Open Access Article
Open Access ArticleDevelopment of a high-throughput and sensitive assay of fusion genes in lung cancer by array-based MALDI-TOFMS†
Han-Tao Wua,
Kun Lia,
Gang Wanga,
Xue-Xi Yanga,
Anna Zhub,
Xu-Ping Xub,
Ming Lia,
Ying-Song Wu*a and
Tian-Cai Liu *a
*a
aInstitute of Antibody Engineering, School of Laboratory Medicine and Biotechnology, Southern Medical University, Guangzhou 510515, Guangdong, P. R. China. E-mail: wg@smu.edu.cn; liutc@smu.edu.cn; Fax: +86-20-37247604; Tel: +86-20-62789355
bGuangzhou Darui Biotechnology Co. Ltd., Guangzhou 510665, China
First published on 6th August 2018
Abstract
ALK (anaplastic lymphoma kinase gene), ROS1 (ros proto-oncogene 1) and RET (ret proto-oncogene) fusions are oncogenic drivers in non-small cell lung cancer (NSCLC). Methods like fluorescence in situ hybridization (FISH) and immunohistochemistry (IHC) are highly sensitive but subjectively analyzed, labor intensive, expensive and unsuitable for multiple fusion gene screening. This study aimed to establish a high-throughput, sensitive and cost-effective screening method (array-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, array-based MALDI-TOFMS) for ALK, ROS1 and RET fusion detection. This method was established with three fusion gene positive cell lines (H2228, ALK positive; HCC78, ROS1 positive; LC-2/AD, RET positive) and negative samples. Then, 34 clinical samples were selected and detected by Sanger sequencing, next generation sequencing (NGS) and array-based MALDI-TOFMS. The results were compared and analyzed and Sanger sequencing was considered the standard. 7 cases showed ALK fusions, 1 case showed ROS1 fusions, no case showed RET fusions and 4 cases were both ALK and ROS1 fusions. Results showed that array-based MALDI-TOFMS was 100% concordant with Sanger sequencing and NGS 82.3%. In this study, we reported the utility of array-based MALDI-TOFMS in the assessment of ALK, ROS1 and RET fusions in routine lung biopsies of FFPE and fresh tissue specimens. Besides, this method may also be applied to the diagnosis, monitoring and prognosis of illness.
1. Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide,1,2 with non-small cell lung cancer accounting for 85% of all lung cancer cases, and its prognosis remains very poor.3,4The anaplastic lymphoma kinase gene (ALK) encodes a receptor tyrosine kinase that is aberrant in a variety of malignancies. Approximately 3–7% of NSCLC patients harbor ALK fusions and they are more commonly found in light smokers (<10 pack years) and/or never-smokers.5,6 DNA rearrangement leads to expression of a constitutively active protein kinase. Tumors harboring ALK rearrangements rely specifically on the chimeric oncoprotein for progression and are sensitive to ALK inhibitors, such as crizotinib, a drug approved by US Food and Drug Administration for the treatment of ALK fusion-positive non-small cell lung cancer (NSCLC).5,7
The ROS proto-oncogene 1, receptor tyrosine kinase (ROS1) is a member of the insulin receptor family. Approximately 2% of lung tumors harbor ROS1 fusions and, like ALK fusions, ROS1 fusions are more commonly found in light smokers (<10 pack years) and/or never-smokers.8 In an expansion cohort of a phase I study, 50 patients with ROS1-positive NSCLC demonstrated a 72% response rate and 19.2 month median progression-free survival interval when treated with crizotinib.9
The ret proto-oncogene (RET) gene, located on chromosome 10, encodes a receptor tyrosine kinase belonging to the RET family. Several recent cancer genome sequencing studies identified RET fusions in 1–2% of NSCLC, most frequently involving kinesin family member 5B (KIF5B) and coiled-coil domain containing 6 (CCDC6) as fusion partners.10 In addition, cells with RET fusions may be sensitive to multitargeted kinase inhibitors such as sunitinib, sorafenib and vandetanib.11
Currently, the gold standard method for gene fusion detection in lung cancer is fluorescence in situ hybridization (FISH).12 However, this technique is expensive, labor intensive and requires assessment by a pathologist meaning that the results are subjective.13 Other methods like immunohistochemistry (IHC) and reverse transcription polymerase chain reaction (RT-PCR) also have their disadvantages, like the false positive rate of RT-PCR14 and the low and partial expression of ALK oncoproteins in IHC.15
NanoString technology is a promising new method for detecting fusions. Adriane and co-workers reported the usefulness of NanoString in the assessment of ALK fusions in 43 lung biopsies of FFPE specimens, which were validated previously by FISH/IHC.16 However, a larger number of samples still need to be assessed for this to be validated.
Consequently, a high-throughput, sensitive and cost-effective method that detects and evaluates ALK, ROS1 and RET fusion gene is urgently needed. MALDI-TOF was widely applied in the detection of protein and nucleic acid. Based on the conservation protein peak, this method can distinguish different species of microorganism.17 David and his co-workers made a proof of principle of their MALDI-TOFMS approach in the clinical setting using recently isolated Fusarium strains demonstrated its validity.18 Joyner utilized MALDI-TOFMS to monitor the kinetics and products of RNA cleavage and proved the method is a rapid, accurate, highly-detailed and semi-quantitative analysis of RNA cleavage.19 In this study, we combined a transcriptomic platform to handle RNA extracted from samples and array to perform high-throughput. Then we validated this method with three fusion gene positive cell lines and negative samples and tested this method with 34 formalin-fixed paraffin-embedded (FFPE) and fresh tissue samples from Chinese population and compared the results with Sanger sequencing and NGS.
2. Experimental
2.1 Reagents and instruments
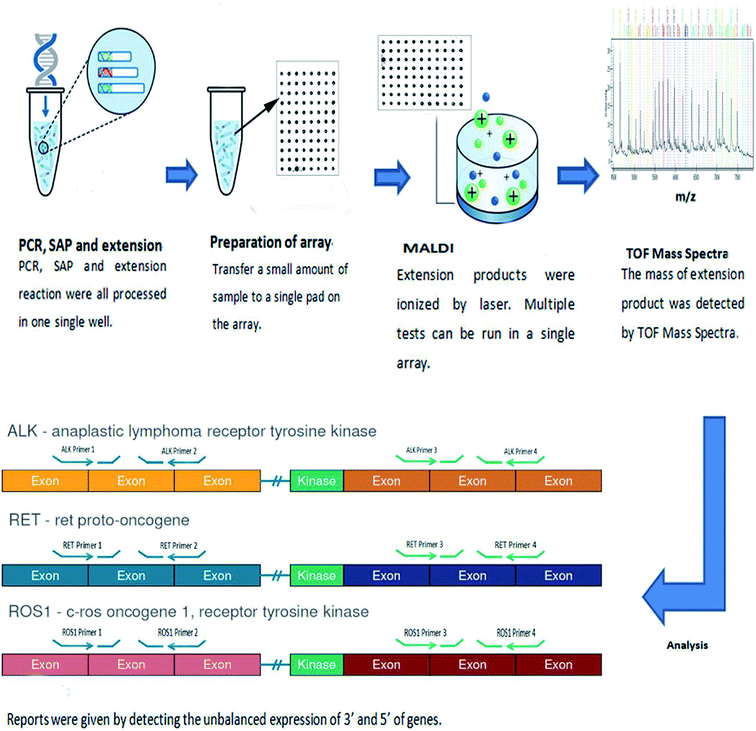
The following materials were used in this study: lung cancer ALK/ROS1/RET gene Fusion Detection Kit (Guangzhou Darui Biotechnology Co. Ltd, Guangzhou, China), Recover All™ Total Nucleic Acid Isolation Kit (Thermo, MA, USA), Qiagen RNeasy Mini kit (Qiagen, Valencia, Germany), Invitrogen SuperScript® VILO cDNA synthesis kit (Thermo, MA, USA), H2O (Sigma-Aldrich, USA), Qubit 3.0™ fluorimeter (Thermo, MA, USA). Veriti Dx Thermal 96 well Cycler (Life Technologies, New York, USA), time of flight mass spectrometry detection system (DR MassARRAY) (Guangzhou Darui Biotechnology Co. Ltd, Guangzhou, China). All processes were followed by the manufacturer recommendations (Fig. 1).2.2 Preparation of samples
2.3 Establishment of array-based MALDI-TOFMS
| a Primers for 3′ and 5′ fusion genes were to ensure fusion types. Primers for EML4 were to ensure the samples resource and the primers designed for GAPDH were to evaluate gDNA quantity. | ||||||||
|---|---|---|---|---|---|---|---|---|
| Primer position | ALK | ROS1 | RET | EML4 | GAPDH | |||
| 3′ | 5′ | 3′ | 5′ | 3′ | 5′ | |||
| Primer number | 2 | 3 | 2 | 2 | 2 | 2 | 2 | 2 |
This method contained 5 steps, PCR, Shrimp alkaline phosphatase reaction (SAP), extension reaction, preparation of array and analysis. The thermocycling cocktail and condition are listed at Tables S1–S3.†
Array was maintained at room temperature. After liquid handling, 40 μL resin was added into extension product to adsorb saline ions, then it was sealed and spotted six dots a time on the array by dispenser. And 15 nL extension product was spotted on the array. While finishing spotting, array was sent into MALDI-TOFMS. The intensity of laser is 500 and the whole process was maintained at 4 °C.
The results were analyzed by Type 4.0, the report was given by comparing the unbalanced expression of 3′ and 5′ of genes.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 reads per sample.
000 reads per sample.Torrent Suite software server for alignment was used to transfer sample data and Ion Reporter software v4.4 for variant calling. After sequencing, reads are aligned and for samples with more than 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mapped reads, fusions were detected when the sequence of one gene was found relocated to that of another gene at the breakpoint. A fusion was called when more than 40 fusion reads supported the breakpoint. Fusions were also inferred qualitatively from differential expression data, where expression of the 5′ and 3′ ends of a transcript were significantly imbalanced in fusion samples compared with normal samples.
000 mapped reads, fusions were detected when the sequence of one gene was found relocated to that of another gene at the breakpoint. A fusion was called when more than 40 fusion reads supported the breakpoint. Fusions were also inferred qualitatively from differential expression data, where expression of the 5′ and 3′ ends of a transcript were significantly imbalanced in fusion samples compared with normal samples.
2.4 Establishment of baseline
128 negative samples were collected to establish baseline. Every extend primer was along with 250 copies competitive sequences which were synthesized to evaluate the expression of fusion gene. After calculating the median and variance of 128 samples, Z scores of each fusion type were deduced by following formula and the median and variance of ALK, ROS1 and RET are listed in Table 2.
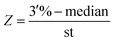 | (1) |
| ALK median | ALK variance | ROS1 median | ROS1 variance | RET median | RET variance |
|---|---|---|---|---|---|
| 0.519 | 0.126 | 0.504 | 0.092 | 0.518 | 0.105 |
2.5 Validation of clinical samples
A total of 34 clinical samples were tested by array-based MALDI-TOFMS, Sanger sequencing and NGS and the results were compared.3. Results
3.1 Results of three cell lines and negative samples
Three cell lines (H2228, ALK positive; HCC78, ROS1 positive; LC-2/AD, RET positive) and negative samples were used as positive and negative standard to establish DR MassARRAY and all these samples were also tested by Sanger sequencing and NGS. The results are listed in Table 3.| Array-based MALDI-TOF | NGS | Sanger sequencing | |||||||
|---|---|---|---|---|---|---|---|---|---|
| ALK | ROS1 | RET | ALK | ROS1 | RET | ALK | ROS1 | ALK | |
| H2228 | P | N | N | EML4-ALK (E13, A20) | N | N | EML4-ALK (E13, A20) | N | N |
| HCC78 | N | P | N | N | SLC34A2-ROS1 (S4, R34) | N | N | SLC34A2-ROS1 (S4, R34) | N |
| LC-2/AD | N | N | P | N | N | KIF5B-RET (K15, R12) | N | N | KIF5B-RET (K15, R12) |
| Negative samples | N | N | N | N | N | N | N | N | N |
As showed in Table 3, the results of array-based MALDI are totally in accordance with Sanger sequencing and NGS.
3.2 Results of 34 clinical samples
After the establishment of array-based MALDI-TOFMS, a total of 34 samples were performed fusion types detecting with three methods and the results are listed in Table 4.| Sample ID | Array-based MALDI-TOF | Sanger sequencing | NGS |
|---|---|---|---|
| 01 | ALK | EML4-ALK(E13, A20) | EML4-ALK (E13, A20) |
| 02 | ALK | EML4-ALK (E13, A20) | EML4-ALK (E13, A20) |
| EML4-ALK (E20,A20) | EML4-ALK (E20, A20) | ||
| 03 | ALK&ROS1 | EML4-ALK (E13, A20) | EML4-ALK (E18, A20) |
| SLC34A2-ROS1(S4-R34) | SLC34A2-ROS1(S4-R32) | ||
| SLC34A2-ROS1(S4-R32) | |||
| 04 | ALK&ROS1 | EML4-ALK (E13, A20) | EML4-ALK (E13, A20) |
| SLC34A2-ROS1(S4-R34) | SLC34A2-ROS1(S4-R34) | ||
| 05 | ALK | EML4-ALK (E13, A20) | EML4-ALK (E13, A20) |
| 06 | ALK | EML4-ALK (E13, A20) | EML4-ALK (E13, A20) |
| 07 | ALK | EML4-ALK (E13, A20) | EML4-ALK (E13, A20) |
| 08 | ROS1 | SLC34A2-ROS1(S4-R34) | SLC34A2-ROS1(S4-R34) |
| SLC34A2-ROS1(S4-R32) | SLC34A2-ROS1(S4-R32) | ||
| 09 | ALK | EML4-ALK (E13, A20) | N |
| 10 | N | N | EML4-ALK (E13, A20) |
| 11 | N | N | N |
| 12 | N | N | N |
| 13 | N | N | N |
| 14 | ALK&ROS1 | EML4-ALK (E13, A20) | EML4-ALK (E6, A20) |
| SLC34A2-ROS1(S4-R34) | |||
| 15 | ALK | EML4-ALK (E13, A20) | N |
| 16 | ALK&ROS1 | EML4-ALK (E13, A20) | N |
| SLC34A2-ROS1(S4-R34) | |||
| 17 | N | N | N |
| 18 | N | N | N |
| 19 | N | N | N |
| 20 | N | N | N |
| 21 | N | N | N |
| 22 | N | N | N |
| 23 | N | N | N |
| 24 | N | N | N |
| 25 | N | N | N |
| 26 | N | N | N |
| 27 | N | N | N |
| 28 | N | N | N |
| 29 | N | N | N |
| 30 | N | N | N |
| 31 | N | N | N |
| 32 | N | N | N |
| 33 | N | N | N |
| 34 | N | N | N |
As shown in Table 4, a total of 34 samples were analyzed by array-based MALDI-TOFMS, Sanger sequencing and NGS. Of these, 12 cases contain fusions gene. 7 samples were ALK positive, 1 sample was ROS1 positive, 4 samples had both ALK and ROS1 fusions gene and no RET fusions gene were detected. Comparison of the three methods showed that the results of array-based MALDI-TOFMS were fully concordant with Sanger sequencing and 82.3% with NGS.
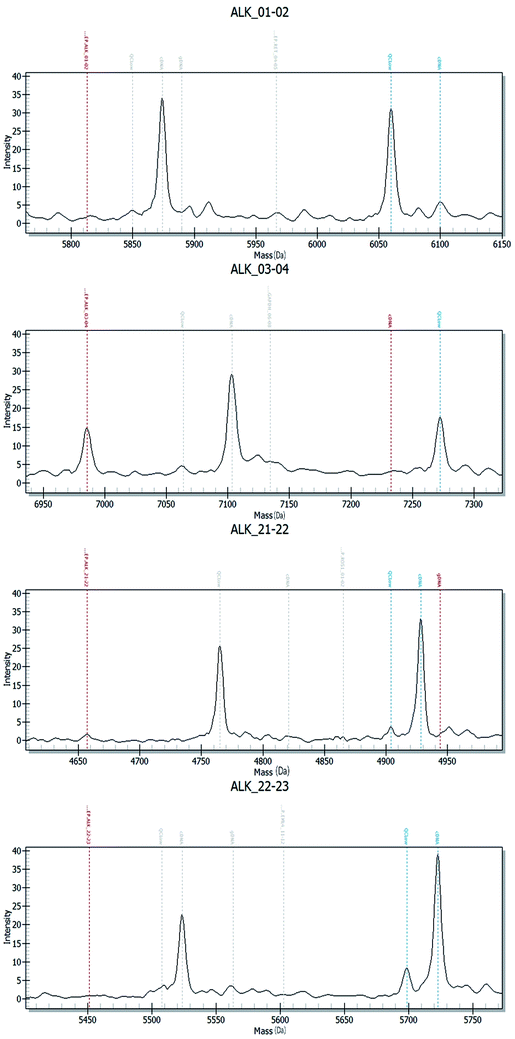
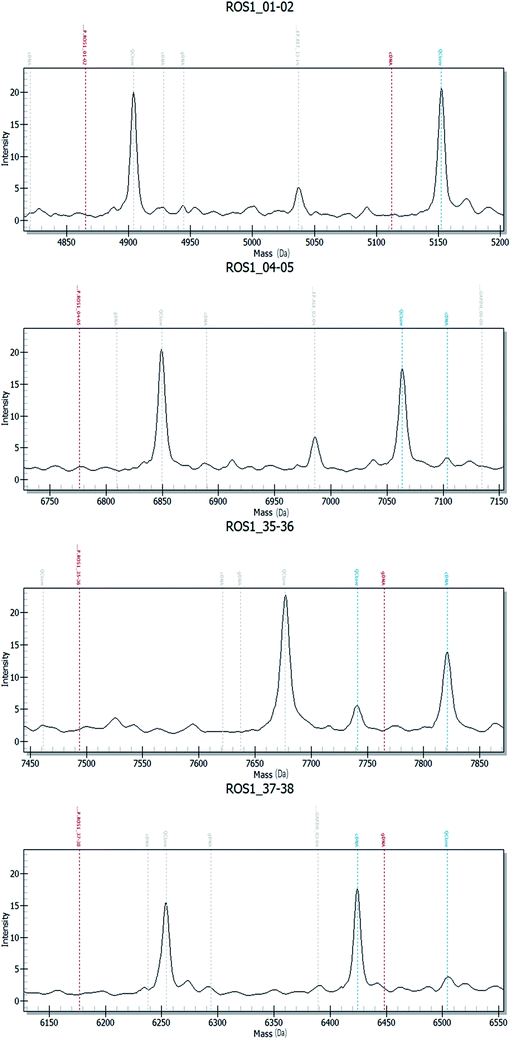
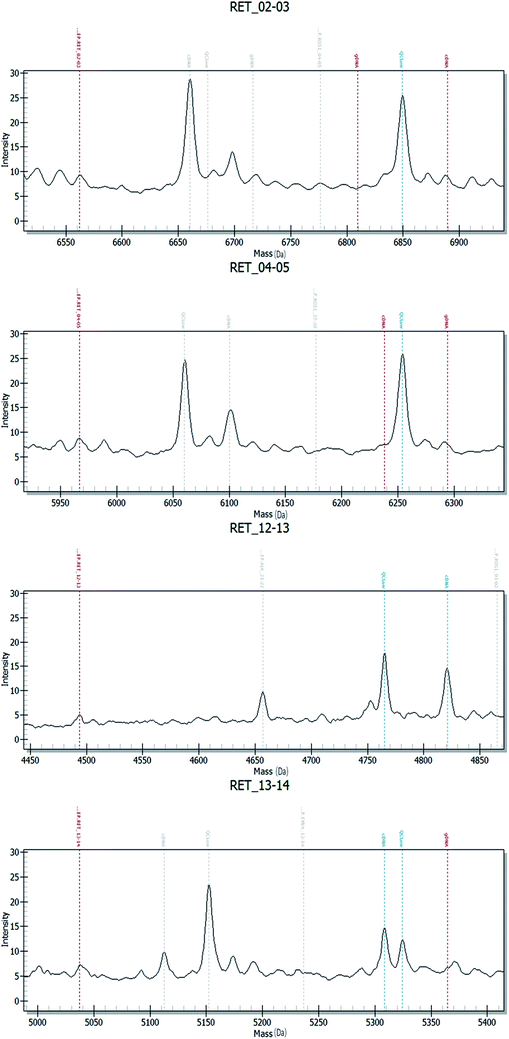
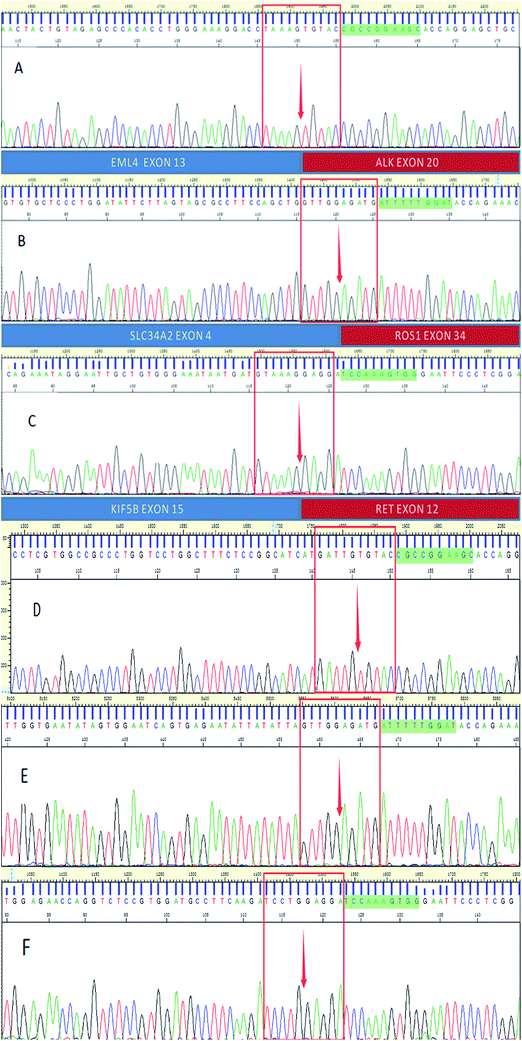
Fig. 2–4 show the results of ALK, ROS1 and RET fusion positive tested by array-based MALDI-TOFMS. We tested 34 clinical samples of which 12 samples were positive for fusions. 7 were ALK positive, 1 was ROS1 positive and 4 were both ALK and ROS1 positive, which agreed with the results from the Sanger sequencing, but was 82.3% concordant with NGS.
Fig. 5 shows the results of ALK, ROS1 and RET fusion positive and negative tested by Sanger sequencing. A total of 34 samples were tested and gave the same results as array-based MALDI-TOFMS. Sanger sequencing provided more information than array-based MALDI-TOFMS, showing that among the 7 ALK fusion samples, six ALK fusions were an EML4-ALK (E13, A20) and 1 sample was an EML4-ALK (E13, A20) (E20, A20), and sample 8 was ROS1 (S4, R32) (S4, R34) fusion. 4 samples were both ALK and ROS1 fusions, no RET fusion was detected.
4. Discussion
ALK, ROS1 and RET fusions are important biomarkers for the detection of fusions in cancer,22–24 and a cost-effective, rapid and sensitive method that are capable of screening multiple fusions is required.In the present study, we established a method to detect ALK, ROS1 and RET fusions gene in one single tube, three cell lines (H2228, ALK positive; HCC78, ROS1 positive; LC-2/AD, RET positive) and negative samples were used as standard. At the meantime, 128 negative samples were collected to establish baseline by giving median and variance, Z score would be calculated to determine fusion types. Then we collected 34 FFPE samples which were not randomly collected but partly validated by FISH previously to test this method and compared results with Sanger sequencing and NGS.
The blinded evaluation of these three methods revealed that array-based MALDI-TOFMS was fully concordant with Sanger sequencing but 82.3% concordant with NGS. However, with the limited cohort size, our statistical power for the analysis of all the three fusions was insufficient. A further assessment of these platforms with a larger number of samples is warranted.
Array-based MALDI-TOFMS requires a cDNA synthesis step, an amplification step, an annealing and single base extension step followed by MALDI-TOFMS whereby the masses of extended primers are obtained. Each extension primer was along with 250 copies competitive sequence to evaluate the 3′ and 5′ section expression of fusion gene. The Z scores of each fusion would be calculated to determine fusion type of samples.
In this study, this method provides a high-throughput, sensitive and cost-effective way to detect ALK, ROS1 and RET fusions. Array-based MALDI-TOFMS is an efficient method because it detects three fusion types in one tube, but Sanger sequencing needs at least 10 tubes. In addition, it requires no professional analysis which means the results are more objective. The whole process takes only one day to get results which is much faster compared with three or four days of NGS. However, this method is unable to identify specific fusion types and it is therefore limited for fundamental research. In this way, the method can not only be time and cost saving but also reduced the need of samples. However, we did not find RET fusion samples as this fusion only accounts for less than 1% and are hard to collect.25
The NGS fusion panel provides not only different fusion types but also the sequence information. Furthermore, it requires only 10 ng of total RNA, 10-fold less RNA than array-based MALDI-TOFMS requires. Ali and co-authors suggested that NGS could be especially useful for detecting ALK rearrangements.26 However, the disadvantages, such as expense and more effort for analysis it required are two major problems for large-scale application of NGS.27 Besides, because of the short sequence of fusion genes, NGS may not reach a precise result.
As the gold standard sequencing method, Sanger sequencing has been applied in many laboratories. Though this method did not approach a high precision in mutation detecting, it still cannot be replaced by new methods like NGS, especially when involves low-quality single-nucleotide variants and insertions or deletions <10 bp.28–30 The reasons why Sanger sequencing has not been used in large-scale application are the high cost and professional results analysis.31
Progression-free survival was significantly longer with targeted medicine than chemotherapy. For examples, patients harboring ALK positive lung cancer treated with crizotinib got a longer progression-free survival than chemotherapy.32 And crizotinib to ROS1 fusion,33 alectinib for RET fusion also approached good effect.34 Fusion gene detecting is vital for patients to get a reasonable treatment. Besides, it can also optimize the clinical resource, reduce the pain and save cost. Methods for detecting fusion gene largely applicated in clinical are FISH and IHC, however, these two methods are limited in throughput, sensitive and cost. FISH and IHC detect one fusion of samples a time, and according to the study of Yi-cheng Wu, these two methods are less sensitive.35 Besides, these two methods are expensive, about 7200yuan for FISH and 1950yuan for IHC.36 Array-based MALDI-TOF is quite a method to overcome the three problems. It detects three fusion gene of 96 samples a time and according to our study, it reached a high accordance with Sanger sequencing, lastly, it costs less than 100yuan per sample.
5. Conclusion
We established a high-throughput, sensitive and cost-effective screening method for detecting ALK, ROS1 and RET fusions which expended the application of mass spectra. Our findings validate this promising method that is a favorable alternative to FISH testing. Although the further evaluation with a larger number of samples would be carried. The developed method showed the potential in single-nucleotide variants, ctDNA detecting and may also be applied to the diagnosing, monitoring and prognosis of illness.Conflict of interest
None.Acknowledgements
The work was supported by the National Natural Science Foundation of China (grant number 21575058, 30901382), 2014 Guangzhou Healthy Care Synergy Innovate Major Special Project (2014) and 2014 Guangzhou Technology Project Cooperation Foundation (2014). The Special Project of International Scientific and Technological Cooperation in Guangzhou Development District (2017GH01).References
- C. S. Dela Cruz, L. T. Tanoue and R. A. Matthay, Lung Cancer: Epidemiology, Etiology, and Prevention, Clin. Chest Med., 2011, 32(4), 605–644 CrossRef PubMed.
- R. Siegel, D. Naishadham and A. Jemal, Cancer statistics, 2013, Ca-Cancer J. Clin., 2013, 63(1), 11–30 CrossRef PubMed.
- Y. Chen, S. Han, M. Zheng, Y. Xue and W. Liu, Clinical Characteristics of 274 Non-Small Cell Lung Cancer Patients in China, Onkologie, 2013, 36(5), 3 CrossRef PubMed.
- H. Wakelee, K. Kelly, M. J. Edelman, 50 Years of Progress in the Systemic Therapy of Non-Small Cell Lung Cancer, American Society of Clinical Oncology Educational Book, 2014, vol. 34, pp. 177–189 Search PubMed.
- E. L. Kwak, Y. Bang, D. R. Camidge, A. T. Shaw, B. Solomon and R. G. Maki, et al., Anaplastic Lymphoma Kinase Inhibition in Non-Small-Cell Lung Cancer, N. Engl. J. Med., 2010, 363(18), 1693–1703 CrossRef PubMed.
- D. W. Wong, E. L. Leung, K. K. So, I. Y. Tam, A. D. Sihoe and L. Cheng, et al., TheEML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-typeEGFR andKRAS, Cancer-Am Cancer Soc, 2009, 115(8), 1723–1733 Search PubMed.
- D. R. Camidge, Y. Bang, E. L. Kwak, A. J. Iafrate, M. Varella-Garcia and S. B. Fox, et al., Activity and safety of crizotinib in patients with ALK-positive non-small-cell lung cancer: updated results from a phase 1 study, Lancet Oncol., 2012, 13(10), 1011–1019 CrossRef PubMed.
- K. Bergethon, A. T. Shaw, S. Ou, R. Katayama and C. M. Lovly, ROS1 Rearrangements Define a Unique Molecular Class of Lung Cancers, J. Clin. Oncol., 2012, 30(8), 863–870 CrossRef PubMed.
- A. T. Shaw, S. I. Ou, Y. Bang, D. R. Camidge, B. J. Solomon and R. Salgia, et al., Crizotinib inROS1 -Rearranged Non–Small-Cell Lung Cancer, N. Engl. J. Med., 2014, 371(21), 1963–1971 CrossRef PubMed.
- J. S. Seo, Y. S. Ju, W. C. Lee, J. Y. Shin, J. K. Lee and T. Bleazard, et al., The transcriptional landscape and mutational profile of lung adenocarcinoma, Genome Res., 2012, 22(11), 2109–2119 CrossRef PubMed.
- D. Lipson, M. Capelletti, R. Yelensky, G. Otto, A. Parker and M. Jarosz, et al., Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies, Nat. Med., 2012, 18(3), 382–384 CrossRef PubMed.
- A. T. Shaw, D. Kim, K. Nakagawa, T. Seto, L. Crinó and M. Ahn, et al., Crizotinib versus Chemotherapy in AdvancedALK -Positive Lung Cancer, N. Engl. J. Med., 2013, 368(25), 2385–2394 CrossRef PubMed.
- T. Rogers, G. M. Arnau, G. L. Ryland, S. Huang, M. E. Lira and Y. Emmanuel, et al., Multiplexed transcriptome analysis to detect ALK, ROS1 and RET rearrangements in lung cancer, Sci. Rep., 2017, 7, 42259 CrossRef PubMed.
- M. Soda, S. Takada, K. Takeuchi, Y. L. Choi and M. Enomoto, A mouse model for EML4-ALK-positive lung cancer, Proc. Natl. Acad. Sci. U. S. A., 2008, 105(50), 19893 CrossRef PubMed.
- J. M. Boland, S. Erdogan, G. Vasmatzis, P. Yang, L. Tillmans and M. Erickson-Johnson, et al., Anaplastic lymphoma kinase immunoreactivity correlates with ALK gene rearrangement and transcriptional up-regulation in non-small cell lung carcinomas, Hum. Pathol., 2009, 40(8), 1152–1158 CrossRef PubMed.
- A. F. Evangelista, M. F. Zanon, A. C. Carloni, F. E. de Paula, M. A. Morini and M. Ferreira-Neto, et al., Detection of ALK fusion transcripts in FFPE lung cancer samples by NanoString technology, BMC Pulm. Med., 2017, 17(1), 86 CrossRef PubMed.
- S. Jadhav, M. Bhave and E. A. Palombo, Methods used for the detection and subtyping of Listeria monocytogenes, J. Microbiol. Methods, 2012, 88(3), 327–341 CrossRef PubMed.
- D. Triest, D. Stubbe, K. De Cremer, D. Piérard, A. Normand and R. Piarroux, et al., Use of Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry for Identification of Molds of the Fusarium Genus, J. Clin. Microbiol., 2015, 53(2), 465–476 CrossRef PubMed.
- J. C. Joyner, K. D. Keuper and J. A. Cowan, Analysis of RNA cleavage by MALDI-TOF mass spectrometry, Nucleic Acids Res., 2013, 41(1), e2 CrossRef PubMed.
- N. Kanaji, S. Bandoh, T. Ishii, A. Tadokoro, N. Watanabe and T. Takahama, et al., Detection of EML4-ALK fusion genes in a few cancer cells from transbronchial cytological specimens utilizing immediate cytology during bronchoscopy, Lung Cancer, 2012, 77(2), 293–298 CrossRef PubMed.
- P. Wijesinghe, G. Bepler and A. Bollig-Fischer, A Mass Spectrometry Assay to Simultaneously Analyze ROS1 and RET Fusion Gene Expression in Non–Small-Cell Lung Cancer, J. Thorac. Oncol., 2015, 10(2), 381–386 CrossRef PubMed.
- K. Takeuchi, Y. L. Choi, Y. Togashi, M. Soda, S. Hatano and K. Inamura, et al., KIF5B-ALK, a Novel Fusion Oncokinase Identified by an Immunohistochemistry-based Diagnostic System for ALK-positive Lung Cancer, Clin. Cancer Res., 2009, 15(9), 3143–3149 CrossRef PubMed.
- K. Rikova, A. Guo and Q. Zeng, et al., Global Survey of Phosphotyrosine Signaling Identifies Oncogenic Kinases in Lung Cancer, Cell, 2007, 131(6), 1190–1203 CrossRef PubMed.
- C. Tomuschat and P. Puri, RET gene is a major risk factor for Hirschsprung's disease: a meta-analysis, Pediatr. Surg. Int., 2015, 31(8), 701–710 CrossRef PubMed.
- K. Takeuchi, M. Soda, Y. Togashi, R. Suzuki, S. Sakata and S. Hatano, et al., RET, ROS1 and ALK fusions in lung cancer, Nat. Med., 2012, 18(3), 378–381 CrossRef PubMed.
- S. M. Ali, T. Hensing, A. B. Schrock, J. Allen, E. Sanford and K. Gowen, et al., Comprehensive Genomic Profiling Identifies a Subset of Crizotinib-ResponsiveALK -Rearranged Non-Small Cell Lung Cancer Not Detected by Fluorescence In Situ Hybridization, Oncologist, 2016, 21(6), 762–770 CrossRef PubMed.
- I. Dagogo-Jack and A. T. Shaw, Screening forALK Rearrangements in Lung Cancer: Time for a New Generation of Diagnostics?, Oncologist, 2016, 21(6), 662–663 CrossRef PubMed.
- E. Vuttariello, M. Borra, C. Calise, E. Mauriello, S. Greggi and A. Vecchione, et al., A New Rapid Methodological Strategy to Assess BRCA Mutational Status, Mol. Biotechnol., 2013, 54(3), 954–960 CrossRef PubMed.
- S. P. Strom, H. Lee, K. Das, E. Vilain, S. F. Nelson and W. W. Grody, et al., Assessing the necessity of confirmatory testing for exome-sequencing results in a clinical molecular diagnostic laboratory, Genet. Med., 2014, 16(7), 510–515 CrossRef PubMed.
- D. Dacheva, R. Dodova, I. Popov, T. Goranova, A. Mitkova and V. Mitev, et al., Validation of an NGS approach for diagnostic BRCA1/BRCA2 mutation testing, Mol. Diagn. Ther., 2015, 19(2), 119–130 CrossRef PubMed.
- S. Lee, S. Zhou, T. Zhou and G. Hong, Sanger Sequencing for BRCA1 c.68_69del, BRCA1 c.5266dup and BRCA2 c.5946del Mutation Screen on Pap Smear Cytology Samples, Int. J. Mol. Sci., 2016, 17(2), 229 CrossRef PubMed.
- B. J. Solomon, T. Mok, D. Kim, Y. Wu, K. Nakagawa and T. Mekhail, et al., First-Line Crizotinib versus Chemotherapy inALK-Positive Lung Cancer, N. Engl. J. Med., 2014, 371(23), 2167–2177 CrossRef PubMed.
- A. T. Shaw, S. I. Ou, Y. Bang, D. R. Camidge, B. J. Solomon and R. Salgia, et al., Crizotinib inROS1-Rearranged Non–Small-Cell Lung Cancer, N. Engl. J. Med., 2014, 371(21), 1963–1971 CrossRef PubMed.
- T. Kodama, T. Tsukaguchi, Y. Satoh, M. Yoshida, Y. Watanabe and O. Kondoh, et al., Alectinib Shows Potent Antitumor Activity against RET-Rearranged Non-Small Cell Lung Cancer, Mol. Cancer Ther., 2014, 13(12), 2910–2918 CrossRef PubMed.
- Y. Wu, Comparison of IHC, FISH and RT-PCR Methods for Detection of ALK Rearrangements in 312 Non-Small Cell Lung Cancer Patients in Taiwan, PLoS One, 2013, 8(8), e70839 CrossRef PubMed.
- H. Jiang-jiang, L. Xian-zhong and H. Da, Economic Analysis on Anaplastic Lymphoma Kinase Related Companion Diagnosis Methods before the Targeted Therapy for Non-small Cell Lung Cancer, Chinese Health Economics, 2017, 36(1), 40–43 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra05165h |
| This journal is © The Royal Society of Chemistry 2018 |