obtained, when peptone was employed as the nitrogen source.
Variation in the nitrogen source can affect the metabolic processes of the microbes significantly to produce different cellulase production. Previous studies have shown that organic nitrogen sources can induce cellulase secretion better than inorganic nitrogen sources.34,35 Peptone was the suitable organic nitrogen source for most cellulases for P. oxalicum HC6.
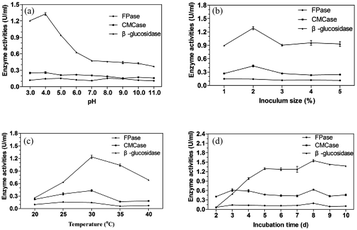 | ||
| Fig. 4 Effects of pH (a), inoculation size (b), temperature (c) and incubation time (d) on the cellulase production. | ||
Many enzyme systems and the transport of several enzyme species across the cell membrane are influenced by the initial pH of medium.36 Moreover, the synthesis and expression of certain genes, and the microbial metabolic activities, are influenced by the internal pH, in response to that of the external environment.37 The preference for acidic pH for cellulase producing microbes has been reported for ascomycetes fungi e.g. T. reesei,38 Talaromyces verruculosus,39 and Chaetomium globosum.40 It was reported that good CMCase activity was observed with Penicillium funiculosum (pH 4.82), Penicillium citrinum (pH 5.5), Penicillium chrysogenum (pH 5.0), and Aspergillus niger (pH 5.0).41–44 As reported, filamentous fungi have reasonably good growth over a broad range of pH 2–9, with an optimal range of 3.8 to 6.0.45 Each microorganism possesses a pH range for its growth and activity with an optimum value within the range. The acidic pH 4 was favorable for the P. oxalicum HC6.
The investigation of inoculation size was performed at 30 °C and pH 4.0 for 5 days and the result was shown in Fig. 4b. The enzyme production was increased with the inoculation size from 1% to 2% and reduced from 2% to 3%, which was then stable from 3% to 5%. Good CMCase, FPase and β-glucosidase activities were obtained with the inoculation size of 2%. Lower inoculation size requires longer time for fungal multiplication and substrate utilization, whereas higher inoculation size increases the spore density as well as the water content in the medium causing hindrance in oxygen penetration resulting in the inhibited fungal growth and enzyme production.46 A balance between nutrients and growing microbes is necessary for optimum enzyme production.
As presented in Fig. 3c, the maximum CMCase and β-glucosidase activities were found at 30 °C with inoculation size of 2% at pH 4.0 for 5 days. Good FPase activity was obtained when the fermentation temperature was 25 °C. When the temperature was increased more than 30 °C, the enzyme activities were reduced significantly. At high temperature, a lower amount of enzyme will be formed due to the thermal denaturation of the metabolic pathway of enzymes.47 However, at low temperature, metabolic activities of microorganisms slow down resulting in low enzyme yield.48
The effect of fermentation time on the cellulase production of P. oxalicum HC6 was illustrated in Fig. 4d. As the fermentation time was increased from 2 days to 8 days, nutrients were efficiently used by microbes that resulted in high enzyme production. After 8 days, the enzyme yield was reduced due to the depletion of nutrients. In addition, higher β-glucosidase/FPase ratio has been previously reported to reduce enzyme inhibition due to lesser cellobiose accumulation during hydrolysis.49,50 Therefore, the crude enzyme from P. oxalicum HC6 with high β-glucosidase/FPase ratio possibly help to decrease enzyme inhibition and to improve glucose production instead of cellobiose.
Based on the optimum conditions for cellulase production of P. oxalicum HC6, the activities of FPase, CMCase and β-glucosidase were 0.20 U mL−1, 0.63 U mL−1, 1.55 U mL−1, when cron stover and peptone were used as the carbon source and nitrogen source, at pH 4.0 and 30 °C with inoculation size of 2% for 8 days.
3.3 Analysis of salt tolerant capability of P. oxalicum HC6
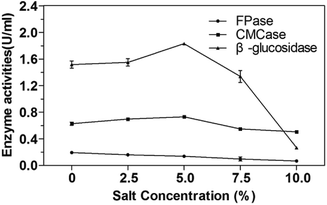
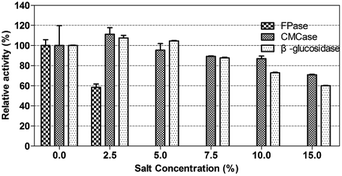
The salt tolerance of P. oxalicum HC6 was determined by selecting different NaCl concentrations of 0–10.0% (w/v). The results were shown in Fig. 5. It was obvious that the activities of CMCase and β-glucosidase of P. oxalicum HC6 were increased with an increase of NaCl concentration in the range of 0–5.0%. High activities of CMCase and β-glucosidase were observed at 0.73 U mL−1 and 1.83 U mL−1 with 5.0% NaCl, respectively, which were increased to 115.87% and 118.06% compared with the enzyme activities in the absence of NaCl. The results showed that CMCase and β-glucosidase could be promoted at an appropriate NaCl concentration. However, FPase activity was inhibited at the presence of NaCl. Smolyanyuk et al. described that salt tolerant fungi could grow at the salt concentration of more than 0.5 mol L−1.51 P. oxalicum HC6 with the salt tolerance of more than 0.7 mol L−1 in this study has an effective salt tolerance capability with high cellulase activity.3.4 Optimization of the conditions for enzyme activities for enzymatic hydrolysis
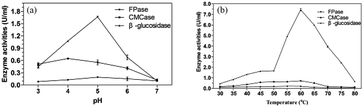
The study for enzyme activities for enzymatic hydrolysis under different pH was carried out at 50 °C (Fig. 6a). CMCase and β-glucosidase showed good activities over a broad acidic pH range of 3.0–6.0. The optimal pH was obtained at pH 4.0 for CMCase (0.65 U mL−1) and pH 5.0 for β-glucosidase (1.67 U mL−1). It was reported that the highest activity for CMCase of Penicillium sp. was 0.409 U mL−1 at the optimal pH 4.2 and highest activity of β-glucosidase was 0.038 U mL−1 at pH 6.0.52 The degradation of lignocellulose requires the cooperation of several cellulases and their synergistic effect. Considering this, pH 5.0 was selected for the following studies.In addition, the thermostable cellulase is an ideal candidate for bioprocessing industry, as a relatively high temperature is favorable for the solubility of the substrate and the efficiency of the enzymatic hydrolysis.53 High temperature could also reduce the risk of contamination by undesirable microorganisms.54 Castro et al. reported an optimum temperature of 60 °C for both endoglucanase and β-glucosidase activities with Penicillium funiculosum ATCC 11797.55
Generally, thermophilic microorganisms having growth temperature ranges from 50 to 80 °C are potential candidates to produce highly thermostable cellulase. Interestingly, several mesophilic microorganisms were reported to produce a considerable thermostable cellulases.56 In this study, although the high enzyme production of P. oxalicum HC6 were obtained at 30 °C, CMCase and β-glucosidase reached their maximum activities at 60 °C, which were 0.71 U mL−1 and 7.44 U mL−1, respectively. It is worth to note that when the temperature was increased to 60 °C, the activities of β-glucosidase and CMCase were increased to 445.51% and 129.09%, respectively. The cellulase of P. oxalicum HC6 with high pH and thermal stability has great potential for the industrial application.
3.5 Stability of enzyme activity of P. oxalicum HC6 under NaCl and ILs
As reported, the endoglucanase from Bacillus agaradhaerens JAM-KU023 was a salt-activated endoglucanase by adding 0.2–2.0 mol L−1 NaCl.57 The cellulase from Aspergillus terreus UniMAP AA-6, was reported to be stable under NaCl and [EMIM]CH3COOH solution. The highest FPase activity of A. terreus UniMAP AA-6 was 0.0625 U mL−1 in the presence of 7.70% NaCl (w/v).58The crude enzyme of P. oxalicum HC6 in this work was determined under different NaCl solution (0–15.0%, w/v) at 60 °C with initial pH 5.0 for 1 h. Every enzyme activity (FPase, CMCase and β-glucosidase) was considered as 100% at NaCl concentration of 0%. As shown in Fig. 7, β-glucosidase activity was increased as NaCl concentration was increased from 0% to 2.5%. The optimal β-glucosidase activity reached 8.00 U mL−1, corresponding to 107.53% of the enzyme activity. At a higher NaCl solution of 15.0%, β-glucosidase activity still remained 4.47 U mL−1, corresponding to 60% of the enzyme activity. The optimal CMCase activity was achieved at 0.79 U mL−1, corresponding to 111.27% of the enzyme activity. At a higher NaCl solution of 15.0%, CMCase activity still remained 0.50 U mL−1, corresponding to 70.42% of the enzyme activity.
It was described that the excess acidic amino acids on the cellulases could provide a negatively charge on its surface. The negative charges keep the protein soluble in a high salt solution either by forming a hydrated ion network with cations or by inhibiting the formation of protein aggregation through electrostatic repulsive charges at the protein surface.59
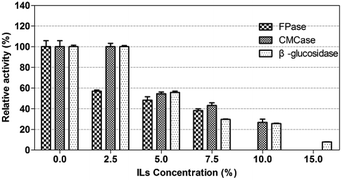
The crude enzyme from P. oxalicum HC6 was further incubated in [EMIM]CH3COOH with different concentrations. The result was shown in Fig. 8. Every enzyme activity (CMCase, FPase and β-glucosidase) was considered as 100% at ILs concentration of 0%. It was found that CMCase and β-glucosidase showed good stability under the ILs concentration of 0–2.5% (w/v), which maintained almost 100% of the enzyme activity. However, β-glucosidase activity was reduced from 100.02% to 25.67%, when the concentration of ILs was increased from 2.5% to 10.0%. CMCase activity was reduced from 100% to 26.76%, when the concentration of ILs was increased from 2.5% to 10.0%. A relatively lower stability was observed for FPase in the presence of [EMIM]CH3COOH.
ILs as effective pretreated method for lignocelluloses was reported widely. However, numbers of extensive washing with water after ILs-pretreatment were required, because the commercial fungal cellulases were inhibited by some ILs,60,61 which increased the production cost. In order to decrease the number of washing steps and increase the yields of sugars, it is necessary to identify ILs-tolerant enzymes for enzymatic hydrolysis of the regenerated cellulose in the presence of ILs. The cellulase from P. oxalicum HC6 in this work was potentially ILs-tolerant enzymes for further application.
It has been suggested that ILs inhibit enzymatic activity by disrupting hydrogen bonding and hydrophobic interactions and depriving the water hydration shell of the protein.62,63 This is similar to the denaturing effect caused by salt on mesophilic proteins. High ionic strength could be the possible reason for both salt and ILs that caused the denaturing effect of enzymes.
3.6 Analysis of scanning electron microscopy
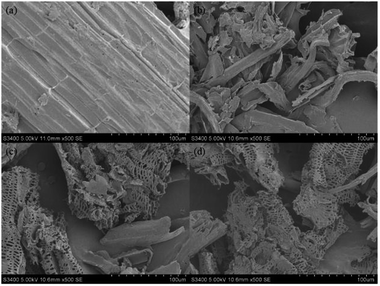
The structural changes in morphology of the corn stover that was pretreated by [EMIM]CH3COOH and the following crude enzyme of P. oxalicum HC6 were investigated. SEM images were taken on a Hitachi S–3400N microscope. The original corn stover showed the compact ordered and rigid structures (Fig. 9a). The significant morphological modification was observed after [EMIM]CH3COOH-pretreatment, which was loose and less compact (Fig. 9b). After the treatment by the crude enzyme (Fig. 9c) and the enzyme from T. reesei ATCC26921 (Fig. 9d), the surface of the residue become more loose and porous. The alteration of morphology indicated the disruption of linkages in lignocellulose, providing a more accessible surface to cellulase, leading to enhanced enzymatic digestibility.3.7 Enzymatic saccharification of the crude enzyme of P. oxalicum HC6
FPase activity was added as 1 U for both the commercial enzyme and the crude enzyme of P. oxalicum HC6. After enzymatic hydrolysis, a high yield of reducing sugar (12.29 mg) was obtained after the saccharification by the crude enzyme of P. oxalicum HC6. The total saccharification degree of cellulose-rich materials was up to 24.58%. Sugars were determined by HPLC (Agilent 1200 Series, USA) analysis. There are 8.93 mg of glucose and 1.64 mg of xylose. In addition, a good yield of reducing sugar (10.70 mg) was obtained after the saccharification by the commercial cellulase and the total saccharification degree was up to 21.40%. 3.46 mg of glucose, 1.73 mg of xylose and 5.05 mg of cellobiose were achieved. No significant difference was observed between P. oxalicum HC6 and commercial cellulase with the same FPase activity.Specifically, higher yield of glucose was obtained, but no cellobiose was observed for the treatment by the crude enzyme of P. oxalicum HC6 compared with the commercial cellulase T. reesei ATCC26921, because of the capability of P. oxalicum HC6 to produce a high β-glucosidase activity. An optimal ratio of β-glucosidase/FPase is critical. Although Trichoderma sp. has been reported to produce good quantities of cellulolytic enzymes, it possess low β-glucosidase activity, which reduce the hydrolysis rate of lignocellulose to glucose.10 Moreover, the lack of β-glucosidase activity of T. reesei cellulase could result in inhibition of the cellulase by accumulated cellobiose in the reaction.64 As a result, the addition of external β-glucosidase enzymes from different microbial origins was always required. Penicillium sp. have the ability to produce complete enzyme mixture with a better β-glucosidase/FPase ratio than Trichoderma sp.65
It was reported that the crude enzyme from Penicillium citrinum CBMAI 1186, Aspergillus sydowii CBMAI 934, Aspergillus sydowii CBMAI 935 and Mucor racemosus CBMAI 847d produced 19%, 19%, 78% and 24% of the saccharification with the addition of FPase (1 U mL−1) for sugarcane bagasse after the alkaline treatment.66 The crude enzyme from Geobacillus stearothermophilus resulted in 71.03% of the saccharification with the addition of FPase (30 U mL−1) for the date palm wastes after the alkaline treatment.67 P. oxalicum HC6 in this study has a potential for its application in enzymatic saccharification.
4. Conclusions
In this study, a fungal strain was newly isolated and identified as Penicillium oxalicum HC6. Under the optimal conditions using corn stover and peptone as the carbon source and nitrogen source at pH 4.0 and 30 °C with inoculation size of 2% (v/v) for 8 days, a relatively high production of FPase, CMCase and β-glucosidase were obtained as 0.20 U mL−1, 0.63 U mL−1 and 1.55 U mL−1, respectively. It was found that the enzyme production of CMCase and β-glucosidase was increased to 0.73 U mL−1 and 1.83 U mL−1 at the presence of NaCl (5.0%, w/v). Good activities of FPase, CMCase and β-glucosidase were observed at 0.20 U mL−1, 0.71 U mL−1 and 7.44 U mL−1 at the optimal pH 5.0 and the optimal temperature of 60 °C. The enzyme activity was stable in the presence of salt and ILs. Hydrolytic efficiency of Penicillium oxalicum HC6 was comparable to the commercial cellulase preparation, which indicated its great potential for its application in the conversion of lignocelluloses to biofuels.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by University Nursing Program for Young Scholars with Creative Talents in Heilongjiang Province (UNPYSCT-2017021).References
- Z. Anwar, M. Gulfraz and M. Irshad, J. Radiat. Res. Appl. Sci., 2014, 7, 163–173 CrossRef.
- A. Pandey, C. R. Soccol, P. Nigam and V. T. Soccol, Bioresour. Technol., 2000, 74, 69–80 CrossRef.
- S. S. Behera and R. C. Ray, Int. J. Biol. Macromol., 2016, 86, 656–669 CrossRef PubMed.
- O. Kirk, T. V. Borchert and C. C. Fuglsang, Curr. Opin. Biotechnol., 2002, 13, 345–351 CrossRef PubMed.
- M. Knauf and M. Moniruzzaman, Int. Sugar J., 2004, 106, 147–150 Search PubMed.
- M. Chandra, A. Kalra, P. K. Sharma, H. Kumar and R. S. Sangwan, Biomass Bioenergy, 2010, 34, 805–811 CrossRef.
- R. L. Howard, E. Abotsi, E. L. J. van Rensburg and S. Y. Howard, Afr. J. Biotechnol., 2003, 2, 602–619 CrossRef.
- N. Srivastava, M. Srivastava, P. K. Mishra, V. K. Gupta, G. Molina, S. Rodriguez-Couto, A. Manikanta and P. W. Ramteke, Renewable Sustainable Energy Rev., 2018, 82, 2379–2386 CrossRef.
- R. Rawat, N. Srivastava, B. S. Chadha and H. S. Oberoi, Energy Fuels, 2014, 28, 5067–5075 CrossRef.
- H. Jorgensen, A. Morkeberg, K. B. R. Krogh and L. Olsson, Enzyme Microb. Technol., 2005, 36, 42–48 CrossRef.
- P. Zhang, S. J. Dong, H. H. Ma, B. X. Zhang, Y. F. Wang and X. M. Hu, Ind. Crops Prod., 2015, 76, 688–696 CrossRef.
- P. Mäki-Arvela, I. Anugwom, P. Virtanen, R. Sjöholm and J. P. Mikkola, Ind. Crops Prod., 2010, 32, 175–201 CrossRef.
- C. G. Liu, J. C. Qin, L. Y. Liu, B. W. Jin and F. W. Bai, ACS Sustainable Chem. Eng., 2016, S1, 577–582 CrossRef.
- S. D. Wang, J. C. Chen, G. H. Yang, W. H. Gao and K. F. Chen, Bioresour. Technol., 2017, 243, 335–338 CrossRef PubMed.
- S. M. Raeisi, M. Tabatabaei, A. Bita, A. Ghafari and S. H. Mood, Waste Biomass Valorization, 2016, 7, 97–107 CrossRef.
- F. Xu, Y. C. Shi and D. Wang, Bioresour. Technol., 2012, 114, 720–724 CrossRef PubMed.
- P. Engel, R. Mladenov, H. Wulfhorst, G. Jager and A. C. Spiess, Green Chem., 2010, 12, 1959–1966 RSC.
- T. Zhang, S. Datta, J. Eichler, N. Ivanova, S. D. Axen, C. A. Kerfeld, F. Chen, N. Kyrpides, P. Hugenholtz and J. F. Cheng, Green Chem., 2011, 13, 2083–2090 RSC.
- R. M. Teather and P. J. Wood, Appl. Environ. Microbiol., 1982, 43, 777–780 Search PubMed.
- V. K. Ghose, Pure Appl. Chem., 1987, 59, 257–268 Search PubMed.
- G. L. Miller, Anal. Chem., 1959, 31, 426–428 CrossRef.
- J. Felsenstein, Evolution, 1985, 39, 739–791 CrossRef PubMed.
- C. Wang, Y. Hsieh, C. Ng, H. Chan, H. Lin, W. Tzeng and Y. Shyu, Enzyme Microb. Technol., 2009, 44, 373–379 CrossRef.
- K. Taechapoempol, T. Sreethawong, P. Rangsunvigit, W. Namprohm, B. Thamprajamchit, S. Rengpipat and S. Chavadej, Appl. Biochem. Biotechnol., 2011, 164, 204–219 CrossRef PubMed.
- P. Wasserscheid and T. Welton, Ionic liquids in synthesis, WILEY–VCH, Federal republic of Germany, 2003, vol. 2, ISBN:3–527–30515–7 Search PubMed.
- A. A. N. Gunny, D. Arbain, R. E. Gumba, B. C. Jong and P. Jamal, Bioresour. Technol., 2014, 155, 177–181 CrossRef PubMed.
- A. Sluiter, B. Hames, R. Ruiz, C. Scarlata, J. Sluitter, D. Templeton and D. Crocker, National Renewable Energy Laboratory, 2011, 1–15 Search PubMed.
- M. G. Adsul, K. B. Bastawde, A. J. Varma and D. V. Gokhale, Bioresour. Technol., 2007, 98, 1467–1473 CrossRef PubMed.
- Y. Cheng, X. Song, Y. Qin and Y. Qu, J. Appl. Microbiol., 2009, 107, 1837–1846 CrossRef PubMed.
- J. Zhang, J. L. Liu, J. Y. Lan, C. J. Duan, Q. S. Ma and J. X. Feng, Biotechnol. Biofuels., 2014, 7, 107 CrossRef.
- H. P. Liao, X. T. Fan, X. L. Mei, Z. Wei, W. Raza, Q. Shen and Y. C. Xu, Biomass. Bioenerg., 2015, 74, 122–134 CrossRef.
- D. M. Kim, E. J. Cho, J. W. Kim, Y. W. Lee and H. J. Chung, Afr. J. Biotechnol., 2014, 13, 145–155 CrossRef.
- N. Kulkarni, A. Shendye and M. Rao, FEMS Microbiol. Rev., 1999, 23, 411–456 CrossRef PubMed.
- M. Sakthivel, N. Karthikeyan, R. Jayaveny and P. Palani, J. Ecobiotechnol., 2010, 2, 6–13 Search PubMed.
- D. J. M. Kumar, P. D. Poovai, C. L. P. Kumar, Y. S. Saroja, A. Manimaran and P. T. Kalaichelvan, Der Pharmacia Lett., 2012, 4, 881–888 Search PubMed.
- U. Gupta and R. Kar, Jordan J. Biosci., 2008, 1, 129–134 Search PubMed.
- E. R. Olson, Mol. Microbiol., 1993, 8, 5–14 CrossRef PubMed.
- C. Li, Z. Yang, R. Z. C. He, D. Zhang, S. Chen and L. Ma, J. Biotechnol., 2013, 168, 470–477 CrossRef PubMed.
- S. Goyari, S. H. Devi, L. Bengyella, M. Khan, C. K. Sharma, M. C. Kalita and N. C. Talukdar, J. Appl. Microbiol., 2015, 119, 88–98 CrossRef PubMed.
- A. H. M. El-Said and A. Saleem, Mycobiology, 2008, 36, 1–9 CrossRef PubMed.
- T. Dutta, R. Sahoo, R. Sengupta, S. S. Ray, A. Bhattacharjee and S. Ghosh, J. Ind. Microbiol. Biotechnol., 2008, 35, 275–282 CrossRef PubMed.
- F. El Bergadi, F. Laachari, M. Sadiki, S. Elabed, M. H. Iraqui and S. K. Ibnsouda, Microbiology, 2016, 85, 47–55 Search PubMed.
- S. Ahmed, M. I. Ghori, M. A. Malana and A. Jamil, Afr. J. Biotechnol., 2012, 11, 7227–7231 Search PubMed.
- A. M. Castro, M. L. Carvalho, S. G. F. Leite and N. Pereira, J. Ind. Microbiol. Biotechnol., 2010, 37, 151–158 CrossRef PubMed.
- M. K. Gowthaman, C. Krishna and M. Moo-Young, Appl. Mycol. Biotechnol., 2001, 1, 305–352 Search PubMed.
- V. H. Vu, T. A. Pham and K. Kim, Mycobiology, 2011, 39, 20–25 CrossRef PubMed.
- S. P. Shah, K. S. Kalia and J. S. Patel, J. Gen. Appl. Microbiol., 2015, 61, 35–43 CrossRef PubMed.
- P. F. Omojasola and O. P. Jilani, Res. J. Microbiol., 2009, 4, 67–74 CrossRef.
- M. Galbe and G. Zacchi, Appl. Microbiol. Biotechnol., 2002, 59, 618–628 CrossRef PubMed.
- M. Camassola, L. R. de Bittencourt, N. T. Shenem, J. Andreaus and A. J. P. Dillon, Biocatal. Biotransform., 2004, 22, 391–396 CrossRef.
- E. V. Smolyanyuk and E. N. Bilanenko, Microbiology, 2011, 80, 877–883 Search PubMed.
- P. S. Santa-Rosa, A. L. Souza, R. A. Roque, E. V. Andrade, S. Astolfi-Filho, A. J. Mota and C. G. Nunes-Silva, Electron. J. Biotechnol., 2018, 31, 84–92 CrossRef.
- O. Khelila and B. Cheba, Procedia. Technol., 2014, 12, 519–528 CrossRef.
- M. J. Liszka, M. E. Clark, E. Schneider and D. S. Clark, Annu. Rev. Chem. Biomol. Eng., 2012, 3, 77–102 CrossRef PubMed.
- V. P. Zambare, A. Bhalla, K. Muthukumarappan, R. K. Sani and L. P. Christophe, Extremophiles, 2011, 15, 611–618 CrossRef PubMed.
- A. M. de Castro, M. L. D. de Carvalho, S. G. F. Leite and N. Pereira, J. Ind. Microbiol. Biotechnol., 2010, 37, 151–158 CrossRef PubMed.
- K. Hirasawa, K. Uchimura, M. Kashiwa, W. D. Grant, S. Ito, T. Kobayashi and K. Horikoshi, Antonie van Leeuwenhoek, 2006, 89, 211–219 CrossRef PubMed.
- A. A. N. Gunny, D. Arbain, P. Jamal and R. E. Gumba, Saudi J. Biol. Sci., 2015, 22, 476–483 CrossRef PubMed.
- A. Ventosa, A. Oren and Y. Ma, Halophiles and Hypersaline Environments Current Research and Future Trends, Springer-Verlag, Berlin, Heidelberg, 2011, vol. 18, ISBN: 978–3–642–20197–4 Search PubMed.
- N. Kamiya, Y. Matsushita, M. Hanaki, K. Nakashima, M. Narita, M. Goto and H. Takahashi, Biotechnol. Lett., 2008, 30, 1037–1040 CrossRef PubMed.
- M. B. Turner, S. K. Spear, J. G. Huddleston, J. D. Holbrey and R. D. Rogers, Green Chem., 2003, 5, 443–447 RSC.
- S. Bose, D. W. Armstrong and J. W. Petrich, J. Phys. Chem. B, 2010, 114, 8221–8227 CrossRef PubMed.
- M. Moniruzzaman, N. Kamiya and M. Goto, Org. Biomol. Chem., 2010, 8, 2887–2899 RSC.
- M. Dashtban and W. Qin, Microb. Cell Fact., 2012, 11, 63 CrossRef PubMed.
- H. Jorgensen and L. Olsson, Enzyme Microb. Technol., 2006, 38, 381–390 CrossRef.
- D. A. Santos, M. M. Oliveira, A. A. S. Curvelo, L. P. Fonseca and A. L. M. Porto, Int. Biodeterior. Biodegrad., 2017, 121, 66–78 CrossRef.
- S. A. Alrumman, Braz. J. Microbiol., 2016, 47, 110–119 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra05729j |
| This journal is © The Royal Society of Chemistry 2018 |