Open Access Article
Open Access ArticleEco-friendly synthesis of CuInS2 and CuInS2@ZnS quantum dots and their effect on enzyme activity of lysozyme†
Irshad Ahmad Mir a,
Kishan Dasa,
Tabasum Akhterb,
Rahul Ranjana,
Rajan Patel
a,
Kishan Dasa,
Tabasum Akhterb,
Rahul Ranjana,
Rajan Patel c and
H. B. Bohidar*ad
c and
H. B. Bohidar*ad
aSchool of Physical Sciences, Jawaharlal Nehru University, New Delhi, India. E-mail: bohi0700@mail.jnu.ac.in
bDepartment of Botany, University of Delhi, New Delhi, India
cCentre for Interdisciplinary Research in Basic Sciences, Jamia Millia Islamia, New Delhi, India
dSpecial Center for Nanosciences, Jawaharlal Nehru University, New Delhi, India
First published on 30th August 2018
Abstract
We report on the green and facile aqueous microwave synthesis of glutathione (GSH) stabilized luminescent CuInS2 (CIS, size = 2.9 nm) and CuInS2@ZnS core–shell (CIS@ZnS, size = 3.5 nm) quantum dots (QDs). The core–shell nanostructures exhibited excellent photo- and water/buffer stability, a long photoluminescence (PL) lifetime (463 ns) and high PL quantum yield (PLQY = 26%). We have evaluated the comparative enzyme kinetics of these hydrophilic QDs by interacting them with the model enzyme lysozyme, which was probed by static and synchronous fluorescence spectroscopy. The quantification of the QD–lysozyme binding isotherm, exchange rate, and critical flocculation concentration was carried out. The core–shell QDs exhibited higher binding with lysozyme yielding a binding constant of K = 5.04 × 109 L mol−1 compared to the core-only structures (K = 6.16 × 107 L mol−1), and the main cause of binding was identified as being due to hydrophobic forces. In addition to the enzyme activity being dose dependent, it was also found that core–shell structures caused an enhancement in activity. Since binary QDs like CdSe also show a change in the lysozyme enzyme activity, therefore, a clear differential between binary and ternary QDs was required to be established which clearly revealed the relevance of surface chemistry on the QD–lysozyme interaction.
Introduction
Quantum dots of type I–III–VI semiconductor nanocrystals such as CuInS2 (CIS) have been investigated for over a decade as valuable alternatives to cadmium and lead based binary toxic quantum dots for applications, like solar cells, light emitting diodes, and biodiagnostic tools.1–3 Compared with traditional fluorescent organic dyes and fluorescent proteins, QDs are brighter and considerably more resistant to photobleaching. Their broad absorption spectra enable the simultaneous excitation of multiple fluorescence colors. However, the toxic nature of Cd-containing QDs remains a major concern for in vitro diagnostics and in vivo imaging. Also, a major disadvantage of a binary system is the color dependence of hydrodynamic size, making multicolor comparisons difficult. Therefore, recently explored Cd- and toxic free2,3 ternary systems such as AgInS2 and CuInS2 nanocrystals, in which the fluorescence wavelength is tuned through chemical composition without altering the size, are gaining importance. Some recent studies have shown possible threats to human health with increasing exposure and uncontrolled application of nanomaterials.4–6 Therefore, it is important to understand how protein interacts with these nanostructures which depends on many factors like protein conformation and orientation. Moreover, it is necessary to comprehend the effects of all nanoparticles on different cells and their possible interaction mechanisms. Despite the astonishing development in the field of nanoscience, little is known quantitatively about the interaction of QDs with cells. For instance, no results in the literature are available on the synthesis of CIS and CIS based core–shell derivatives, and their interactions with model proteins (lysozyme) and nanocrystal effect on enzymatic activity of lysozyme.Enzymes are essential for maintaining biochemical reactions in the human body, and the disorder of enzymatic activities may cause metabolic imbalance and different diseases. Enzymatic activity is a function of a variety of parameters. For instance, the effects of the water activity and the solvent ordering can have profound impact on the enzyme kinetics. Matsue and Miyawaki7 have shown that water activity and aqueous solvent ordering considerably alters the enzymatic activities of alcohol dehydrogenase, lysozyme, and b-galactosidase in various aqueous solutions. Chipman8 performed the kinetic analysis of the reaction of lysozyme with oligosaccharides from bacterial cell walls to map the various ways in which any oligomer can associate with a given enzyme. Interestingly, it was found that nonproductive binding was also seen to have implications for the mechanism of lysozyme action. Since the advent of nanoscience and nanotechnology in the past few decades, many nanomaterial based formulations and drugs have entered the pharmaceutical industry in a big way. This has made nanomedicine and nanomaterial based diagnostics a forefront area for research and development. In any such application, the fundamental challenge is to probe and understand the complexity of protein-nanomaterial conjugation which must depend on the surface chemistry of the nanoparticles. Of the available nanomaterials, Au, Ag and silica nanoparticles, and semiconductor quantum dots like CdSe are shown to have significant effect on protein structure upon binding thereby altering its biological activity.9–11 Vertegel et al.9 have extensively studied the interaction of silica nanoparticles of size between 9-40 nm with common enzymes like, lysozyme, horseradish peroxidase, catalase, and trypsin to find that these get adsorbed onto the surface of silica nanoparticles, which in turn alters the secondary structure of the proteins (loss in the α-helix content). A second consequence was the significant loss in the enzyme activity.9,12–14 A consensus has emerged in the nanoscience community that nanoparticle size and surface morphology does influence the protein activity upon conjugation.
The interaction of low-dimensional nanomaterials, commonly called quantum dots, with proteins is of significance because the first phase of interaction between nanoparticles and biopolymers occurs in a protein-rich environment in the physiological fluids. One of the issues that concerns biological sciences is the effect of quantum dots on enzyme activity. The superior optical properties, high PL quantum yield, stable brightness, tunable and narrow emissions, and long PL lifetime are few of the attributes of semiconductor nanocrystals, also known as quantum dots (QDs). Studies have shown that nanosensors based on QDs are capable of probing multiple enzyme activities simultaneously. Knudsen et al.11 have reviewed the current development of QD-based nanosensors for enzyme detection recently. This review substantiates that the enzyme-QD hybrid system, equipped with unique electronic, optical and catalytic properties which may be used in addressing the specific challenges in the diagnostics and therapeutics. As far as the effect on enzyme activity is concerned, lysozyme is normally opted as a model enzyme since its properties are well studied and it happens to be the first enzyme to have its three-dimensional structure determined by X-ray diffraction technique. Size dependent lysozyme–nanoparticle interaction has been studied in the past by Vertegel et al.9 and Das et al.10 who used silica and CdSe nanomaterials, respectively, to establish effect of lysozyme adsorption governed secondary structure alterations. Clear correlation between surface curvature of nanoparticles and protein α-helix structure, and its activity could be established by Vertegel et al.9 Hydrophilic CdSe quantum dots of size 2.5 and 6.3 nm were interacted with lysozyme by Das et al.10 and the results revealed that upon binding with QDs conformational changes in lysozyme occurred and smaller size QDs were found to bind poorly to lysozyme, but produced a greater change in enzyme activity compared to larger QDs.
A pertinent question arises here, how similar are the binary and ternary QDs as far as their influence on enzyme activity is concerned? Clearly, these two types of nanocrystals are associated with different surface chemistry. In order to resolve this issue, herein, we study the enzyme activity of lysozyme bound to a ternary QD (CIS), and its core–shell derivative (CIS@ZnS). Bindings were characterized in terms of binding isotherm, exchange rate, and critical flocculation concentration. The binary and ternary QDs were profiled for their influence on enzyme activity on model enzyme lysozyme.
Materials and methods
Materials
Copper nitrate trihydrate (Cu(NO3)·3H2O, 99.99%), reduced glutathione (GSH, 98%), indium nitrate (In(NO3)3·xH2O, 99%), zinc acetate dehydrate (Zn(Ac)2·2H2O, 98%), and sodium sulfide nonahydrate (Na2S·9H2O, 99.99%) were purchased from sigma-Aldrich, USA. These were used as received. All procedures were performed at room temperature 25 °C unless otherwise stated. HEK-293 cells were obtained from National centre for Cell Science, Pune, India. These cells were cultured at 37 °C temperature in the presence of 5% Co2 in DMEM (Dulbecco's Modified Eagle's Medium) additional supplemented with 100 μg ml−1 streptomycin, 100 U ml−1 penicillin and 10% FCS (Fetal calf serum).Synthesis of CIS quantum dots
The synthesis procedure has been reported in ref. 15 In a typical synthesis, 0.02 mmol of Cu(NO3), 0.08 mmol of In(NO3)3·xH2O, and 0.4 mmol of GSH was added to of 40 ml deionized water in a of 200 ml reaction flask. The appearance of the solution changed from turbid to clear under vigorous stirring when adjusted to pH 8.5 in 1.0 M NaOH solution. Then, a freshly prepared Na2S solution (0.05 M) was added to the reaction flask. High-quality CuInS2 QDs were prepared after microwave irradiation of the reactant solution for 5 min. Then, the mixture was allowed to cool to a temperature lower than 50 °C. The aliquots of this preparation contained the CIS QDs. The color of the obtained dispersion was orange.Synthesis of core–shell quantum dots
For the deposition of ZnS shell around the CIS QDs, the reaction solution was cooled down to room temperature 25 °C. Typically, 0.1 M Zn(Ac)2 and 0.05 M Na2S solutions were injected into the freshly made CIS QD dispersion described earlier for the preparation of the core–shell QDs. This mixture solution was further microwave irradiated at 100 °C for 5 min that produced a high quality CIS@ZnS QDs of light orange color. Immediately, after the synthesis the aliquots were treated with excess of ethanol followed by centrifugation at 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, and redispersion in DI water. This cycle was repeated 5 times and the final product was dispersed in (DI) water/buffer for storage. The XRD data of the CIS@ZnS core–shell QD samples exhibit diffraction peaks shifted towards higher 2θ values with respect to the corresponding peaks of core-only CIS QDs. This shift in 2θ values, non-existence of extra peaks which for ZnS would be expected at 2θ = 29.33°, 48.80° and 57.61°(JCPDS no. 05-0566),22 clearly indicates that these samples were not a mixture of CIS and ZnS. Rather a fraction of Zn2+ ions had diffused into the nanocrystal structure of AIS, thus, widening and distorting it.21,23 Additionally, in the fluorescence spectra there were not extra peaks, which for ZnS and CIS would be expected around 400 and 600 nm, respectively. Therefore, it was clear that the targeted core–shell structures were formed.
000 rpm, and redispersion in DI water. This cycle was repeated 5 times and the final product was dispersed in (DI) water/buffer for storage. The XRD data of the CIS@ZnS core–shell QD samples exhibit diffraction peaks shifted towards higher 2θ values with respect to the corresponding peaks of core-only CIS QDs. This shift in 2θ values, non-existence of extra peaks which for ZnS would be expected at 2θ = 29.33°, 48.80° and 57.61°(JCPDS no. 05-0566),22 clearly indicates that these samples were not a mixture of CIS and ZnS. Rather a fraction of Zn2+ ions had diffused into the nanocrystal structure of AIS, thus, widening and distorting it.21,23 Additionally, in the fluorescence spectra there were not extra peaks, which for ZnS and CIS would be expected around 400 and 600 nm, respectively. Therefore, it was clear that the targeted core–shell structures were formed.
Preparation of lysozyme solution
The aqueous solution of lysozyme were prepared at a concentration of 0.01% (w/v) by dissolving known amount of the protein powder in 10 mM phosphate buffer solution (pH 6.4 ± 0.1) at 25 °C using a magnetic stirrer for almost 1 hour. This produced optically clear and transparent solutions. These were stored in sterilized air tight glass bottles for future use. The pH of the quantum dot dispersion was 8.5 ± 0.1. Keeping the protein concentration fixed different protein-QD complexes are generated by varying the QD concentration.Characterizations
UV-Vis absorption spectra were obtained using spectrophotometer (CE-7300, Cecil Instruments, U. K.) operating in the wavelength range from 190 to 900 nm. Zeta potential measurement was performed on an electrophoresis instrument (ZC-2000, Microtec, Japan). The average values reported were based on triplicate measurements. Crystallographic and size characterizations were done by using XRD (Rigaku D/Max 2200 diffractometer) and TEM (JEOL 2100F) respectively. Further details about the XRD and TEM can be obtained from ref. 18. Surface morphology was obtained using FESEM with FIB and EBL (Tescan Model LYRA 3 XMU) instrument. The compositions analysis and elemental purity of QDs were determined from the spectra obtained by energy dispersive analysis of X-rays (EDAX) using BRUKER X-Flash detector 4010 (Germany) attached with EVO, 40 ZEISS Scanning electron microscopy (SEM) instrument. The steady state fluorescence and time-resolved fluorescence measurements (TRFS) were performed using Varian Cary eclipse fluorescence spectrophotometer having a spectral range of 190 to 1000 nm and Edinburgh FL920 fluorescence Life Time Spectrometer respectively. More details are given in ref. 18 and 19 circular dichroism (CD) experiments were carried out with Applied Photo physics Chirascan instrument (USA) to estimate the secondary structure of proteins using the standard operation procedure.20 Synchronous fluorescence spectra were measured using the same spectrofluorometer at two different wavelength intervals (Δλ) 15 and 60 nm, which give the characteristic information on tyrosine (Tyr) and tryptophan (Trp) residues, respectively. Temperature was controlled during experiments using constant-temperature cell holder connected to temperature controlled water bath (Varian, USA). FTIR data was obtained on Varian 7000 FTIR. The details of PL quantum yield analysis are given in Mir et.al.21Cytotoxicity assay
MTT assay was used to evaluate the cytotoxicity of CuInS2 and CuInS2@ZnS QDs, 1 × 105 cells per well of HEK-293 cells were seeded in a 96-well plate and left overnight for attachment. The following day fresh media was added and cells were treated with different concentration of CuInS2 and CuInS2@ZnS QDs (12.5–3200 μM). The cells were allowed to grow for 24 h. 20 μl of 5 mg ml−1 of MTT 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide was added to each well four hours prior to completion of incubation. On completion of incubation, media was removed and to each well 200 μl of DMSO was added and it was incubated for further period of 5–10 min at room temperature, which gave a purple color to the solution. OD was taken in ELISA reader at 595 nm. The % of cytotoxicity was calculated using the following formulaThe data so obtained thus used to calculate the IC50 values for compounds from the curves obtained by plotting percentage cell viability against concentration of compounds.
Enzyme activity determination
The activity of lysozyme was measured using spectrophotometric turbidity assay on a Shimadzu UV-Vis 2500 spectrophotometer at 30 °C. Lysozyme was dissolved in 50 mM sodium phosphate buffer (pH 6.4 ± 0.1). The concentration of lysozyme was 50 mg ml−1. A stock substrate solution of Micrococcus lysodeikticus was prepared in 50 mM sodium phosphate buffer with a concentration of 3 mg ml−1. A portion (0.15 ml) of the lysozyme solution was mixed with 3.0 ml of the Micrococcus lysodeikticus solution into a cuvette and the change in absorbance at 450 nm was measured. After that, 50 μl of free lysozyme/mixture (QD–lysozyme) was added to the cuvette and the dynamic absorbance was scanned for 300 s. The initial slope (up to which curve show linearity) of the dynamic absorbance curve (i.e. absorbance vs. time curve) at 450 nm was calculated. The percent relative activity (RA%) was calculated using the following relation
 | (1) |
Results and discussion
Luminescence and absorption properties
The photographs of hydrophilic CIS core-only and CIS@ZnS core–shell QDs in normal day light and under a 365 nm UV illumination are shown in Scheme 1(right side), obtained under optimal experimental conditions (Cu/In 1/4, pH 8.5). The optical visualization of luminescence signature of CIS and CIS@ZnS core–shell QDs are clearly seen from the photographs. Here CIS@ZnS QDs exhibits higher brightness under UV illumination that can be attributed from the formation of core–shell structures.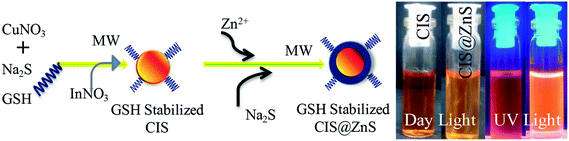 | ||
| Scheme 1 Schematic representation of the synthesis process of CIS and CIS@ZnS core–shell QDs. Right side photographs of CIS and CIS@ZnS QDs under day and UV light. | ||
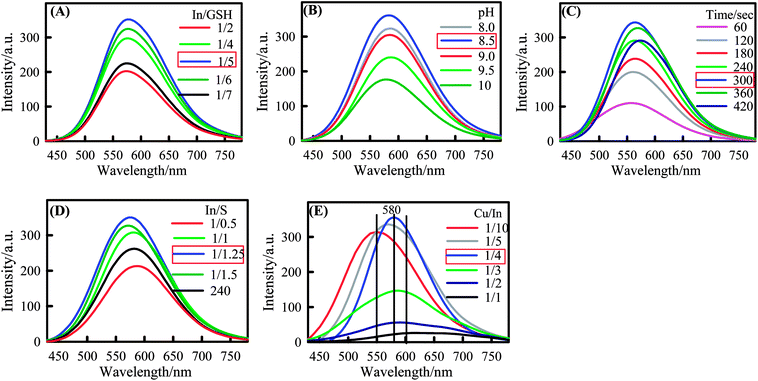
The appraisal of absorption and emission properties of CIS and CIS@ZnS core–shell was investigated during the synthesis. Different optimizations were done during the synthesis of CIS and CIS@ZnS core–shell QDs for the improved stability and photoluminescence quantum yields (PLQY). Influence of pH; irradiation time; In![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GSH; In
GSH; In![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S and Cu
S and Cu![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) In molar ratios, on optical properties were investigated. It is shown in Fig. 1A that In
In molar ratios, on optical properties were investigated. It is shown in Fig. 1A that In![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GSH molar ratio strongly influenced the fluorescence intensity of CIS and CIS@ZnS QDs. As shown in Fig. 1A different In/GSH molar ratios have different emission peak intensity. The maximum fluorescence intensity was obtained when the In/GSH molar ratio was 1/5. The highest PLQY was obtained corresponding to the highest emission peak intensity (at In/GSH ≈1/5). This optimized In/GSH molar ratio corresponding to highest PLQY is highlighted by red box in figure legends. Similarly the pH values strongly affect the PLQY and stability of CIS and CIS@ZnS QDs. It was noticed that the when the pH value was below 8 the solution was turbid. We investigated the effect of pH values from 8 to 10 on QD fluorescence intensity. It was found that fluorescence intensity of CIS@ZnS QDs attains maximum value when the pH value was 8.5 as shown in the Fig. 1B. Thus, the optimized pH (8.5) at which QDs show the highest emission peak intensity resulting the highest PLQY was also highlighted by box in legends of Fig. 1. In addition, In
GSH molar ratio strongly influenced the fluorescence intensity of CIS and CIS@ZnS QDs. As shown in Fig. 1A different In/GSH molar ratios have different emission peak intensity. The maximum fluorescence intensity was obtained when the In/GSH molar ratio was 1/5. The highest PLQY was obtained corresponding to the highest emission peak intensity (at In/GSH ≈1/5). This optimized In/GSH molar ratio corresponding to highest PLQY is highlighted by red box in figure legends. Similarly the pH values strongly affect the PLQY and stability of CIS and CIS@ZnS QDs. It was noticed that the when the pH value was below 8 the solution was turbid. We investigated the effect of pH values from 8 to 10 on QD fluorescence intensity. It was found that fluorescence intensity of CIS@ZnS QDs attains maximum value when the pH value was 8.5 as shown in the Fig. 1B. Thus, the optimized pH (8.5) at which QDs show the highest emission peak intensity resulting the highest PLQY was also highlighted by box in legends of Fig. 1. In addition, In![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S molar ratio also show strong influence on spectroscopic properties. As shown in Fig. 1C maximum fluorescence intensity and PLQY of CIS@ZnS QDs was obtained when the In
S molar ratio also show strong influence on spectroscopic properties. As shown in Fig. 1C maximum fluorescence intensity and PLQY of CIS@ZnS QDs was obtained when the In![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S ratio was 1
S ratio was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.25. Besides the above optimizations the samples ware irradiated for different time periods as shown in Fig. 1D. When the samples were irradiated for 300 s maximum fluorescence intensity was obtained.
1.25. Besides the above optimizations the samples ware irradiated for different time periods as shown in Fig. 1D. When the samples were irradiated for 300 s maximum fluorescence intensity was obtained.
Finally the effect of Cu/In cation ratio on PLQY was critically probed. Fig. 1E shows the optimized Cu/In ratio at which the maximum PLQY was obtained is 1/4. It is clearly seen from the Fig. 1E that emission peak of CIS@ZnS QDs can be tuned from 550 to 600 nm over the Cu/In composition range from 1/10 to 1/1. All the above optimizations were yielded the highest PLQY (see Fig. 1)). The quantum yields (QY) were determined following the procedure given in ref. 20–22.
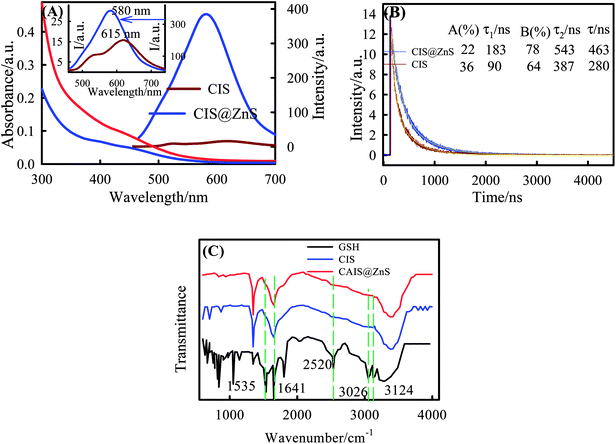
The fluorescence (FL) and absorbance spectra of the CIS QDs (obtained under optimal experimental conditions such as In/GSH ≈ 1/5; pH ≈ 8.5; In/S ≈ 1/1.25; Cu/In ≈1/4 and irradiation time ≈ 300 s) are shown in Fig. 2A. The absorption spectrum shows an absorption edge at 460 nm. The optical bandgap obtained from the Tauc plot shown in Fig.S1A (ESI†) was found to be 2.7 eV. The FL emission spectrum of these QDs shows a peak at 615 nm. To improve the luminescence properties and stability against oxidation/aggregation of core only QDs, these were coated with a protective ZnS shell. The excitation and emission spectra of core–shell QDs show a shift to higher bandgap energy compared to the core- only QDs. This blue shift arises from alloy structure of ZnS.17 Fig. 2A shows the excitation and emission spectra of core-shall QDs, which are broad and nearly Gaussian profile with the peak at 580 nm corresponding to the excitation at 425 nm. Both in the excitation and emission spectra of the core–shell structures Stokes shift was ≈35 nm compared to the core-only data. The respective photoluminescence quantum yields (QY) of CIS and CIS@ZnS QDs were determined as 1.0% and 26%, under optimal condition of Cu![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) In = 1
In = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4.
4.
Time-resolved fluorescence spectroscopy (TRFS) is another important optical tool for the characterization of semiconductor QDs and to obtain the radiative lifetime of FL emission. The excited-state lifetime of the emission shows remarkable change for core-shall QDs compared to the core-only nanocrystals. Fig. 2D shows the decay curves that were well fitted to a bi-exponential function, and average PL lifetime obtained were 280 and 463 ns respectively for CIS and CIS@ZnS nanocrystals.
To confirm the presence of GSH capping on the surface of QD, FTIR spectra of pure GSH and GSH capped QDs are compared which is shown in Fig. 2C. It was found that absorption bands of free GSH at 3124 cm−1 and 3026 cm−1 are assigned to the stretching N–H bands. The characteristic bands at 2520, 1641 and 1535 cm−1 belong to the stretching vibration of –SH group, stretching vibration of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond and distortion vibration of NH bond in amide groups, respectively. In contrast, the disappearance of the –SH vibration observed at 2520 cm−1 of the free glutathione ligand, and the diminishing of the N–H distortion vibration band clearly indicates that the GSH was de-protonated, and may be coordinated to the surface of the QDs through the SH and NHR groups.
O bond and distortion vibration of NH bond in amide groups, respectively. In contrast, the disappearance of the –SH vibration observed at 2520 cm−1 of the free glutathione ligand, and the diminishing of the N–H distortion vibration band clearly indicates that the GSH was de-protonated, and may be coordinated to the surface of the QDs through the SH and NHR groups.
Structural characterization
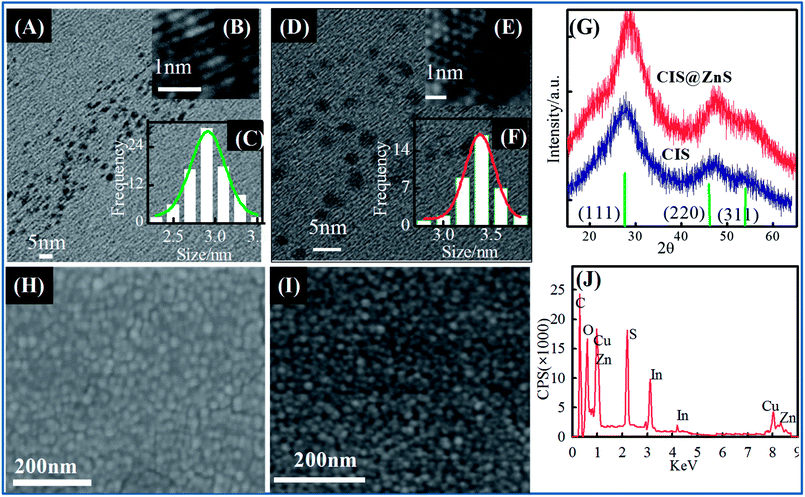
To evaluate the crystalline states, structure, morphology and composition of CIS and CIS@ZnS QDs, X-ray diffraction (XRD) analysis, (TEM), FESEM, SEM-EDX measurements were carried out. Fig. 3(B and E) show the HRTEM images of CIS and CIS@ZnS QDs under optimal experimental conditions (Cu/In 1/4, pH 8.5). HRTEM images were used to characterize the crystalline structure and morphologies of the QDs that exhibit well-resolved lattice planes which is shown in the inset of Fig. 3(B and E). These nanocrystals show a narrow size distribution and with average CIS QD size of about 2.9 ± 0.5 nm and with clearly resolved lattice fringes (Fig. 3(C and E) shows, the average size of the core/shell QDs increased to 3.5 ± 0.5 nm after coating with a ZnS shell, indicating successful coating. XRD patterns of these QDs are shown in Fig. 3G. The broad diffraction in peaks XRD patterns (Fig. 3G) indicates particles have small crystallite size. Three major peaks of CIS QDs were observed having 2θ values of 28.1, 46.5, and 55.1°, that could be indexed to (112), (024), and (116) lattice planes of the tetragonal crystal structure for the CIS nanocrystals (JCPDS no. 47-1372). It is important to note that core–shell QDs exhibits the similar diffraction peaks, but here diffraction peaks shifted towards higher 2θ values with respect to the corresponding peaks of CIS QDs. This 2θ shift values, while not finding any extra peaks, which for ZnS would be expected at 2θ = 29.33°, 48.80° and 57.61° (JCPDS No. 05-0566),23 clearly indicates that these QDs were not a mixture of CIS and ZnS. Rather a fraction of Zn2+ ions diffused into the nanocrystal structure of CIS, thus, widening and distorting it.16Therefore, a notable right shift in the peak positions of the CIS QDs was observed arising from shelling with ZnS because of the diffusion of Zn2+ ions into the copper–indium lattice.17 This shift towards the higher 2θ values suggested the decrease in lattice parameters due to the smaller ionic radius of Zn2+ relative to those of Cu+ and In3+.
FESEM images shown in Fig. 3(H and I) illustrates the core and core–shell structures with high magnification. These images show that the prepared QDs were found to be in spherical in shape with uniform morphology. SEM-EDX spectrum shown in Fig. 3J was carried out for elemental identification. From EDX analysis validation of constituent elements was done and it was found to be as expected (see Table S1 in the ESI†). Zeta potential (ζ) values were found to be −32 and −40 mV for core-only and core–shell structures, respectively. The higher negative value of zeta potential attributes the signature of higher stability. Colloidal theory suggests that particles with zeta potential values of greater than ±30 mV are highly stable in dispersion. These profiles are shown in Fig. S1(B and C) (ESI†).
The low resolution of the TEM instrument used did not allow collection of high contrast TEM images. However, observation of enhancement in PL quantum yield, blue shift in emission peak, shift in XRD peaks towards higher 2θ value, and the increase in average particle size seen from the TEM images clearly indicated the existence of core–shell structures.
Biocompatibility of CIS and CIS@ZnS core–shell QDs
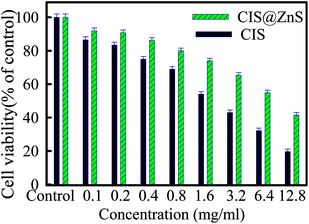
Before use for biological application, we had to evaluate its toxicity to normal cells, in this regards the cytotoxicity of CIS and CIS@ZnS QDs were evaluated on HEK-293 cells. The cell viability decreased in dose dependent manner (Fig. 4). Whereas there were no significant cytotoxicity observed at 100 and 200 μg ml−1 treated groups, shown in Fig. 4. It is evident that the cytotoxicity of QDs was negligible even after treating the cells at high concentration of nanoparticles (3.6 mg ml−1). Core-shell QDs are highly biocompatible than core only QDs. The obtained IC50 values for CIS and CIS@ZnS QDs were found to be 2.2 and 8.5 mg ml−1. This directly signifies the very good biocompatibility of QDs after formation of core–shell structures.Lysozyme–QD binding behavior by UV-Vis spectroscopy
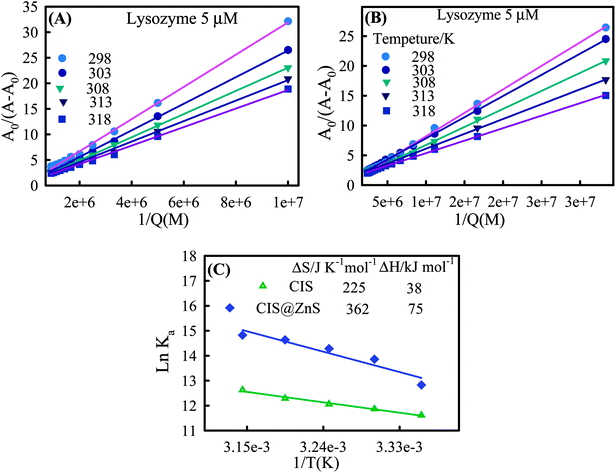
The UV-Vis absorption data were analyzed to determine the nature of the biointeraction. UV absorbance data is shown in Fig. S2† for CIS and CIS@ZnS core–shell QDs that could be used for quantitative comparison of the binding strength of lysozyme with QDs at different temperature, association constant Ka was determined using the Benesi-Hildebrand eqn (2).
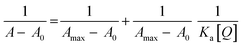 | (2) |
| Temperature (K) | Ka CIS (×105 L mol−1) | ΔGCIS (kJ mol−1) | Ka CIS@ZnS (X×105 L mol−1) | ΔGCIS@ZnS (kJ mol−1) |
|---|---|---|---|---|
| 298 | 1.11 | −29.05 | 3.72 | −32.88 |
| 303 | 1.43 | −30.18 | 10.45 | −34.69 |
| 308 | 1.73 | −31.30 | 15.92 | −36.50 |
| 313 | 2.18 | −32.43 | 22.69 | −38.31 |
| 318 | 3.05 | −33.55 | 27.31 | −40.11 |
Thermodynamic parameters and binding modes
The interaction forces between small molecule and a biological macromolecule are composed of mainly four types: hydrogen bonding, van der Waals force, electrostatic interactions and hydrophobic interactions.24,25 Ross and Subramanian have categorized these forces based on the thermodynamic parameters (ΔH and ΔS).25 According to their conclusions when ΔH < 0, ΔS > 0, electrostatic force plays a major role in the interaction. van der Waals and hydrogen bond interactions are possible if ΔH < 0 and ΔS < 0. If ΔH > 0 and ΔS > 0 the main forces are hydrophobic.26 In addition, they proposed that the interaction is a spontaneous molecular interaction resulting in ΔS > 0 and ΔG < 0. To elucidate the nature of biointeraction between lysozyme and QDs, ΔH and ΔS were calculated from the slope and intercept of the plot of ln(Ka) versus 1/T (Fig. 5C) yielded the enthalpy ΔH, and entropy ΔS of the enzyme reactions, according to the following Van't Hoff equation.
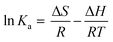 | (3) |
The free energy change (ΔG) is estimated from the following equation
| ΔG = ΔH − TΔS | (4) |
The values of ΔG for the interaction between QD and lysozyme are summarized in Table 1. The values obtained from Fig. 5C for ΔH are 38 and 75 kJ mol−1 and ΔS are 225 and 362 J K−1 mol−1 respectively for CIS and CIS@ZnS QDs. These results implied hydrophobic interactions were important in the binding between QD and lysozyme. Negative values of free energy are the consequences of spontaneous interaction.
Nature of binding
Fluorescence quenching mechanism of lysozyme due to complexation with QDs was monitored at different concentrations of the QDs and at fixed lysozyme concentration. It is shown in Fig. S3 (ESI†) that the emission intensity decreased continuously with increasing concentration of QDs. Fig. S4 (ESI†) demonstrates the linear dependence of the ratio between the emission intensity as a function of QD concentration, consistent with Stern–Volmer equation.27,28| F0/F = 1 + KSV[Q] | (5) |
Here F0 and F are the emission intensities in the absence and presence of the QDs, and [Q] is the concentration of the QDs. The Stern–Volmer constant KSV can be obtained from the slope of the F0/F versus [Q] shown in Fig. S4† and the calculated values are listed in Table 2. From the Stern Volmer constant we can find the quenching constant by using the equation given by
| KSV = Kqτ0 | (6) |
| S. no. | QDs | Ksv (×106 L mol−1) | Kq (×1014 L mol−1 s−1) |
|---|---|---|---|
| 1 | CIS | 3.94 | 3.94 |
| 2 | CIS@ZnS | 28.6 | 28.6 |
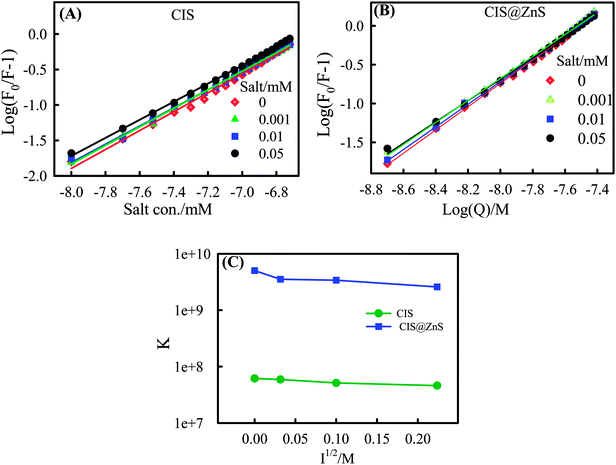
In the fluorescence quenching experiments (Fig. S3 (ESI†)), in the Stern–Volmer model, when QDs bind independently to a set of equivalent binding sites on the lysozyme surface, the binding parameters can be obtained from the formula27,28
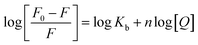 | (7) |
Where, Kb and n are the binding constant and number of binding sites, respectively. The plot (Fig. 6) of intensity ratio represented as log(F0/F − 1) versus log[Q] gives a straight line from which Kb and n could be determined. These values are summarized in Table 3 at different salt concentrations.
| NaCl/M | Kb/CIS (107 M−1) | n | Kb/CIS@ZnS(109 M−1) | n |
|---|---|---|---|---|
| 0 | 6.16 | 1.3 | 5.04 | 1.5 |
| 0.001 | 5.90 | 1.3 | 3.53 | 1.4 |
| 0.01 | 5.10 | 1.3 | 3.39 | 1.5 |
| 0.05 | 4.60 | 1.3 | 2.58 | 1.4 |
All the binding experiments were done in a buffer solution at pH of 6.4 ± 0.5 where the lysozyme possesses approximately +7.5 mV of zeta potential. Also, the zeta potential of the CIS and CIS@ZnS QDs were strongly negative ζ = −32 and −40 mV. Hence, it is important to note the presence of electrostatic interaction. To confirm this we did perform the binding experiments at different ionic strengths (I = 0, 0.001, 0.01, 0.1 N NaCl). There was universal quenching of fluorescence caused by QD-protein binding. Fluorescence quenching data (Fig. S3 (ESI†)) in the presence of QDs at different concentrations, and under different ionic strengths could be used to find the nature of binding. This data was used to make the plot of binding constant versus ionic strength in Fig. 6C. The percentage decrease of binding constant with salt concentration was not so significant, that indicated there was less screening of surface charge due to the mobile ions. These results strongly support the conclusion drawn from the entropy and enthalpy data that the type of binding was largely hydrophobic.
Synchronous fluorescence spectra
Synchronous fluorescence spectroscopy (SFS) simultaneously scans the excitation and emission monochromators while maintaining a fixed wavelength interval between them.27 When the wavelength interval between the excitation and emission is fixed at Δλ = 15 or 60 nm, then SFS gives characteristic information on the Tyr and Trp residues in a protein.27,29,30Fig. S5 (ESI†) reflects SFS profiles of the lysozyme with increasing QDs concentration for Δλ = 15 nm and Δλ = 60 nm. SFS data allows us explore the relative variation of synchronous fluorescence intensity (F/F0) of lysozyme with varying QD concentration for Δλ = 15 and 60 nm (Fig. S6, ESI†). In the case of both the QDs the slope was higher for Δλ = 60 nm than Δλ = 15 nm plot, which implies that QDs are most probably located very closer to Trp compared to the Tyr residue.
Effect of QD on secondary structure and the enzyme activity of lysozyme
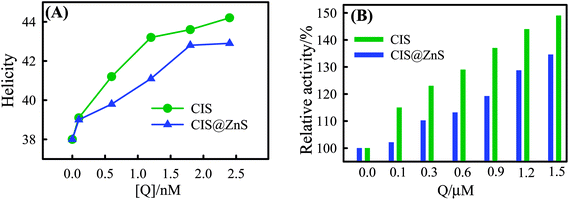
The effect on the secondary structure of lysozyme was studied in the far-UV region (200–260 nm) at a pH of 6.4 ± 1 in 10 mM sodium phosphate buffer. The CD spectra shown in Fig. S7 (ESI†) exhibits two negative bands located at 208 nm (n–π transition) and 222 nm (n–π* transition), which is the characteristic of α-helical structure in protein.31 The peak values of the negative peak decreased with QD concentration, and therefore, considerable increase in the ellipticity was noted (see Fig. 7A), which arises from a folding process undergone by lysozyme. The % helicity of the protein was determined from the calculated mean residue ellipticity (MRE) (deg cm2 dmol−1) values evaluated at 208 nm using the equation
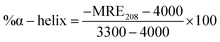 | (8) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 is the MRE value of a helix structure all determined at 208 nm. This process accounts for gain in the α-helix content of the secondary structure of the protein concomitant with a little loss in β-sheet. Protein helicity was found to increase from ≈38 to 43% for both CIS and CIS@ZnS QDs with QD concentration in the range of 0.1 to 2.4 nM (see Fig. 7A). This clearly infers binding of QD to protein surface, which causes protein to reveal its hydrophobic residues. At higher QD concentration, disruption in the α-helix structure of protein was noticed. This results in the appearance of more solvent compatible structures such as β-sheets and random coils.
000 is the MRE value of a helix structure all determined at 208 nm. This process accounts for gain in the α-helix content of the secondary structure of the protein concomitant with a little loss in β-sheet. Protein helicity was found to increase from ≈38 to 43% for both CIS and CIS@ZnS QDs with QD concentration in the range of 0.1 to 2.4 nM (see Fig. 7A). This clearly infers binding of QD to protein surface, which causes protein to reveal its hydrophobic residues. At higher QD concentration, disruption in the α-helix structure of protein was noticed. This results in the appearance of more solvent compatible structures such as β-sheets and random coils.
Fig. S8 (ESI†) shows the effect of the concentration of QD on the enzyme activity of lysozyme. The normalized reading of optical density (OD) of micrococcus lysodeikticus was used to indicate the activity of lysozyme. As can be seen, the higher the concentration, the higher was the activity of lysozyme. The enzyme was highly activated (almost 1 and 1/2 times) when the concentration of QD was greater than 1.5 μM. This is summarized in Fig. 7B. In Table 4 a comprehensive summary of model protein Lysozyme adsorbed on the surface of different types of nanomaterials.
| Nanomaterial | Particle Size/nm | Zeta Potential | Binding Constant/M | Kactivity/% (Retained) | Effect on secondary structure | Ref. |
|---|---|---|---|---|---|---|
| a — Not mentioned. *concentration dependent. | ||||||
| Silica | 4 | — | — | 60 | Strong loss in α-helical structure | 3 |
| 20 | — | — | 70 | Loss in α-helical structure | ||
| 100 | — | — | 40 | Loss in α-helical structure | ||
| Au | — | — | — | 28 | Loss in α-helical structure | 7 |
| Nanodiamond | 5 | −7 | — | 10 | — | 8 |
| 50 | −15 | 15 | ||||
| 100 | −30 | 75 | ||||
| 500 | −35 | 55 | ||||
| TiO2 | 60 | −49 | 4.7 × 106 | 10 | Loss in α-helical structure* | 9 |
| CdSe | 2.5 | −56 | 1.74 × 1010 | 250* | Loss in α-helical structure* | 11 |
| 6.3 | −62 | 40.7 × 1010 | 170* | Loss in α-helical structure* | ||
| CIS | 2.9 | −32 | 6.16 × 107 | 149* | Gain in α-helical structure* | This study |
| CIS@ZnS | 3.5 | −40 | 5.04 × 109 | 138* | Gain in α-helical structure* | |
Conclusions
We have successfully synthesized highly stable and water dispersible monodisperse CIS (core-only) and CIS@ZnS core–shell quantum dots of size 2.9 and 3.5 nm respectively following a facile and green synthesis protocol. American Chemical Society monograph on Green Chemistry stipulates that methods must require minimum energy for a reaction to take place which must occur in a green solvent.32 Thus, for the synthesis to be called green use of safer solvents, and minimum time and energy consumption are a must.33 In our case, the reaction medium was water and reactions were carried out inside a microwave oven that assured low power consumption and uniform heating. The synthesis did not require more than 5 min. Thus, our protocol adequately met all the desired criteria stipulated for green synthesis. The synthesized QDs were associated with long luminescence lifetimes and reasonably high quantum yields. These QDs were interacted with model protein lysozyme in order to evaluate the alteration in the enzyme activity under various ionic strengths, and temperature conditions using fluorescence and UV-absorbance techniques. The core–shell structure was seen to bind preferably to this enzyme exhibiting a binding constant that was at least 80 times higher than the core-only case. This contention was supported by the interaction enthalpy data. Interestingly, concentration dependent secondary structure enhancement in lysozyme was noticed that attributed higher retention of enzyme activity. A broad comparison of lysozyme binding to other nanoparticles is provided that clearly indicates that the particle surface chemistry plays an essential role in modulating the enzyme activity of proteins.Author contributions
IAM performed the synthesis and physical characterization. IAM, KD TA and RR designed of the experimental plan, carried out the data collection, its analysis and interpretation. IAM, RP and HBB contributed to conceptualization of the study, drafted and revised the manuscript critically for important intellectual content. All authors have provided final approval of the manuscript to be published.Conflicts of interest
Authors declare no conflict of interest.Acknowledgements
IAM, TA, KD and RR acknowledge University Grants Commission and Council of Scientific & Industrial Research, Government of India for Research Fellowship. This study was funded by the PURSE-Phase II support of Department of Science and Technology (DST), India. We are thankful to the Advanced Research Instrumentation Facility (AIRF) of the University for allowing us access to their facilities (TEM, TRFS, CD and FESEM). We are thankful to National centre for Cell Science, Pune, India for providing us HEK-293 cells.References
- J. M. Pietryga, Y.-S. Park, J. Lim, A. F. Fidler, W. K. Bae, S. Brovelli and V. I. Klimov, Chem. Rev., 2016, 116, 10513–10622 CrossRef PubMed.
- K. E. Knowles, K. H. Hartstein, T. B. Kilburn, A. Marchioro, H. D. Nelson, P. J. Whitham and D. R. Gamelin, Chem. Rev., 2016, 116, 10820–10851 CrossRef PubMed.
- G. Xu, S. Zeng, B. Zhang, M. T. Swihart, K.-T. Yong and P. N. Prasad, Chem. Rev., 2016, 116, 12234–12327 CrossRef PubMed.
- K. Stollewerk, A. Jofré, J. Comaposada, J. Arnau and M. Garriga, Food Control, 2014, 38, 130–135 CrossRef.
- Y. He, P. Yin, H. Gong, J. Peng, S. Liu, X. Fan and S. Yan, Sens. Actuators, B, 2011, 157, 8–13 CrossRef.
- H.-J. Zhan, P.-J. Zhou, L. Ding, Z.-Y. He and R. Ma, J. Lumin., 2012, 132, 2769–2774 CrossRef.
- S. Matsue and O. Miyawaki, Enzyme Microb. Technol., 2000, 26, 342–347 CrossRef PubMed.
- D. M. Chipman, Biochemistry, 1971, 10, 1714–1722 CrossRef PubMed.
- A. A. Vertegel, R. W. Siegel and J. S. Dordick, Langmuir, 2004, 20, 6800–6807 CrossRef PubMed.
- K. Das, K. Rawat, R. Patel and H. B. Bohidar, RSC Adv., 2016, 6, 46744–46754 RSC.
- B. R. Knudsen, M. L. Jepsen and Y.-P. Ho, Expert Rev. Mol. Diagn., 2013, 13, 367–375 CrossRef PubMed.
- F. Secundo, Chem. Soc. Rev., 2013, 42, 6250–6261 RSC.
- G. A. Petkova, K. Záruba, P. Žvátora and V. Král, Nanoscale Res. Lett., 2012, 7, 287 CrossRef PubMed.
- E. Perevedentseva, P.-J. Cai, Y.-C. Chiu and C.-L. Cheng, Langmuir, 2010, 27, 1085–1091 CrossRef PubMed.
- W.-W. Xiong, G.-H. Yang, X.-C. Wu and J.-J. Zhu, ACS Appl. Mater. Interfaces, 2013, 5, 8210–8216 CrossRef PubMed.
- I. A. Mir, V. S. Radhakrishanan, K. Rawat, T. Prasad and H. B. Bohidar, Sci. Rep., 2018, 8, 9322 CrossRef PubMed.
- X. Tang, W. B. A. Ho and J. M. Xue, J. Phys. Chem. C, 2012, 116, 9769–9773 CrossRef.
- I. A. Mir, K. Rawat and H. B. Bohidar, Colloids Surf., A, 2016, 509, 182–189 CrossRef.
- I. A. Mir, K. Rawat and H. B. Bohidar, Mater. Res. Express, 2016, 3, 105014 CrossRef.
- I. A. Mir, K. Rawat and H. B. Bohidar, Colloids Surf., A, 2017, 520, 131–137 CrossRef.
- I. A. Mir, K. Das, K. Rawat and H. B. Bohidar, Colloids Surf., A, 2016, 494, 162–169 CrossRef.
- I. A. Mir, K. Rawat and H. B. Bohidar, Cryst. Res. Technol., 2016, 51, 561–568 CrossRef.
- M. Madkour and F. Al Sagheer, Opt. Mater. Express, 2017, 7, 158–169 CrossRef.
- M. E. Pacheco and L. Bruzzone, J. Lumin., 2012, 132, 2730–2735 CrossRef.
- P. D. Ross and S. Subramanian, Biochemistry, 1981, 20, 3096–3102 CrossRef PubMed.
- B. Ojha and G. Das, J. Phys. Chem. B, 2010, 114, 3979–3986 CrossRef PubMed.
- J. R. Lakowicz, Principles of Fluorescence Spectroscopy, Springer, New York, 2007 Search PubMed.
- W. Chen, L. J. Young, M. Lu, A. Zaccone, F. Ströhl, N. Yu, G. S. Kaminski Schierle and C. F. Kaminski, Nano Lett., 2016, 17, 143–149 CrossRef PubMed.
- M. Kumari, N. Dohare, N. Maurya, R. Dohare and R. Patel, J. Biomol. Struct. Dyn., 2017, 35, 2016–2030 CrossRef PubMed.
- D. Ray, B. K. Paul and N. Guchhait, Phys. Chem. Chem. Phys., 2012, 14, 12182–12192 RSC.
- J. Fan, X. Chen, Y. Wang, C. Fan and Z. Shang, J. Zhejiang Univ., Sci., B, 2006, 7, 452–458 CrossRef PubMed.
- M. A. Ryan and M. Tinnesand, Introduction to green chemistry, instructional activities for introductory chemistry, American Chemical Society, 2002 Search PubMed.
- D. Dallinger and C. O. Kappe, Chem. Rev., 2007, 107, 2563–2591 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra04866e |
| This journal is © The Royal Society of Chemistry 2018 |