Open Access Article
Open Access ArticleDevelopment of a Pichia pastoris whole-cell biocatalyst with overexpression of mutant lipase I PCLG47I from Penicillium cyclopium for biodiesel production
Yihan Liu*abcd,
Lin Huangabd,
Dong Zhengad,
Yu Fuad,
Mengying Shanbd,
Zehua Xubd,
Jieying Mabd and
Fuping Lu *abcd
*abcd
aKey Laboratory of Industrial Fermentation Microbiology, Ministry of Education, Tianjin 300457, P. R. China. E-mail: lyh@tust.edu.cn; lfp@tust.edu.cn; Fax: +86 022 60602298; Tel: +86 022 60601958
bTianjin Key Laboratory of Industrial Microbiology, Tianjin 300457, P. R. China
cNational Engineering Laboratory for Industrial Enzymes, Tianjin 300457, P. R. China
dThe College of Biotechnology, Tianjin University of Science and Technology, Tianjin 300457, P. R. China
First published on 20th July 2018
Abstract
Penicillium cyclopium lipase I (PCL) is a thermolabile triacylglycerol lipase with very low activity against monoacylglycerols, and there have been no reports on the transesterification of oil to produce biodiesel. A mutant PCLG47I with an improved thermostability was previously obtained through replacing Gly47 with Ile in PCL. In this study, a novel Pichia pastoris whole-cell biocatalyst (WCB) with overexpression of PCLG47I was constructed and characterized for biodiesel production from soybean oil. The optimum conditions for biodiesel preparation were 1 g soybean oil, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 initial oil/methanol molar ratio with 3 times methanol addition of 1
2 initial oil/methanol molar ratio with 3 times methanol addition of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.75 oil/methanol molar ratio at 4 h intervals, 7% water content, 400 U lipase, temperature of 25 °C, and reaction time of 20 h. Under the optimum conditions, the FAME yield reached 60.7% and remained 47.3% after 4 batch cycles, and no glycerol was generated as a byproduct. These findings indicated that this WCB is a promising biocatalyst for biodiesel production in a relatively cost-effective manner. Additionally, the resulting enzymatic process may provide a potential method for biodiesel production at an industrial scale.
0.75 oil/methanol molar ratio at 4 h intervals, 7% water content, 400 U lipase, temperature of 25 °C, and reaction time of 20 h. Under the optimum conditions, the FAME yield reached 60.7% and remained 47.3% after 4 batch cycles, and no glycerol was generated as a byproduct. These findings indicated that this WCB is a promising biocatalyst for biodiesel production in a relatively cost-effective manner. Additionally, the resulting enzymatic process may provide a potential method for biodiesel production at an industrial scale.
Introduction
Recently, biodiesel has received much attention as a promising alternative to replace the current fossil fuels as a mixture of fatty acid methyl esters (FAMEs). It has many advantages over traditional fossil fuels, including renewability, sustainability, biodegradation, nontoxicity, and environmental friendliness.1 Many studies have shown that biodiesel could be produced in vegetable oils with an alkaline catalyst or the esterification of fatty acids with an acid catalyst by the transesterification of triacylglycerols.2,3 However, chemical processes present various drawbacks, such as the formation of soap, difficulties in the recovery of glycerol and the removal of chemical catalysts, requirement of high energy, and production of wastewater.1,4To address these problems, lipases-catalyzed biodiesel production has been studied as an substitute to overcome the limitations of chemical-catalyzed methods.5 Lipases (triacylglycerol hydrolases, EC 3.1.1.3) exhibit the catalytic activity under mild conditions, resulting in lower energy consumption,6,7 which will lower the cost of biodiesel production. Additionally, lipases have a wide diversity of substrates able to catalyze transesterification of triglycerides and esterification of free fatty acids (FFAs) simultaneously in vegetable oils,8,9 leading to a more efficient strategy for biodiesel production. In addition, the separation and purification of biodiesel is a simple and environmentally friendly process.10 However, lipases-catalyzed biodiesel production is limited in the scale-up process at present because of the high cost and low operational stability of free lipase.9 An effective solution is to recycle lipases for batch reaction to reduce the cost. The use of immobilized lipase (e.g., Novozym 435 and Lipozyme RM IM) is beneficial to improve the reusability and stability of lipases.11 However, these lipases are immobilized by complicated processed including extraction, purification, and immobilization, which is an expensive method.1 Moreover, the conformation of lipases are altered when they are bound to the carriers, which may change the specificity and reduce the activity of lipases.7,12 These features are the main obstacles in the industrialization of lipase-catalyzed biodiesel production. Recent reports13,14 indicated that the whole-cell biocatalyst (WCB) of lipases can eliminate the requirement for enzyme purification and immobilization, thus reducing the overall cost of lipases-catalyzed biodiesel production. In addition, WCB provides a natural environment for enzyme localization and conformation,15 which can improve the stability and reusability of lipases without affecting their specificity and activity.16–19
Yeast surface display systems, including Pichia pastoris, Yarrowia lipolytic, and Saccharomyces cerevisiae, are widely used to prepare WCB for the expression of many heterologous proteins and display many advantages, such as simple genetic manipulation, standard fermentation for the production of target proteins, and use as a biocatalyst without purification and immobilization.13,16 Nevertheless, studies of the lipases displayed on the yeast cell surfaces as WCBs for biodiesel production remain limited. Rhizopus oryzae lipase displayed on S. cerevisiae was employed for biodiesel production with a yield of 78.3% after 72 h.20 Rhizomucor miehei lipase-displaying P. pastoris was used as the biocatalyst, reaching a biodiesel yield of 83.1% after 72 h.21 Displayed Candida antarctica lipase B and R. miehei lipase, which were expressed on the cell surface of P. pastoris, afforded biodiesel yield over 90% in 12 h.16,22 A biocatalyst through coexpressing of C. antarctica lipase B and Thermomyces lanuginosus lipase on the cell surface of P. pastoris was used in biodiesel production with biodiesel conversion of 95.45% in 12.6 h.23
During lipases-catalyzed biodiesel production, glycerol, the main byproduct of transesterification reactions, inhibits lipases activity by covering them, preventing the accessibility of lipases active sites to the substrates.24 1,3-Specific lipases can simultaneously synthesize biodiesel and monoacylglycerols (MAGs) to avoid the glycerol production. As is known, the obtained MAGs are considered valuable products widely used in food, pharmaceutical, and cosmetic industries.25 Previous studies have reported that most lipases from Penicillium have a hydrolytic preference for the 1- and 3-positions of triacylglycerols (TAGs),26 and Penicillium cyclopium lipase I (PCL) shows preferential specificity toward TAGs and low activity against diacylglycerols (DAGs) but almost no activity toward MAGs.26,27 Compared with the commonly used and widely investigated lipases for biodiesel production, PCL has not received enough attention for its potential applications. PCL has an optimum activity at 25 °C and thereby being considered as a cold-active lipase. Cold-active lipases exhibit high activity at low temperatures compared to their mesophilic or thermophilic counterparts that drastically reduce their activities at low temperatures, which is beneficial to the energy saving for biodiesel production because the biodiesel synthesis by most other lipases was conducted at higher temperatures.28 However, the poor thermostability prevented PCL from being applied in long-term processes of industrial applications. Compared with those mesophilic and thermophilic lipases, PCL displayed more difficulty in the enhancement of the thermostability because of its structural flexibility. In the previous study, PCL was engineered through the direct evolution with error-prone polymerase chain reaction (PCR), and a novel mutant PCLG47I with significantly improved thermostability was obtained.29 PCLG47I could efficiently maintain its activity at 35 °C, 40 °C, and 45 °C with the time extension, leading to its potential long-term application at low temperatures in the process of biodiesel production.
Based on the above, the goals of this study was (i) to exploit the new preparation of PCLG47I as the form of WCB using the P. pastoris displayed system, (ii) to optimize the conditions for biodiesel production, such as reaction temperature, oil/methanol molar ratio, enzyme loading amount, water content, methanol additional strategy, and reaction time, and (iii) to evaluate the reusability of WCB for efficient biodiesel production, thereby assessing the potential of using the PCLG47I WCB as a biocatalyst for biodiesel production by the transesterification of oil with methanol. To our knowledge, this study is the first report on the use of recombinant P. pastoris yeast to functionally overexpress lipase from P. cyclopium as a WCB for biodiesel production.
Materials and methods
Chemicals and enzymes
Standard FAMEs were purchased from Sigma-Aldrich (St. Louis, MO, USA). Restriction endonucleases, T4 DNA ligase, Pyrobest DNA polymerase, and genetic manipulation kits were provided by Takara Bio (Dalian, China). Refined soybean oil was purchased from a commercial source. All other chemicals and reagents were obtained from standard sources and were of analytical grade. The recombinant PCL (rPCL) and PCLG47I (rPCLG47I) were produced by the expression of the corresponding genes in Escherichia coli and purified according to Liu et al.29Plasmid, strains, and media
The Pichia pastoris surface display vector pKFS was provided by Professor Ying Lin (South China University of Technology). The flocculation functional domain (FFD) of lectin-like cell-wall protein (Flo1p) with its own secretion signal sequence was cloned into the N-terminal of vector pPIC9K without signal peptide to get the pKFS, which was used as the yeast cell surface display vector. The codon-optimized pclmG47I was synthesized by replacing the rare codon in the pclG47I sequence according to the preferred codon usage of Pichia pastoris GS115 (BGI Co., Beijing, China). E. coli DH5α was used for pclmG47I cloning and sequencing, and P. pastoris GS115 was used for the expression of the PCLMG47I. E. coli strains were cultured in Luria–Bertani (LB) medium. The medium for yeast culture were prepared as described Liu et al.30Construction of the expression plasmid on the yeast cell surface
The pclmG47I fragment was digested with MluI and NotI and cloned into the MluI/NotI site of pKFS to produce the recombinant plasmid pKFS-pclmG47I. Then, the resulting plasmid was transformed into E. coli DH5α competent cells. After restriction enzymes digestion analysis and DNA sequencing confirmation, the plasmid pKFS-pclmG47I was linearized by SacI and transformed into P. pastoris GS115 competent cells. The positive transformants harboring multicopy integration of fdd-pclmG47I were screened on MD plates with G418 (0.5–2.5 mg mL−1) for the production of the displayed PCLMG47I (dPCLMG47I).Preparation of dPCLMG47I-displaying whole-cell biocatalyst
The selected recombinant strain, GS115/pKFS-pclmG47I, was inoculated into 5 mL of YPD medium and cultured at 30 °C for 24 h in 50 mL flask at 220 rpm shaking. Then, 1 mL of culture was transferred to a 250 mL flask containing 50 mL of BMGY medium and cultured at 30 °C at 220 rpm shaking until the optical density at 600 nm reached 2.0. The cells were harvested by centrifugation at 8000 × g for 20 min and re-suspended in a 250 mL flask with 50 mL BMMY medium. In addition, 0.5% methanol (final concentration, v/v) was added to the culture medium every 12 h to maintain the induction of the fusion protein FDD-PCLMG47I expression with a total induction time of 120 h at 30 °C at 220 rpm shaking. In addition to the aforementioned operation, the fermentation conditions, including the methanol concentration, pH value, culture volume, temperature, and shaking speed, were optimized to improve the expression level of dPCLMG47I. The induced cells were centrifugally harvested at 8000 × g for 10 min and washed twice with 50 mmol L−1 sodium phosphate buffer (pH 8.0) to remove media components, and then the harvested cells were lyophilized for 24 h. The obtained dPCLMG47I-expressing cells were used as WCB for the following study.PCL activity assay
The activity assay of lipase was carried out according to the method of Liu et al.29 One unit of lipase activity was defined as the amount of enzyme required to hydrolyze olive oil to release 1 μmol of fatty acid per minute.Transesterification of soybean oil with methanol
The enzymatic transesterification reactions were performed in a 20 mL screw-cap glass vial containing 1 g of soybean oil, dPCLMG47I WCB, and distilled water with constant shaking at 200 rpm. The effects of different parameters, such as temperature, oil/methanol molar ratio, enzyme loading, water content, methanol addition, and reaction time, on biodiesel production were investigated using single-factor experimental design. After the reaction, the reaction mixture was collected and centrifuged at 5000 × g for 10 min, and the upper layer was analyzed via gas chromatography to determine the contents of FAMEs.Gas chromatography analysis of biodiesel
The sample analysis was performed on Agilent 7890A gas chromatography (GC) system (Agilent Technologies, Santa Clara, California, USA) equipped with a flame-ionization detector, and a HP-5MS column (Agilent Technologies) [60 m (length) × 320 μm (inner diameter), 0.25 μm (film thickness)]. Injector and detector temperatures were set at 245 °C and 360 °C, respectively. Nitrogen was used as the carrier gas with the flow rate of 1 mL min−1. For analyzing the oil samples, the column temperature was held at 90 °C for 0.5 min and then elevated to 170 °C at a rate of 10 °C min−1 and maintained at 170 °C for 8 min and then from 170 to 250 °C at a rate of 4 °C min−1, and then the temperature was maintained at 250 °C for 4 min. For the analysis of glycerol samples, the column temperature was held at 120 °C for 1 min and then elevated to 180 °C at a rate of 10 °C min−1 and maintained at 180 °C for 1 min. The contents of methyl palmitate, methyl stearate, methyl oleate, methyl linoleate, and methyl linolenate were calculated based on the standard curves derived from standard solutions of each. FAMEs yield (%) was defined as the ratio between the amount of FAMEs produced, and the total amount of FAMEs when all soybean oil was converted.Reusability of dPCLMG47I
To evaluate the reusability of dPCLMG47I, the biocatalyst was reused for the transesterification of soybean oil under the optimal conditions. After the reaction completion, the biocatalyst was collected from the reaction mixture by centrifugation at 5000 × g for 5 min. The recovered biocatalyst was washed with 50 mmol L−1 sodium phosphate buffer (pH 8.0) and remixed in fresh reactants to initiate the next new batch reaction under the same conditions. In each batch of reaction, the FAMEs yield from the reaction mixture was determined.Results and discussion
Cloning of the pclmG47I gene in P. pastoris
To maximize PCLG47I expression, the codon optimized pclG47I gene (pclmG47I) was synthesized based on the codon bias of P. pastoris. Notably, many heterologous proteins could be expressed at a higher level in P. pastoris through this approach.19,30,31 After optimization, pclmG47I gene had 155 nucleotide variations corresponding to 135 of 258 codons switch to P. pastoris's favorite, which could enhance the PCLMG47I expression level in P. pastoris. In addition, FFD, which is located near the N-terminus of Flo1p, can recognize and link to α-mannan carbohydrates of the yeast cell-wall by noncovalent bonds. It was reported that FFD was successfully used as anchor to immobilize the heterologous proteins on the yeast cell surface in many studies.13,32,33 In this study, to display PCLMG47I on the cell surface of P. pastoris, the PCLMG47I was fused to the C-terminus of the FFD by cloning the pclmG47I gene into the vector pKFS to engineer the plasmid pKFS-pclmG47I. Then, the resulting plasmid was transformed into P. pastoris GS115, and a recombinant P. pastoris strain containing multiple integrated PCLMG47I expression cassettes (P. pastoris/pKFS-pclmG47I) was selected on MD medium plates with G418 at concentrations of 2.0 mg mL−1.Preparation of dPCLMG47I WCB
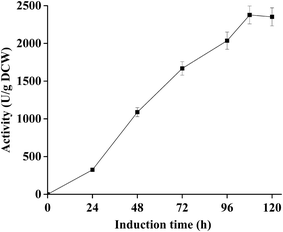
As shown in Fig. 1, the hydrolysis activity of dPCLMG47I on the P. pastoris GS115 cell surface expressed by P. pastoris/pKFS-pclmG47I dramatically increased in a time course manner. After methanol induction for 108 h in a flask, the highest activity of dPCLMG47I was 2377 U g−1 dry cell weight (DCW) under optimal conditions [0.5% (v/v) methanol in BMMY medium, pH 6.0, 32 °C, culture volume of 30 mL, and shaking at 260 rpm]. The culture supernatant displayed low hydrolysis activity (66 U mL−1), indicating that most of the FFD-PCLMG47I was successfully immobilized on the cell wall of P. pastoris GS115. After freezing at −80 °C for 12 h and drying with a lyophilizer for 24 h, the dPCLMG47I powder with activity of 2377 U g−1 DCW was obtained.Currently, the main limitations for the scale-up industrial implementation of lipase catalyzed biodiesel production include (i) the high cost of most commercial lipases due to their low production efficiency and (ii) the relatively weak reusability of these lipases. So far, E. coli has been widely used for the expression of many recombinant proteins, however, the heterogeneous proteins are expressed at relatively low levels34 and obtained with inconvenient preparation process because they are intracellularly expressed. In contrast, P. pastoris, as an established protein expression host, has been successfully applied for the overexpression of recombinant proteins by high cell densities in economical culture media.35 At present, the P. pastoris cell surface display system has been used for the expression of heterogeneous proteins on the yeast cells as WCB with great success,13 which can combine high-level gene expression, protein purification, and enzyme immobilization. In fact, various lipases have been effectively expressed on the surfaces of yeast cells for biodiesel production.13,14 However, to our knowledge, the present study is the first report on the display of a lipase from P. cyclopium on the yeast cell surface using P. pastoris.
To date, most studies have focused on the lipases expression by yeast display systems for biodiesel production from soybean oil rather than from waste oils,14 since waste oils have more undesired properties than refined oils, and refined soybean oil is a high-quality substrate for biodiesel production, which was beneficial to the evaluation of a novel lipase for biodiesel production. Hence, to determine whether biodiesel production through transesterification is feasibly catalyzed by dPCLMG47I WCB, the soybean oil was used as the feedstock substrate. In this research, we designed a series of experimental to study the optimal reaction conditions for biodiesel production with dPCLMG47I WCB.
Effect of reaction temperature
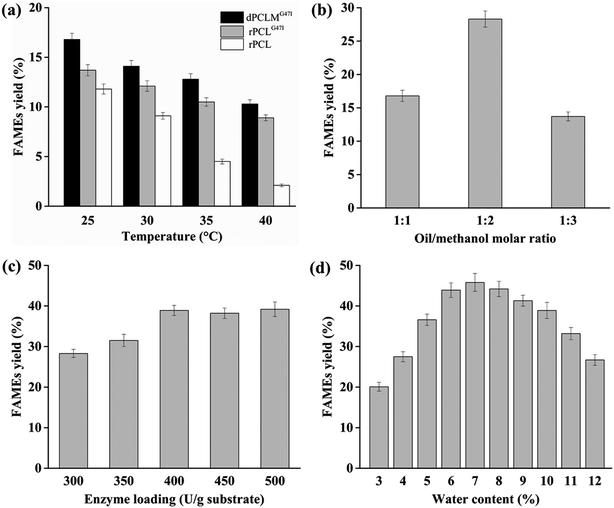
Generally, the higher temperature can enhance the lipases activity and help substrates absorb required energy to break through the energy barrier, resulting in the improvement of the reaction rate.36 However, the higher temperature has potential risk to denature the lipases, thereby decreasing the reaction rate. In addition, the lipase-catalyzed transesterification for biodiesel production was often performed for a long reaction time, which makes it necessary to maintain the lipase activity for a prolonged time. Hence, the optimum temperature and thermostability of the lipases are important for high biodiesel yield.The effects of temperatures from 25 °C to 40 °C on the biodiesel production using rPCL, rPCLG47I, and dPCLMG47I were investigated when the other variables were set as follows: water content 10% (v/w), oil/methanol molar ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, lipase loading 300 U g−1 substrate, and a reaction time of 24 h. The results are shown in Fig. 2(a). When the reaction temperature raised from 25 °C to 40 °C, the FAMEs yields obtained by rPCL, rPCLG47I, and dPCLMG47I-catalyzed transesterification gradually decreased, indicating that the optimum temperature for the stability of rPCL, rPCLG47I, and dPCLMG47I was 25 °C. Additionally, the dPCLMG47I WCB exhibited higher yield of biodiesel than rPCL and rPCLG47I at temperatures between 25 °C and 40 °C, suggesting that the thermostability of dPCLMG47I was significantly improved in the form of the WCB engineered with the P. pastoris cell surface display system. Our findings were consistent with the findings of other studies.33,37,38 The possible reason for this finding may be that the WCB provides a natural environment for enzyme localization and conformation, protecting cellular proteins against heat denaturation and helping them maintain their activity at higher temperature.15 In our study, the maximum FAMEs yield (16.8%) was observed by dPCLMG47I WCB catalysis at 25 °C. In many studies, the optimal temperature of the most lipases for biodiesel synthesis was between 30 and 55 °C.4,36,39,40 The PCL is most active at 25 °C and stable at temperatures below 25 °C, suggesting that this enzyme cannot maintain its catalytic activity for a long time when the temperature is above 25 °C. In a previous study, the thermostability of PCLG47I at temperatures above 25 °C was significantly improved by the substitution of Gly with Ile at position 47 in PCL.29 The presented results showed that by immobilization of PCLMG47I onto P. pastoris cells, dPCLMG47I exhibited more stability than PCLG47I, which made it more suitable for biodiesel production at 25 °C for a prolonged reaction time. It has been reported that lower temperatures could reduce the reaction rate, causing the prolongation of the reaction time for a high biodiesel yield.39 These results showed that biodiesel production at a lower temperature and less reaction time had many advantages, such as energy saving, milder reaction conditions, and higher efficiency, and thus reducing the production cost of biodiesel. Therefore, the reaction temperature was fixed at 25 °C in the subsequent experiments.
1, lipase loading 300 U g−1 substrate, and a reaction time of 24 h. The results are shown in Fig. 2(a). When the reaction temperature raised from 25 °C to 40 °C, the FAMEs yields obtained by rPCL, rPCLG47I, and dPCLMG47I-catalyzed transesterification gradually decreased, indicating that the optimum temperature for the stability of rPCL, rPCLG47I, and dPCLMG47I was 25 °C. Additionally, the dPCLMG47I WCB exhibited higher yield of biodiesel than rPCL and rPCLG47I at temperatures between 25 °C and 40 °C, suggesting that the thermostability of dPCLMG47I was significantly improved in the form of the WCB engineered with the P. pastoris cell surface display system. Our findings were consistent with the findings of other studies.33,37,38 The possible reason for this finding may be that the WCB provides a natural environment for enzyme localization and conformation, protecting cellular proteins against heat denaturation and helping them maintain their activity at higher temperature.15 In our study, the maximum FAMEs yield (16.8%) was observed by dPCLMG47I WCB catalysis at 25 °C. In many studies, the optimal temperature of the most lipases for biodiesel synthesis was between 30 and 55 °C.4,36,39,40 The PCL is most active at 25 °C and stable at temperatures below 25 °C, suggesting that this enzyme cannot maintain its catalytic activity for a long time when the temperature is above 25 °C. In a previous study, the thermostability of PCLG47I at temperatures above 25 °C was significantly improved by the substitution of Gly with Ile at position 47 in PCL.29 The presented results showed that by immobilization of PCLMG47I onto P. pastoris cells, dPCLMG47I exhibited more stability than PCLG47I, which made it more suitable for biodiesel production at 25 °C for a prolonged reaction time. It has been reported that lower temperatures could reduce the reaction rate, causing the prolongation of the reaction time for a high biodiesel yield.39 These results showed that biodiesel production at a lower temperature and less reaction time had many advantages, such as energy saving, milder reaction conditions, and higher efficiency, and thus reducing the production cost of biodiesel. Therefore, the reaction temperature was fixed at 25 °C in the subsequent experiments.
Effect of oil to methanol molar ratio
High methanol concentration can shift the chemical equilibrium towards biodiesel synthesis, leading to an increased biodiesel yield.41 However, on the other hand, an excessive amount of methanol can inhibit lipase activity, resulting in a decreased yield.4Experiments were designed to examine the effect of the molar ratio of oil to methanol in the range of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 on biodiesel production, while the other variables were set as follows: water content 10% (v/w), lipase loading 300 U g−1 substrate, temperature of 25 °C, and reaction time of 24 h. The results are shown in Fig. 2(b). Our data indicated that FAMEs yield increased when the molar ratio of oil to methanol varied from 1
3 on biodiesel production, while the other variables were set as follows: water content 10% (v/w), lipase loading 300 U g−1 substrate, temperature of 25 °C, and reaction time of 24 h. The results are shown in Fig. 2(b). Our data indicated that FAMEs yield increased when the molar ratio of oil to methanol varied from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, and the maximum FAMEs yield (28.3%) was obtained at an oil to methanol molar ratio of 1
2, and the maximum FAMEs yield (28.3%) was obtained at an oil to methanol molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. When the molar ratio of oil to methanol exceeded 1
2. When the molar ratio of oil to methanol exceeded 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, the FAMEs yield decreased. The higher ratios (1
2, the FAMEs yield decreased. The higher ratios (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) of oil to methanol could deactivate the dPCLMG47I because of the significant excess of methanol in the reaction mixture, and the lower oil/methanol molar ratios (1
3) of oil to methanol could deactivate the dPCLMG47I because of the significant excess of methanol in the reaction mixture, and the lower oil/methanol molar ratios (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were not sufficient to perform a more complete equilibrium displacement towards the biodiesel. Consequently, the oil to methanol molar ratio was set at 1
1) were not sufficient to perform a more complete equilibrium displacement towards the biodiesel. Consequently, the oil to methanol molar ratio was set at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 in the subsequent experiments.
2 in the subsequent experiments.
Effect of dPCLMG47I loading
The amount of dPCLMG47I is a key factor that affects the cost and yield of biodiesel production. To determine the effect of the amount of dPCLMG47I on the biodiesel reaction, the loading of dPCLMG47I varied from 300 to 500 U g−1 substrate were tested, while the other factors were fixed as follows: water content 10% (v/w), oil/methanol molar ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, temperature of 25 °C, and reaction time of 24 h. The results are shown in Fig. 2(c). FAMEs yield increased from 28.3% to 38.9% when dPCLMG47I loading amount ranged from 300 to 400 U g−1. Subsequently, the FAMEs yield remained constant with an increase in dPCLMG47I loading. More substrate molecules could be accessed by the active center of the lipase when the amount of available lipase was enhanced,42 which should improve the yield of biodiesel. However, the presence of excess lipase in the reaction did not improve biodiesel yield as expected. The possible reason may be that not all active sites of lipase could be exposed to the substrates.4 Thus, the lipase should be used with a minimum amount that can be consumed by the reaction since overdose of lipase does not effectively enhance the biodiesel yield. Hence, 400 U g−1 of dPCLMG47I was selected as the optimum enzyme loading applied in the subsequent experiments.
2, temperature of 25 °C, and reaction time of 24 h. The results are shown in Fig. 2(c). FAMEs yield increased from 28.3% to 38.9% when dPCLMG47I loading amount ranged from 300 to 400 U g−1. Subsequently, the FAMEs yield remained constant with an increase in dPCLMG47I loading. More substrate molecules could be accessed by the active center of the lipase when the amount of available lipase was enhanced,42 which should improve the yield of biodiesel. However, the presence of excess lipase in the reaction did not improve biodiesel yield as expected. The possible reason may be that not all active sites of lipase could be exposed to the substrates.4 Thus, the lipase should be used with a minimum amount that can be consumed by the reaction since overdose of lipase does not effectively enhance the biodiesel yield. Hence, 400 U g−1 of dPCLMG47I was selected as the optimum enzyme loading applied in the subsequent experiments.
Effect of water content
Water plays an important role in biphasic systems for biodiesel production. The appropriate water content is essential for maintaining the three-dimensional structure of lipase for its activity and increasing the available interface between the aqueous and organic phases for lipase catalysis.41,42 However, excessive water content can increase the flexibility of lipase and activate the hydrolytic activity of lipase in aqueous phase, leading to a decrease in transesterification yields.41,42 The optimum water content can help the lipase maintain maximal transesterification activity while only minimal hydrolysis activity in the reaction.The effect of different water contents from 3% to 12% (v/w, based on soybean oil) on biodiesel production was determined when other variables were set as follows: oil/methanol molar ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, lipase loading 400 U g−1 substrate, temperature of 25 °C, and reaction time of 24 h. The results are shown in Fig. 2(d). The FAMEs yield increased when the water contents varied from 3 to 7%, and the maximum yield reached 45.8% with 7% water content. When the water content was higher than 7%, the production significantly decreased. These results suggested that the optimum water content could enhance the efficiency of lipase on the transesterification reaction, leading to a higher production biodiesel yield. Many studies have shown that the best water content varied depending on the feedstock, the lipase type, the immobilized support, and the organic solvent employed.43,44 Studies indicated that the optimum water content required to maintain the highest biodiesel yield is 1.15–20% based on the substrate weight.1,2,4,36,42,45 Subsequently, the water content of the reaction was set at 7% in the remaining experiments.
2, lipase loading 400 U g−1 substrate, temperature of 25 °C, and reaction time of 24 h. The results are shown in Fig. 2(d). The FAMEs yield increased when the water contents varied from 3 to 7%, and the maximum yield reached 45.8% with 7% water content. When the water content was higher than 7%, the production significantly decreased. These results suggested that the optimum water content could enhance the efficiency of lipase on the transesterification reaction, leading to a higher production biodiesel yield. Many studies have shown that the best water content varied depending on the feedstock, the lipase type, the immobilized support, and the organic solvent employed.43,44 Studies indicated that the optimum water content required to maintain the highest biodiesel yield is 1.15–20% based on the substrate weight.1,2,4,36,42,45 Subsequently, the water content of the reaction was set at 7% in the remaining experiments.
Effect of methanol additional strategy
The addition of appropriate amounts of methanol as a reaction substrate to the reaction mixture can shift the reaction to biodiesel synthesis and improve the biodiesel yield, whereas excess amounts of methanol will lead to the denaturation and inactivation of lipase activity. Hence, it is necessary to reach the balance point of methanol concentration between catalysis and toxicity for the high yield of biodiesel. Until recently, many studies have reported that a strategy of step-wise addition of methanol into the reaction system could be used to successfully promote biodiesel production with high efficiency.4,41,42,46 However, the detailed methods of every strategy were different, depending on the feedstocks variety, lipases property, and alcohols type.Based on the optimal initial oil/methanol molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, we designed different strategies to introduce additional methanol to the reaction mixture (extra methanol with oil/methanol molar ratio of 1
2, we designed different strategies to introduce additional methanol to the reaction mixture (extra methanol with oil/methanol molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5, 1
0.5, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.75, and 1
0.75, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, respectively, was added at different reaction time points of 4, 8, and 12 h). Meanwhile, the other variables were set as follows: water content 7% (v/w), initial oil/methanol molar ratio 1
1, respectively, was added at different reaction time points of 4, 8, and 12 h). Meanwhile, the other variables were set as follows: water content 7% (v/w), initial oil/methanol molar ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
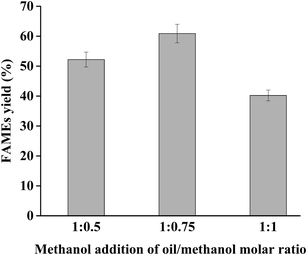
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, lipase loading 400 U g−1 substrate, temperature of 25 °C, and reaction time of 24 h. The results are shown in Fig. 3. The maximum and second FAMEs yield of 60.9% and 52.2% were obtained using an oil to methanol molar ratio of 1
2, lipase loading 400 U g−1 substrate, temperature of 25 °C, and reaction time of 24 h. The results are shown in Fig. 3. The maximum and second FAMEs yield of 60.9% and 52.2% were obtained using an oil to methanol molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.75 and 1
0.75 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5, respectively, which were all higher than the yield from no additional methanol. These findings indicated that the accumulated amount of new additional methanol and remaining methanol in reaction mixture at 4, 8, and 12 h could promote the transesterification reaction without inactivating dPCLMG47I, causing an improved FAMEs yield. Compared with the 1
0.5, respectively, which were all higher than the yield from no additional methanol. These findings indicated that the accumulated amount of new additional methanol and remaining methanol in reaction mixture at 4, 8, and 12 h could promote the transesterification reaction without inactivating dPCLMG47I, causing an improved FAMEs yield. Compared with the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 molar ratio, the 1
0.5 molar ratio, the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.75 molar ratio showed a higher yield because of the extra methanol content. However, FAMEs yield (40.2%) decreased when the extra methanol of oil to methanol molar ratio of 1
0.75 molar ratio showed a higher yield because of the extra methanol content. However, FAMEs yield (40.2%) decreased when the extra methanol of oil to methanol molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was added to the reaction mixture compared with that obtained without the addition of methanol. This result may be caused by the accumulation of methanol beyond the tolerance of dPCLMG47I, leading to lipase denaturation. Therefore, the best methanol-adding strategy was methanol addition with oil/methanol molar ratio of 1
1 was added to the reaction mixture compared with that obtained without the addition of methanol. This result may be caused by the accumulation of methanol beyond the tolerance of dPCLMG47I, leading to lipase denaturation. Therefore, the best methanol-adding strategy was methanol addition with oil/methanol molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.75 at 4 h intervals with initial molar ratios of oil
0.75 at 4 h intervals with initial molar ratios of oil![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol of 1
methanol of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 in the remaining experiments.
2 in the remaining experiments.
Effect of reaction time
Extended reaction time typically results in higher biodiesel yield, but this may consume excessive energy, leading to higher cost for biodiesel production. The effect of reaction times from 0–24 h on the biodiesel production was investigated when the other variables were set as follows: water content 7% (v/w), lipase loading 400 U g−1 substrate, the initial oil/methanol molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, the addition of methanol with oil/methanol molar ratio of 1
2, the addition of methanol with oil/methanol molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
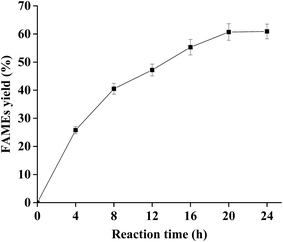
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.75 at various reaction times of 4, 8, and 12 h, and temperature of 25 °C. The results are shown in Fig. 4. The FAMEs yield increased in a time course manner from 0 to 20 h. Generally, 2 moles of FAMEs and 1 mole of MAGs are produced by 2 mol of methanol and 1 mole of TAGs at a stoichiometric ratio when sn-1,3 regioselective lipases are used as catalysts, which can give the theoretical biodiesel yield of 66.7%.9 In this study, the maximum yield reached 60.7% after 20 h, which was close to the theoretical maximum for sn-1,3 regioselective lipases (66.7%). When the reaction time exceeded 20 h, FAMEs yield maintained a constant with the increasing reaction time because PCL shows very low activity against MAGs.47 Most importantly, glycerol, the by-product that had negative effect on the biodiesel production, was not detected along the reactions. The previous studies reported that it was approximately 6–72 h for the suitable reaction time of biodiesel production,13,14 and a shorter reaction time was preferred to save energy in industrial scale applications. Based on these results, a reaction time of 20 h was selected in the subsequent experiments.
0.75 at various reaction times of 4, 8, and 12 h, and temperature of 25 °C. The results are shown in Fig. 4. The FAMEs yield increased in a time course manner from 0 to 20 h. Generally, 2 moles of FAMEs and 1 mole of MAGs are produced by 2 mol of methanol and 1 mole of TAGs at a stoichiometric ratio when sn-1,3 regioselective lipases are used as catalysts, which can give the theoretical biodiesel yield of 66.7%.9 In this study, the maximum yield reached 60.7% after 20 h, which was close to the theoretical maximum for sn-1,3 regioselective lipases (66.7%). When the reaction time exceeded 20 h, FAMEs yield maintained a constant with the increasing reaction time because PCL shows very low activity against MAGs.47 Most importantly, glycerol, the by-product that had negative effect on the biodiesel production, was not detected along the reactions. The previous studies reported that it was approximately 6–72 h for the suitable reaction time of biodiesel production,13,14 and a shorter reaction time was preferred to save energy in industrial scale applications. Based on these results, a reaction time of 20 h was selected in the subsequent experiments.
Reusability of dPCLMG47I
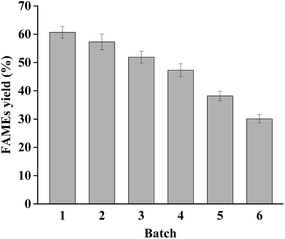
It is crucial to reuse the biocatalyst in most bioprocesses, and biocatalysts with good reusability can reduce the production costs effectively. To evaluate the reusability of dPCLMG47I, the reaction was performed under the optimal reaction conditions. Generally, a considerable amount of by-product glycerol catalyzed by non-region specific lipase can attach onto the yeast cell surface, resulting in cell agglutination, forming an obstacle refraining the substrates from accessing the active sites of lipases.14 In addition, the use of WCB absorbing the glycerol is inconvenient for subsequent reactions, and an additional step is required to remove the adsorbed glycerol, increasing the recycling process complexity and cost. The lipases having very low activity against MAGs can simultaneously produce biodiesel and MAGs, which is beneficial to biodiesel production by avoiding the generation of glycerol. Therefore, dPCLMG47I can maintain its catalytic activity during the reaction process and can be simply recycled by centrifuging the reaction mixture after each batch for reuse.The dPCLMG47I was recovered from the reaction mixture by centrifugation after each batch reaction (20 h per batch), washed with 50 mmol L−1 sodium phosphate buffer (pH 8.0), and added to the fresh reactants for the next batch. As shown in Fig. 5, the FAMEs yield gradually decreased with increasing recycle number. The FAMEs yield in the first batch was 60.7%, and after 4 batches, the FAMEs yield was 47.3%, which was approximately 80% of the initial batch yield. From a practical perspective, dPCLMG47I can be reused at least 4 times. This result indicated that WCB provides an excellent environment for displayed lipases to maintain their conformation. The high reusability of the biocatalysts may significantly reduce the costs of industrial biodiesel production. Recently, various heterologous lipases have been successfully displayed by yeast cell surface display systems as recyclable WCB for biodiesel production, and lipase WCBs could be reused from two to as high as twenty batches.14 The reuse frequency difference between different studies might be caused by many factors such as the feedstock source, enzyme property, enzyme loading, oil/methanol molar ratio, and methanol adding strategy. Our data indicated that dPCLMG47I had relatively good reusability and is a promising biocatalyst for commercial biodiesel production. Furthermore, dPCLMG47I, could be combined with mono- and diacylglycerol lipases instead of one lipase, providing a route to increase the biodiesel yield and reduce the cost of biodiesel production.
Conclusions
This study is the first investigation of a novel P. pastoris WCB with the overexpression of PCLMG47I for biodiesel production at a low temperature. The highest FAMEs yield (60.7%) was achieved under optimum conditions, and the biodiesel yield remained 47.3% after 4 batch cycles. The dPCLMG47I WCB has many advantages, including convenient preparation, high catalytic activity and stability at low temperature, good tolerance to methanol, easy recovery, and relatively high reusability which would be further improved in the future. These features demonstrated its bright perspective as a biocatalyst for biodiesel production in a relatively cost-effective manner. Moreover, the resulting enzymatic process may provide a potential method for biodiesel production at an industrial scale.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Fund of China (31571805 and 31461143026); the Tianjin Support Plan Program of Science and Technology (16YFZCSY01040); and the Tianjin Natural Science Fund (17JCYBJC23700).References
- T. W. Tan, J. K. Lu, K. L. Nie, L. Deng and F. Wang, Biotechnol. Adv., 2010, 28, 628–634 CrossRef PubMed.
- K. H. Kim, O. K. Lee, C. H. Kim, J. W. Seo, B. R. Oh and E. Y. Lee, Bioresour. Technol., 2016, 211, 472–477 CrossRef PubMed.
- S. B. Velasquez-Orta, J. G. M. Lee and A. Harvey, Fuel, 2012, 94, 544–550 CrossRef.
- T. C. Kuo, J. F. Shaw and G. C. Lee, Bioresour. Technol., 2015, 192, 54–59 CrossRef PubMed.
- S. Hama and A. Kondo, Bioresour. Technol., 2013, 135, 386–395 CrossRef PubMed.
- M. Aarthy, P. Saravanan, M. K. Gowthaman, C. Rose and N. R. Kamini, Chem. Eng. Res. Des., 2014, 92, 1591–1601 CrossRef.
- L. P. Christopher, H. Kumar and V. P. Zambare, Appl. Energy, 2014, 119, 497–520 CrossRef.
- J. Y. Yan, Y. J. Yan, S. X. Liu, J. Hu and G. L. Wang, Bioresour. Technol., 2011, 102, 4755–4758 CrossRef PubMed.
- J. Rodrigues, V. Perrier, J. Lecomte, E. Dubreucq and S. Ferreira-Dias, Bioresour. Technol., 2016, 218, 1224–1229 CrossRef PubMed.
- A. Gog, M. Roman, M. Tosa, C. Piz and F. D. Irimie, Renewable Energy, 2012, 39, 10–16 CrossRef.
- M. L. Verma, C. J. Barrow and M. Puri, Appl. Microbiol. Biotechnol., 2013, 97, 23–39 CrossRef PubMed.
- A. Guldhe, B. Singh, T. Mutanda, K. Permaul and F. Bux, Renewable Sustainable Energy Rev., 2015, 41, 1447–1464 CrossRef.
- Y. Liu, R. Zhang, Z. S. Lian, S. H. Wang and A. T. Wright, J. Mol. Catal. B: Enzym., 2014, 106, 17–25 CrossRef.
- Z. Liu, S. H. Ho, T. Hasunuma, J. S. Chang, N. Q. Ren and A. Kondo, Bioresour. Technol., 2016, 215, 324–333 CrossRef PubMed.
- W. A. Duetz, J. B. V. Beilen and B. Witholt, Curr. Opin. Biotechnol., 2001, 12, 419–425 CrossRef PubMed.
- Z. Jin, S. Y. Han, L. Zhang, S. P. Zheng, Y. Wang and Y. Lin, Bioresour. Technol., 2013, 130, 102–109 CrossRef PubMed.
- R. Koda, T. Numata, S. Hama, S. Tamalampudi, K. Nakashima, T. Tanaka, C. Ogino, H. Fukuda and A. Kondo, J. Mol. Catal. B: Enzym., 2010, 66, 101–104 CrossRef.
- T. Takaya, R. Koda, D. Adachi, K. Nakashima, J. Wada, T. Bogaki, C. Ogino and A. Kondo, Appl. Microbiol. Biotechnol., 2011, 90, 1171–1177 CrossRef PubMed.
- J. Y. Yan, X. L. Zheng and S. Y. Li, Bioresour. Technol., 2014, 151, 43–48 CrossRef PubMed.
- T. Matsumoto, S. Takahashi, M. Kaieda, M. Ueda, A. Tanaka, H. Fukuda and A. Kondo, Appl. Microbiol. Biotechnol., 2001, 57, 515–520 CrossRef PubMed.
- D. F. Huang, S. Y. Han, Z. L. Han and Y. Lin, Biochem. Eng. J., 2012, 63, 10–14 CrossRef.
- Z. L. Han, S. Y. Han, S. P. Zheng and Y. Lin, Appl. Microbiol. Biotechnol., 2009, 85, 117–126 CrossRef PubMed.
- Y. J. Yan, L. Xu and M. Dai, RSC Adv., 2012, 2, 6170–6173 RSC.
- A. Robles-Medina, P. A. González-Moreno, L. Esteban-Cerdán and E. Molina-Grima, Biotechnol. Adv., 2009, 27, 398–408 CrossRef PubMed.
- N. J. Zhong, L. Z. Cheong and X. B. Xu, Eur. J. Lipid Sci. Technol., 2014, 116, 97–107 CrossRef.
- N. Li and M. H. Zong, J. Mol. Catal. B: Enzym., 2010, 66, 43–54 CrossRef.
- H. Chahinian, L. Nini, E. Boitard, J. P. Dubès, L. Sarda and L. C. Comeau, Lipids, 2000, 35, 919–925 CrossRef PubMed.
- Q. J. Yan, X. J. Duan, Y. Liu, Z. Q. Jiang and S. Q. Yang, Biotechnol. Biofuels, 2016, 86, 1–13 Search PubMed.
- Y. H. Liu, H. Liu, L. Huang, S. Gui, D. Zheng, L. B. Jia, Y. Fu and F. P. Lu, RSC Adv., 2017, 7, 38538–38548 RSC.
- Y. H. Liu, L. Huang, M. J. Li, H. Liu, W. Guo, S. Gui, J. L. Niu and F. P. Lu, Process Biochem., 2016, 51, 1472–1478 CrossRef.
- S. W. Chang, G. C. Lee and J. F. Shaw, J. Agric. Food Chem., 2006, 54, 815–822 CrossRef PubMed.
- Z. B. Jiang, B. Gao, R. Ren, X. Y. Tao, Y. S. Ma and D. Z. Wei, BMC Biotechnol., 2008, 8, 1–7 CrossRef PubMed.
- Y. H. Liu, T. Zhang, J. Qiao, X. G. Liu, J. X. Bo, J. L. Wang and F. P. Lu, J. Agric. Food Chem., 2014, 62, 5354–5360 CrossRef PubMed.
- G. Potvin, A. Ahmad and Z. Zhang, Biochem. Eng. J., 2012, 64, 91–105 CrossRef.
- M. Ahmad, M. Hirz, H. Pichler and H. Schwab, Appl. Microbiol. Biotechnol., 2014, 98, 5301–5317 CrossRef PubMed.
- S. Kuepethkaew, K. Sangkharak, S. Benjakul and S. Klomklao, Renewable Energy, 2017, 104, 139–147 CrossRef.
- T. Tanino, T. Aoki, W. Y. Chung, Y. Watanabe, C. Ogino, H. Fukuda and A. Kondo, Appl. Microbiol. Biotechnol., 2009, 82, 59–66 CrossRef PubMed.
- H. Fukuda, S. Hama, S. Tamalampudi and H. Noda, Trends Biotechnol., 2008, 26, 668–673 CrossRef PubMed.
- A. Arumugam, D. Thulasidharan and G. B. Jegadeesan, Renewable Energy, 2018, 116, 755–761 CrossRef.
- X. M. Wang, X. L. Qin, D. M. Li, B. Yang and Y. H. Wang, Bioresour. Technol., 2017, 235, 18–24 CrossRef PubMed.
- J. J. Huang, J. Xia, W. Jiang, Y. Li and J. L. Li, Bioresour. Technol., 2015, 180, 47–53 CrossRef PubMed.
- Q. H. You, X. L. Yin, Y. P. Zhao and Y. Zhang, Bioresour. Technol., 2013, 148, 202–207 CrossRef PubMed.
- K. R. Jegannathan, S. Abang, D. Poncelet, E. S. Chan and P. Ravindra, Crit. Rev. Biotechnol., 2008, 28, 253–264 CrossRef PubMed.
- J. Lu, Y. Chen, F. Wang and T. Tan, J. Mol. Catal. B: Enzym., 2009, 56, 122–125 CrossRef.
- T. Rakkan, S. Suwanno, N. Paichid, T. Yunu, S. Klomklao and K. Sangkharak, Fuel, 2017, 209, 309–314 CrossRef.
- J. Amoah, S. H. Ho, S. Hama, A. Yoshida, A. Nakanishi, T. Hasunuma, C. Ogino and A. Kondo, Bioresour. Technol., 2016, 211, 224–230 CrossRef PubMed.
- A. Ibrik, H. Chahinian, N. Rugani, L. Sarda and L. C. Comeau, Lipids, 1998, 33, 377–384 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2018 |