Open Access Article
Open Access ArticleReconstitution properties of biologically active polymersomes after cryogenic freezing and a freeze-drying process†
Robert Ccorahua‡
 a,
Silvia Moreno‡b,
Hannes Gumzbc,
Karin Sahreb,
Brigitte Voit*bc and
Dietmar Appelhans
a,
Silvia Moreno‡b,
Hannes Gumzbc,
Karin Sahreb,
Brigitte Voit*bc and
Dietmar Appelhans *b
*b
aDepartment of Mechanical Engineering, Pontificia Universidad Católica del Peru, Lima 32, Peru
bLeibniz-Institut für Polymerforschung Dresden e.V., Hohe Straße 6, 01069 Dresden, Germany. E-mail: applhans@ipfdd.de; voit@ipfdd.de
cTechnische Universität Dresden, Faculty of Science, Department of Chemistry and Food Chemistry, 01069 Dresden, Germany
First published on 17th July 2018
Abstract
Reconstitution of biologically active polymersomes from the frozen or solid state into any fluid state is still a challenging issue for the design of new biological experiments and for the formulation of therapeutic agents. To gain knowledge about the reconstitution of pH-responsive and photo-crosslinked polymersomes, surface-functionalized and enzyme-containing polymersomers were cryogenically frozen (−20 °C) or freeze-dried with inulin as the lyoprotectant (0.1% w/v) and stored for a defined time period. Reconstituting those polymersomes in solution by thawing or a re-dispersing process revealed their original physical properties as well as their function as a pH-switchable enzymatic nanoreactor.
Introduction
Over the last two decades, polymersomes have been used as a supramolecular platform to mimic artificial organelles and cells and to imitate cell membrane functions for the communication/transport between compartments. Moreover, they are also usable as versatile materials in the field of drug delivery, bio-imaging, diagnostics, and theranostics.1–11 The great interest in them, fabricated by the assembly of amphiphilic block copolymers, is derived from the higher mechanical stability of polymeric membranes compared to their natural counterparts, the phospholipid bilayers, also named liposomes.1,2,10,11Generally, these polymeric vesicles possess versatile properties for successful use in biomedical applications. Studies on the surface functionalization of polymersomes have been carried out, while the loading of drugs, proteins and enzymes by polymersomes' cavity and membrane is also possible.1–20 Furthermore, various kinds of polymersomes address the required biological and pharmacological properties for in vitro and in vivo studies. However, the encapsulation of enzymes without deactivation by polymersomes and their long-term use for biomedical applications are still challenging issues.2,10,11,15,17,20 For example, the process of lyophilization is often used to prepare pharmaceutical formulations with easy handling, allowing reconstitution into solution when it is necessary.21 It is important to have a biologically active material of desirable quality that retains its physicochemical characteristics after lyophilization, storage and reconstitution. During the freezing step of some samples, undesired processes can happen such as membrane rupture of polymeric vesicles, deformation of shapes or tight sticking of several polymeric vesicles, which may result in non-completely re-dispersion or uncontrolled aggregation processes afterwards. To avoid such lyophilization stresses, lyoprotectant molecules are added to solutions before freeze-dried process starts.22,23 This also increases the long-term stability upon storage. Molecules such as inulin and mannitol have been previously explored as lyoprotectant molecules for the re-dispersion of polymersomes.19,20 The lyoprotectant molecules have shown to retain the stability and diameter of previously formed polymersomes after lyophilization.
Until now, in our group several enzymes (e.g., myoglobin or glucose oxidase) have been encapsulated by pH-switchable and photo-crosslinked polymersomes and their multicompartments.14,17 It was shown that the encapsulation provides an enhanced enzyme stability over time. Moreover, those polymersomes are also addressable for host–guest interactions with glycodendrimers as protein mimics (∅ ≤ 6 nm) at the inner and outer sphere of polymersome membrane. These supramolecular assemblies and hybrid architectures are excellent candidates for different bio-related studies.16,17,20,24 Therefore, the goal of this study was to investigate the reconstitution of biologically active, pH-responsive and photo-crosslinked polymersomes from frozen or solid state to fluid state (Fig. 1, 2 and 4). For this cryogenic freezing (−20 °C) and lyophilization processes, combined with long/short-term storage, have been applied and the influence of lyoprotectant molecules on the reconstitution of pure polymersomes (Fig. 1). This also included the study on enzyme loaded polymersomes as nanoreactors (Fig. 2). Moreover, biohybrid architectures, composed of human serum albumin-(HSA)-modified polymersomes (Psome-HSA), were also used and validated (Fig. 4).
Experimental part
Materials
If not stated otherwise, all chemicals were used as received. All chemicals, anhydrous tetrahydrofurane (THF, Sigma-Aldrich), anhydrous 2-butanone (Fluka) and triethylamine (Fluka) were stored over a molecular sieve. Poly(ethylene glycol) methyl ether (MeO-PEG-OH; Mn = 2000 g mol−1; Mw/Mn = 1.05), albumin from human serum (HSA), diethylaminoethyl methacrylate (DEAEM), 2,2′-bipyridine, 2-bromoisobutyryl bromide, 2-aminoethanol, 4-aminobutanol, methacryloylic chloride, copper-I-bromide, aluminum oxide (neutral, activated), myoglobin from equine skeletal muscle (Myo, essentially salt-free, lyophilized powder), guaiacol, phosphate buffered saline (tablet), (1R,8S,9s)-bicyclo[6.1.0]non-4-yn-9-ylmethyl N-succinimidyl carbonate (COD), inulin from chicory, D-mannitol, fluorescein-5(6)-isothiocyanate (FITC), albumin from human serum (HSA), sodium hydroxide and magnesium sulfate were purchased from Sigma-Aldrich. 3,4-Dimethylmaleic acid anhydride, THF, toluene, chloroform-d and ethyl acetate were purchased from Acros Organics. From Merck (Germany) n-hexane, hydrochloric acid (37%) and silica gel were purchased. 6-Monodeoxy-6-monoamino-β-cyclodextrin was purchased from Cyclodextrin Shop (Division of AraChem, Netherlands).Devices
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 g mol−1.
400 g mol−1.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 by volume. The preparation was done without salt.
1 by volume. The preparation was done without salt.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000× magnification ahead. The average was calculated by analyzing 250 particles for each sample.
000× magnification ahead. The average was calculated by analyzing 250 particles for each sample.Synthesis of the compounds
Stock solutions. (1R,8S,9s)-bicyclo[6.1.0]non-4-yn-9-ylmethyl N-succinimidyl carbonate (COD) (2 mg in 200 μL DMSO), fluorescein-5(6)-isothiocyanate (FITC) (2 mg in 200 μL DMSO).
HSA-COD. Albumin from human serum (HSA) (10 mg, 0.15 μmol) was dissolved in 500 μL carbonate buffer (pH 10) and stirred for 30 min before adding COD solution in DMSO (14 μL, 0.47 μmol).26 After 20 hours of stirring at room temperature, the mixture was extensively dialyzed against 1 mM PBS for 2 days (dialysis membrane: 3.5–5 kDa MWCO) to remove all unbounded molecules. Finally, the purified mixture was freeze dried overnight and the white product was isolated. MALDI-TOF-MS: 67
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 g mol−1 (3–4 COD groups attached to HSA).
500 g mol−1 (3–4 COD groups attached to HSA).
HSA-βCD. HSA-COD (10 mg, 0.15 μmol) was dissolved in 500 μL in carbonate buffer (pH 10) and stirred for 30 min before adding mono-(6-azido-6-desoxy)-β-cyclodextrin (1 mg, 0.9 μmol).27 After 48 hours of stirring at room temperature, the mixture was extensively dialyzed against 1 mM PBS for 2 days (dialysis membrane: 3.5–5 kDa MWCO) to remove all unbounded molecules. Finally, the purified mixture was freeze dried overnight and the white product was isolated. MALDI-TOF-MS: 69
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 g mol−1 (2COD groups + 2βCD groups attached to HSA).
900 g mol−1 (2COD groups + 2βCD groups attached to HSA).
HSA-βCD-FITC. HSA-βCD (10 mg, 0.15 μmol) was dissolved in 500 μL in carbonate buffer (pH 10) and stirred for 30 min before adding fluorescein 5(6)-isothiocyanate solution in DMSO (35 μL, 0.9 μmol). After 20 hours of stirring at room temperature, the mixture was extensively dialyzed against 1 mM PBS for 2 days (dialysis membrane: 3.5–5 kDa MWCO) to remove all unbounded molecules. Finally, the purified mixture was freeze-dried overnight and the yellow product was isolated. MALDI-TOF-MS: 71
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 g mol−1 (2COD groups + 2βCD groups + 3 FITC groups attached to HSA). This sample is stored at a known concentration in PBS at 4 °C (Cstock = 1.8 mg mL−1).
200 g mol−1 (2COD groups + 2βCD groups + 3 FITC groups attached to HSA). This sample is stored at a known concentration in PBS at 4 °C (Cstock = 1.8 mg mL−1).
Formation and characterization of polymersomes
Photo-crosslinking of polymersomes was performed using Omnicure S2000 (Lumen Dynamics Group Inc., Canada) device equipped with a high pressure mercury lamp (0.35 W cm−2, UV light between 320–500 nm). The polymersome solution was passed through a 0.8 μm nylon filter and then irradiated for 3 min to trigger the crosslinking. The irradiation procedure was carried out with 2 mL of polymersome solution.16,20,28
Psome frozen. Samples were frozen at −20 °C for different time periods (1, 7, 14 and 30 days) and slowly thawed to room temperature for carrying out DLS measurements, pH-titration experiments, cyclic pH switches and cryo-TEM study.
Psome-FD. Empty and loaded-polymersomes solutions (1 mg mL−1) were freeze-dried in presence of either mannitol (0.1% wt) or inulin (0.1% wt). First, the aqueous polymersome samples were frozen with liquid nitrogen. Then, they were placed in a freeze-drier overnight, and after freeze-drying process the samples were stored at 4 °C until re-dissolving. For re-dissolving procedure ultrapure water was added to the samples, reaching the same previous polymersome solution concentration before freeze-drying. Then, the pH is slightly lowered to pH 6.5 with PBS buffer while the sample is stirred at 200 rpm. After stirring for 30 min the pH was raised again to pH 8 and the characterization was carried out.
The same procedures were carried out with non-crosslinked polymersomes and were stored for 8 days. These samples were analysed by DLS to study the influence of crosslinking on the stability of the polymersomes.
Functionalized polymersomes
Myoglobin activity assay. The assay for determining enzymatic activity of myoglobin was adopted from Gräfe et al. with very slight modifications.20 Thus, 300 μL of sample solution are transferred into a micro-cuvette and 8 μL of hydrogen peroxide solution (1 M in PBS) and 8 μL of guaiacol solution (0.1 M in PBS) are added. A lid is placed on top of the micro-cuvette, the mixture is vigorously shaken and placed in the UV/Vis spectrometer. The absorbance at 470 nm is measured every 12 s for a duration of 400 s. Each assay is executed in triplicates.
Results and discussion
Reconstitution properties of empty polymersomes after cryogenic freezing and freeze-drying process
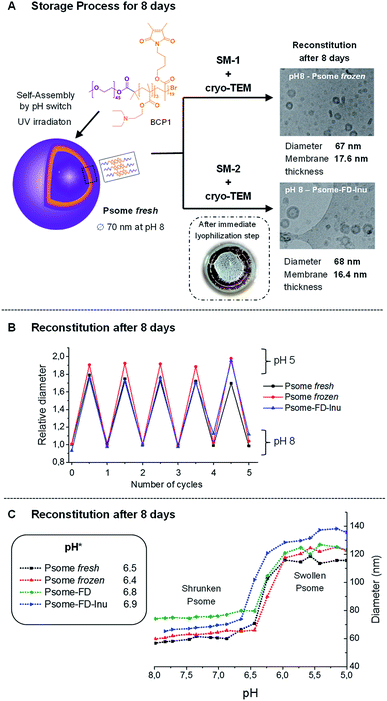
To carry out the above-mentioned study, our well established block copolymer system (BCP1, Fig. 1A) was synthesized by atom transfer radical polymerization (ATRP),25 using the previously published approach.15,20 This block copolymer has a methoxy end groups at the hydrophilic poly(ethylene glycol) (PEG) segment, while the hydrophobic part consists of pH-sensitive 2-(diethylamino)ethyl methacrylate (DEAEM) and photo-crosslinker, 3,4-dimethyl maleic imidobutyl methacrylate (DMIBMA). Then, polymersomes were fabricated by the self-assembly of BCP1, using the so-called pH switch method.29 Afterwards polymersomes were crosslinked by UV irradiation for 180 s. The final goal of our work was to optimize storage methods and stability of functionalized polymersomes loaded with proteins of biological interest. However, first experiments with empty polymersomes have been carried out.Depending on the type of polymersome application that is aimed for at a later stage, two storage methods SM-1 and SM-2 have been investigated: SM-1 – a cryogenic freezing process of polymersomes at −20 °C for 8 days followed by thawing reconstitution process to room temperature; and SM-2 – a lyophilization process in presence and absence of lyoprotectant molecules followed by a storage at 4 °C for 8 days and a reconstitution in selected solvent. It is important that the reconstitution of any lyophilized formulation, prepared by SM-2, should be easy in practice and should only take a minimal time.30
To apply the SM-2 method, it is important to know the necessary minimum concentration of lyoprotectant molecule which smoothly preserves all desirable characteristics (colloidal stability with defined diameter; cyclic pH switches; pH* = turning point which presents the half power on polymersomes' swelling) of pH-responsive polymersomes. Therefore, an optimization process with lyoprotectant molecules such as inulin (Inu) and mannitol (0.1, 1% and 5% w/v) were carried out. From these preliminary tests, it was not possible to reconstitute the polymersomes at 5% and 1% w/v Inu, high tendency towards aggregation was observed due to the interaction of H-bond-active inunlin as polysaccharide with polymersome entities. However, at 0.1% w/v Inu and 1% w/v mannitol very promising results were obtained (Table 1). Finally, 0.1% w/v Inu was chosen for the following experiments. The results indicated that this concentration is sufficient to protect the colloidal stability of photo-crosslinked polymersomes (=reconstitution) when applying SM-2. However, to dissolve our lyophilized polymersomes completely, it was necessary to lower the pH to 6.5 with PBS buffer and stirring of the corresponding solution for 30 minutes was needed. With this knowledge about the optimum concentration of lyoprotectant molecules (using only Inu with 0.1% w/v) we started our first experiment series with both storage methods SM-1 and SM-2 to study the colloidal stability (=hydrodynamic diameter (Dh)) of reconstituted photo-crosslinked polymersomes (Psome) by DLS and compared them with freshly prepared Psome sample. Fig. 1 presents the characteristics of photo-crosslinked polymersome samples depending on the applied storage methods SM-1 and SM-2, including results from cyclic pH-switches from dynamic light scattering (DLS), cryo-TEM study and pH-dependent titration experiments for determining diameters of various polymersomes. To get a visual impression on lyophilized sample after immediate freeze-drying process (FD), a photograph of the lyophilized sample in the presence of inulin is presented in Fig. 1A (bottom). The lyophilized samples are solid materials.
| Sample | Diametera (nm) | Polydispersitya | Diameterb (nm) | Thicknessb (nm) | ||
|---|---|---|---|---|---|---|
| Shrunkenc | Swollend | Shrunkenc | Swollend | Shrunkend | Shrunkend | |
| a The DLS measurements.b Cryo-TEM measurements.c pH 8 shrunken.d pH 5 swollen. | ||||||
| Psome fresh | 67 | 127 | 0.146 | 0.152 | — | — |
| Psome frozen | 80 | 144 | 0.214 | 0.217 | 67 | 17.6 |
| Psome-FD | 85 | 145 | 0.214 | 0.217 | — | — |
| Psome-FD-Inu (0.1 wt%) | 78 | 132 | 0.221 | 0.168 | 68.4 | 16.4 |
| Psome-FD-Mann (0.1 wt%) | 88 | 145 | 0.213 | 0.200 | — | — |
| Psome-FD-Mann (1 wt%) | 84 | 136 | 0.233 | 0.220 | — | — |
| Psome-FD-Mann (5 wt%) | 120.3 | 142.4 | 0.282 | 0.187 | — | — |
Table 1 summarizes the validated Dhs obtained from SM-1, SM-2 and freshly prepared Psome sample (Psome fresh). For shrunken Psome at pH 8 for Psome fresh Dh of 67 nm has been found, as expected from previous results. For samples after SM-2 with Inu (Psome-FD-Inu) Dh increased slightly to 78 nm and to 89 nm after SM-1 (Psome frozen), and Dh of 85 nm resulted for SM-2 in absence of Inu. For the swollen Psome samples at pH 5 validated Dhs also scatter from 127 nm for Psome fresh up to 144 and 145 nm for Psome-FD without Inu and Psome frozen, respectively. The Dh of swollen Psome-FD-Inu (132 nm) is close to the sample of Psome fresh (127 nm). Generally, the polydispersity (PDI) of all these samples are in the range of 0.15 and 0.22 as found in our previous studies.16,17,20,28 Dh data from DLS always respect the contribution of water shell surrounding Psome and, thus, are always larger than the geometrical diameter of shrunken Psome investigated by cryo-TEM.16,17,20,28 This observation is also made in the present study, exemplified for Psome frozen and Psome-FD-Inu. Data from cryo-TEM study (Table 1) outline that the average Dh (67–68 nm) and membrane thickness (16.4–17.6 nm) for Psome frozen and Psome-FD-Inu is similar, but smaller against data validated by DLS measurements (Table 1). Cryo-TEM images for Psome frozen and Psome-FD-Inu are presented in Fig. 1A.
With these promising results for the colloidal stability of differently stored Psome, the next experiment series of selected samples (Psome fresh, Psome frozen and Psome-FD-Inu) was started. Here the aim was to determine the potential of reconstituted Psomes as enzymatic nanoreactors with pH dependent membrane permeability, needed for the control of the uptake and release of small molecules. Again, the differently stored Psome smoothly show repeatedly swelling and shrinking cycles upon pH switch between pH 5 and 8 (5× times) (Fig. 1B). Moreover, pH switch cycles were also carried out with SM-1 samples stored at −20 °C for 1, 2, 7, 14 and 30 days (Fig. ESI-2†). There is no difference in the pH switches for Psome fresh, Psome frozen and Psome-FD-Inu. A final experiment was performed to compare the pH-dependent titration of differently stored Psome followed by DLS measurements (Fig. 1C). The validated pH-dependent diameters and pH* (half power of Psome swelling) for Psome fresh, Psome frozen, Psome-FD and Psome-FD-Inu confirm the same performance as found in a previous study.28 A slight difference is recognizable that pH* of Psome-FD and Psome-FD-Inu is shifted to slightly higher values (6.8 and 6.9) than of Psome fresh (6.4). This difference is negligible, when comparing the slight variation of pH* for different Psome fresh in a previous study.28
Overall, we can conclude from all presented results that there is no negative influence of the used storage methods SM-1 and SM-2 on the reconstitution of frozen and freeze-dried Psome samples in various solutions. This enables us to retain the pH-switchable characteristics and vesicular shape for polymersomes. Thus, both storage methods may provide a simple handling of our samples for future biological experiments.
This previous study has been carried out with crosslinked polymersomes, therefore, this high stability for reconstituted polymersomes could be due to the crosslinking process.31 To prove this, some experiments were performed with non-crosslinked polymersomes. Once the assembly process was completed, SM-1 and SM-2 methods were applied in the presence and absence of inulin. Afterwards, all these samples were compared with a fresh sample by DLS. The results showed that none of the samples maintained their properties. In fact, in some samples the disassembly process can be observed (Fig. ESI-3†). Finally, it can be concluded the importance of crosslinking process for this type of polymersome fabricated by the assembly of BCP1 (Fig. 1A).
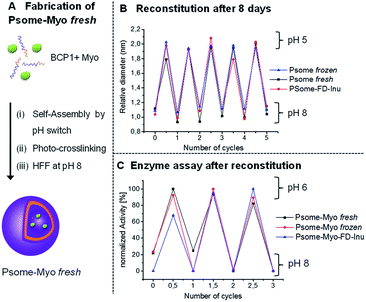
Reconstitution properties of polymersomes-encapsulated myoglobin after cryogenic freezing and freeze-drying process
To further extend the applicability of our long-term stable and pH-switchable Psomes towards biological experiments or the fabrication of complex biohybrid structures,17,32 there is a need to validate the storage, stability and reconstitution of our Psomes loaded with enzymes producing enzymatic nanoreactors.16,17,20 For fabricating polymersome-encapsulated myoglobin (Psome-Myo) our previously reported protocol has been adopted and further modified (Fig. 2A).20 Then, variation in size and enzymatic activity of the enzyme-loaded Psomes after (i) a cryogenic freezing process (at −20 °C) or (ii) a lyophilization process in presence of inulin has been studied. Psome-Myo was stored as described before for SM-1 and SM-2: (i) at −20 °C for 8 days; and (ii) lyophilized in presence of inulin (0.1% w/v) followed up by the storage at 4 °C for 8 days.Results from DLS measurements suggest that the differences in Dh and their resulting polydispersity are insignificant (data points at 0 and 0.5 cycles in Fig. 2B). Moreover, samples Psome-Myo fresh, Psome-Myo frozen and Psome-Myo-FD-Inu show the expected stability in the Psome-Myo diameters for repeated swelling and shrinking cycles upon pH switches (5× times) (Fig. 2B). The main aim of this experiment was to know if the enzyme retains its activity after the cryogenic freezing and lyophilization process, including their storage at different temperatures. Thus, several cycles of enzymatic activity at pH 6 and pH 8 were also studied by UV-Vis spectroscopy, demonstrating a high retention of the enzyme activity after both storage processes SM-1 and SM-2 (Fig. 2C).
Reconstitution properties of HSA-modified polymersomes after cryogenic freezing and freeze-drying process
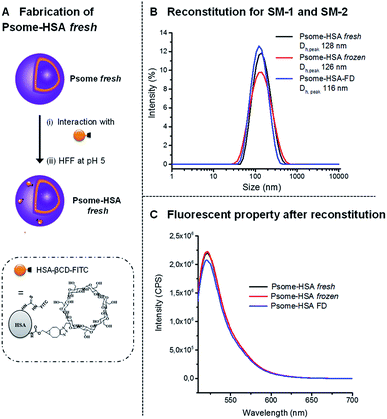
Another possible application of Psomes is as drug delivery systems or as artificial organelles.10,11,17,33 For that it would be interesting to see if also protein-modified Psome can be reconstituted. Therefore, polymersomes were decorated with human serum albumin (HSA), knowing that protein-carriers for the drug delivery are also validated as nontoxic, non-immunogenic, biocompatible and biodegradable.34 In this work, HSA, modified with β-CD and FITC (HSA-βCD-FITC), has been prepared and characterized for the decoration of the polymersome surface (Fig. 3A).27,35 Here we made use of the fact that PEG chains interact with the β-CD cavity. The different HSA derivatives, HSA-βCD and HSA-βCD-FITC, were characterized by matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF). Differences in molecular weights allowed us to determine the number of β-CD and FITC groups on the HSA protein (Fig. 3).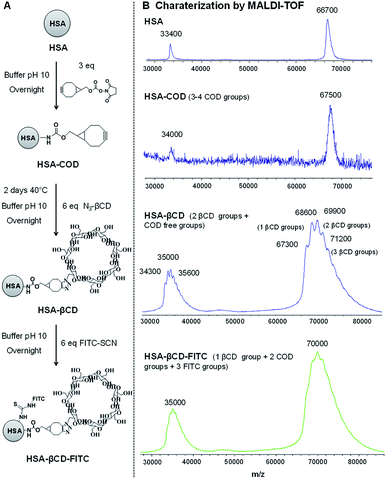 | ||
| Fig. 3 (A) Reaction scheme and (B) MALDI-TOF spectrum of HSA-βCD-FITC used in the preparation of functionalized polymersomes. | ||
The interaction of β-CD modified HSA with the membrane of Psome through the outer PEG shell27 was performed in diluted solution at pH 5 to avoid any aggregation process of HSA-modified polymersomes (Psome-HSA). While we usually work with 1 mg mL−1 of polymersome for conjugating smaller molecules modified with β-CD,27 in the case of Psome-HSA it is necessary to work at 0.25 mg mL−1. Then, the excess non-conjugated HSA-βCD-FITC protein was separated from our biohybrid structure Psome-HSA by HFF method at pH 5 (Fig. 4A).
Psome-HSA, freshly purified at pH 5, were also stored as before, using SM-1 and SM-2: (i) at −20 °C for eight days; and (ii) lyophilized in absence of Inu followed up by the storage at 4 °C for 8 days. The preservation of diameter and fluorescence intensity for Psome-HSA has been studied. By DLS, there is no significant difference in Dhs of the reconstituted Psomes-HSA when compared with freshly prepared Psome-HSA (Psome-HSA fresh) (Fig. 4B). The resulting Dhs are around 125 nm at pH 5 for Psome-HSA fresh and frozen sample, while Dh for freeze-dried sample (Psome-HSA-FD) is 116 nm. The lower value for Psome-HSA-FD can be explained with a densification and/or a reorganisation of the polymersome membrane that may occur during the freeze-drying process. Analysing fluorescence results (Fig. 4C), similar results can be observed for Psome-HSA frozen and Psome-HAS-FD sample in comparison with Psome-HSA fresh sample. Then, it is important to note that the fluorescence is not affected by these storage methods.
Conclusions
In summary, we demonstrated that after reconstitution (biologically active) polymersomes can retain their physico-chemical properties (diameter, membrane thickness, cyclic pH-switches and re-dispersion) and cyclic switch off and on membrane permeability controlling the activity of loaded enzymes after (i) lyophilization or (ii) cryogenic freezing, both combined with long/short-term storage. For the lyophilization and subsequent storage process, the use of inulin as a lyoprotectant (0.1% w/v) was necessary to provide the desired solubility and stability of (biologically active) polymersome solutions after reconstitution. Therefore, this work opens the avenue to store enzymatic nanoreactors or protein-polymersome hybrid architectures in a solid state at a temperature between −20 and 4 °C for at least 8 days. This allows easy handling and storage of polymersomes between experiments, safe transport, and in general this makes them suitable for technical use in a reproducible way. Finally, these reconstitution experiments in aqueous solution may also facilitate the fabrication of complex multicompartments with controllable concentration of biologically active entities.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Robert Ccorahua has been financial supported by Peruvian Council of Science and Technology (Concytec-Cienciactiva). Silvia Moreno has been supported by a grant from Fundación Alfonso Martín Escudero (Spain government).References
- Y. Tu, F. Peng, A. Adawy, Y. Men, L. K. E. A. Abdelmohsen and D. A. Wilson, Chem. Rev., 2016, 116, 2023 CrossRef PubMed.
- M. Marguet, C. Bonduelle and S. Lecommandoux, Chem. Soc. Rev., 2013, 42, 512 RSC.
- J. Li, S. Xiao, Y. Xu, S. Zuo, Z. Zha, W. Ke, C. He and Z. Ge, ACS Appl. Mater. Interfaces, 2017, 9, 17727 CrossRef PubMed.
- A. Joseph, C. Contini, D. Cecchin, S. Nyberg, L. Ruiz-Perez, J. Gaitzsch, G. Fullstone, X. Tian, J. Azizi, J. Preston, G. Volpe and G. Battaglia, Sci. Adv., 2017, 3, e1700362 CrossRef PubMed.
- A. Peyret, E. Ibarboure, N. Pippa and S. Lecommandoux, Langmuir, 2017, 33, 77079 CrossRef PubMed.
- A. Nazemi, F. Martínez, T. J. Scholl and E. R. Gillies, RSC Adv., 2012, 2, 7971 RSC.
- Q. Liu, L. Song, S. Chen, J. Gao, P. Zhao and J. Du, Biomaterials, 2017, 114, 23 CrossRef PubMed.
- J. Leong, J. Y. Teo, V. K. Aakalu, Y. Y. Yang and H. Kong, Adv. Healthcare Mater., 2018, 7, 1701276 CrossRef PubMed.
- F. Fernández-Trillo, L. M. Grover, A. Stephenson-Brown, P. Harrison and P. M. Mendes, Angew. Chem., Int. Ed., 2017, 56, 3142 CrossRef PubMed.
- J. Gaitzsch, X. Huang and B. Voit, Chem. Rev., 2016, 116, 1053 CrossRef PubMed.
- C. G. Palivan, R. Goers, A. Najer, X. Zhang, A. Car and W. Meier, Chem. Soc. Rev., 2016, 45, 377 RSC.
- T. Wang, J. Jiang, Y. Xiao, Y. Zou, J. Gao and J. Du, RSC Adv., 2015, 5, 55602 RSC.
- Z. Deng, Y. Qian, Y. Yu, G. Liu, J. Hu, G. Zhang and S. Liu, J. Am. Chem. Soc., 2016, 138, 10452 CrossRef PubMed.
- Y. Zou, M. Zheng, W. Yang, F. Meng, K. Miyata, H. J. Kim, K. Kataoka and Z. Zhong, Adv. Mater., 2017, 29, 1703285 CrossRef PubMed.
- J. Gaitzsch, D. Appelhans, L. Wang, G. Battaglia and B. Voit, Angew. Chem., Int. Ed., 2012, 51, 4448 CrossRef PubMed.
- B. Iyisan, J. Kluge, P. Formanek, B. Voit and D. Appelhans, Chem. Mater., 2016, 28, 1513 CrossRef.
- X. Liu, P. Formanek, B. Voit and D. Appelhans, Angew. Chem., Int. Ed., 2017, 56, 16233 CrossRef PubMed.
- W. Yang, Y. Xia, Y. Zou, F. Meng, J. Zhang and Z. Zhong, Chem. Rev., 2017, 29, 8757 Search PubMed.
- C. Ken Wong, A. J. Laos, A. H. Soeriyadi, J. Wiedemann, P. M. G. Curmi, J. J. Gooding, C. P. Marquis, M. H. Stenzel and P. Thordarson, Angew. Chem. Int. Ed., 2015, 54, 5317 CrossRef PubMed.
- D. Gräfe, J. Gaitzsch, D. Appelhans and B. Voit, Nanoscale, 2014, 6, 10752 RSC.
- A. Suzuki, T. Kuroiwa, K. Horikoshi, A. Kanazawa and S. Ichikawa, Colloids Surf., B, 2017, 159, 412 CrossRef PubMed.
- W. Y. Ayen and N. Kumar, Eur. J. Pharm. Sci., 2012, 46, 405 CrossRef PubMed.
- J. M. Kelly, E. E. Pearce, D. R. Martin and M. E. Byrne, Polymer, 2016, 87, 316 CrossRef.
- M. A. Yassin, D. Appelhans, R. Wiedemuth, P. Formanek, S. Boye, A. Lederer, A. Temme and B. Voit, Small, 2015, 11, 1580 CrossRef PubMed.
- K. Matyjaszewski, Macromolecules, 2012, 45, 4015 CrossRef.
- A. W.-T. Choi, K. K. S. Tso, V. M. W. Yim, H. W. Liu and K. K. W. Lo, Chem. Commun., 2015, 51, 3442 RSC.
- B. Iyisan, A. C. Siedel, H. Gumz, M. Yassin, J. Kluge, J. Gaitzsch, P. Formanek, S. Moreno, B. Voit and D. Appelhans, Macromol. Rapid Commun., 2017, 38, 1700486 CrossRef PubMed.
- H. Gumz, T. H. Lai, B. Voit and D. Appelhans, Polym. Chem., 2017, 8, 2904 RSC.
- R. T. Pearson, N. J. Warren, A. L. Lewis, S. P. Armes and G. Battaglia, Macromolecules, 2013, 46, 1400 CrossRef.
- N. A. Williams and G. P. Polli, J. Parenter. Sci. Technol., 1984, 38, 48 Search PubMed.
- Y. Anraku, A. Kishimura, M. Oba, Y. Yamasaki and K. Kataoka, J. Am. Chem. Soc., 2010, 132, 1631 CrossRef PubMed.
- X. Liu, D. Appelhans, Q. Wei and B. Voit, Adv. Sci., 2017, 4, 1600308 CrossRef PubMed.
- X. Wang, H. Sun, F. Meng, R. Cheng, C. Deng and Z. Zhong, Biomacromolecules, 2013, 14, 2873 CrossRef PubMed.
- T. Komatsu, Nanoscale, 2012, 4, 1910 RSC.
- C. Iancu, L. Mocan, C. Bele, A. I. Orza, F. A. Tabaran, C. Catoi, R. Stiufiuc, A. Stir, C. Matea, D. Iancu, L. Agoston-Coldea, F. Zaharie and T. Mocan, Int. J. Nanomed., 2011, 6, 129 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra03964j |
| ‡ Equal contribution of both authors. |
| This journal is © The Royal Society of Chemistry 2018 |