Open Access Article
Open Access ArticleA nano-fibrous platform of copolymer patterned surfaces for controlled cell alignment†
Kai Zhanga,
Alexandra Arranjaab,
Hongyu Chenc,
Serhii Mytnyka,
Yiming Wanga,
Sander Oldenhofad,
Jan H. van Esch a and
Eduardo Mendes
a and
Eduardo Mendes *a
*a
aAdvanced Soft Matter Group, Department of Chemical Engineering, Delft University of Technology, Delft, 2629 HZ, The Netherlands. E-mail: E.Mendes@tudelft.nl
bDepartment of Radiology and Nuclear Medicine, University Medical Center Utrecht, Utrecht, 3584 CX, The Netherlands
cDepartment of Chemical Engineering, Virginia Polytechnic Institute and State University, Blacksburg, VA 24061, USA
dNetherlands Forensic Institute, Den Haag, 2497 GB, The Netherlands
First published on 13th June 2018
Abstract
The last decade has witnessed great progress in understanding and manipulating self-assembly of block copolymers in solution. A wide variety of micellar structures can be created and many promising applications in bioscience have been reported. In particular, nano-fibrous micelles provide a great platform to mimic the filamentous structure of native extracellular matrix (ECM). However, the evaluation of this kind of filomicellar system with potential use in tissue engineering is virtually unexplored. The question behind it, such as if the block copolymer nano-fibrous micelles can regulate cellular response, has lingered for many years because of the difficulties in preparation and 3D manipulation of these tiny objects. Here, by using a combination approach of self-assembly of block copolymers and soft lithography, we establish a novel and unique nano-fibrous 2D platform of organized micelles and demonstrate that patterned micelles enable control over the cellular alignment behavior. The area density and orientation of fibrous micelles determine the alignment degree and directionality of cells, respectively. Furthermore, when cells were cultured on multi-directionally aligned micelles, a competitive response was observed. Due to the virtually infinite possibilities of functionalization of the micelle corona, our work opens a new route to further mimic the native fibrous networks with artificial micelles containing various functionalities.
Introduction
Tissue engineering has attracted growing attention to create living constructs that closely resemble native tissues in order to rescue patients from organ failure or tissue malfunction.1 As a crucial element in tissue engineering, the extracellular matrix (ECM) in which cells reside provides a variety of essential biophysical and biochemical milieu for cell growth, proliferation, migration and function.2–4 Therefore, developing a tailored artificial scaffold to mimic the native ECM that consists of complex and organized nano-/micro- fibrous structures is highly desirable.To address this challenge, a wide variety of materials and technologies have been developed.5,6 For example, electrospinning is usually used to create fibrous meshes and even 3D scaffolds with controlled orientation and mechanical properties,7,8 while phase separation methods are applied for fabricating porous scaffolds with minimal setup requirements and easy control of scaffold porosity.9–11 More recently, inspired by an ubiquitous process in nature, self-assembly strategy has become increasingly important not only in biological but also in synthetic systems.12 It provides a facile route to construct complex and hierarchical structures, such as triple-helix morphology of collagen.13 Based on this principle, many synthetic materials with self-assembly properties have been developed.14 Amphiphilic block copolymers that consist of two or more chemically distinct polymers is one important sort of these materials. Due to the micro-phase separation between different blocks, block copolymers can self-assemble into various micellar structures in solution, offering many potential applications in bioscience fields.15,16 Especially, nano-fibrous micelles provide a great platform to mimic the filamentous structure of native ECM.17
However, to our best knowledge, no study has been reported to use this kind of block copolymer assemblies to mimic the ECM due to the challenges in relation to preparation of long enough (hundreds of microns) semi-flexible filomicelles as well as limitations to their physical manipulation. In this work, we give a step in this direction by producing a flat surface as a unique nano-fibrous platform with quenched, ordered, ultra-long block copolymer micelles. Although the copolymer micellar network proposed here cannot fully substitute the extra cellular matrix, it is important to notice the strong similarity between them in relation to the dimensions and mechanical properties we have recently shown that these micelles exhibit diameters of 46 nm, and can easily achieve lengths of ∼200 μm which are comparable to the native ECM structures (collagen fibrils) which have diameters of 40–80 nm and length of 20–200 μm.18,19 Also, and very importantly, such micelles exhibit Young's moduli that can be tuned between 3 and 13 GPa, depending on the diameter of their glassy core20 whereas single collagen fibrils exhibit Young's modulus of around 5.4 GPa.19 Using this platform, we investigate the effects of patterned block copolymer micelles on the alignment behaviour of cells. We find that by adjusting the area density and orientation of micelles, one can control the extent of cellular alignment and orientation degree, respectively. Moreover, competitive guidance was also observed when cells were cultured on substrates with micelles aligned in two or more directions simultaneously. This study demonstrates the ability of block copolymer fibrous micelles to topologically regulate cellular alignment and it also opens the door to the possible use of ultra-long block copolymer filomicelles in tissue engineering.
Results and discussion
Fabrication of micellar platform
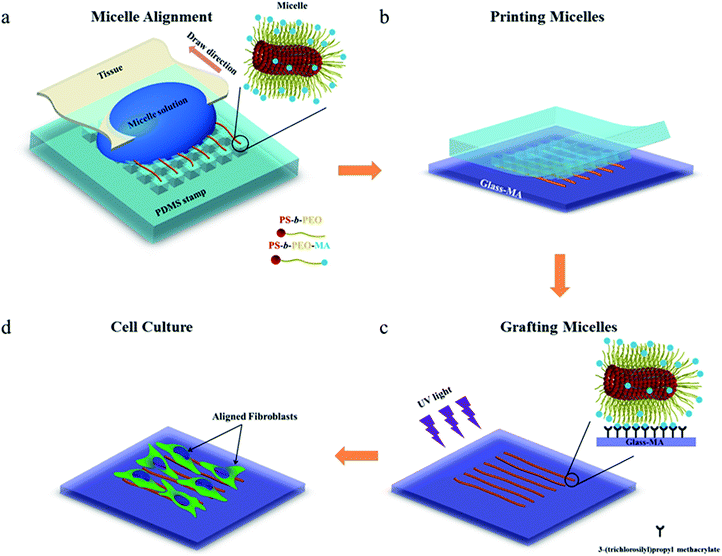
To investigate whether the block copolymer fibrous micelles are capable of regulating cellular response, a key challenge is the directional manipulation of nano-fibril micelles of block copolymers. Here, we combined the quenched, out-of-equilibrium assembly of block copolymers and soft lithography approaches to fabricate patterned micellar structures for this study. A schematic illustration of the manufacturing process of micellar platform for cell alignment is depicted in Scheme 1. First, cross-linkable PS-b-PEO micelles were prepared by evaporation-induced self-assembly method and aligned on micro-pillars of PDMS stamps using our former reported dewetting approach21,22 (Scheme 1a). Next, due to the specific viscoelastic behavior of PDMS, the aligned micelles can be successfully transfer-printed to the acrylated glass coverslides (Scheme 1b). Repeating the first two steps offered an easy way to control the area densities of aligned micelles on the glass surface. To immobilize the patterned micelles, the glass slide with aligned micelles was irradiated under UV light for 20 minutes to bond the micelles on the glass surface (Scheme 1c), followed by water rinsing to remove the residual photo initiator. The final new nanofibrous platform was formed and used for further cell culture experiments (Scheme 1d).As shown in Fig. 1, the density of micelles printed on glass surface increased with increasing number of micelles printing times. The individual aligned micelles were distinguishable in the case of low printing times (Fig. 1a and b), while it became much harder to image the samples with ten and twenty printing times (Fig. 1c and d). In order to analyze the alignment of micelles after multiple printings, Fast Fourier Transform (FFT) of the confocal images were taken. As shown in the inserted FFT images of Fig. 1a–d, all of them have the ellipsoidal shape, which indicates a clear directionality of printed micelles. This suggests that the amount of printing times has no detectable influence on the alignment of micelles.
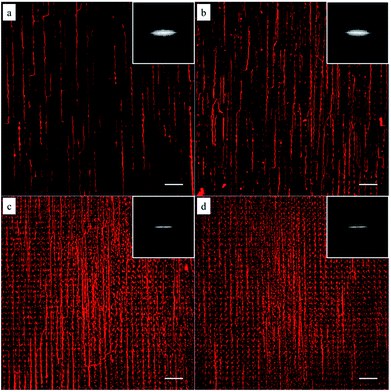 | ||
| Fig. 1 Confocal microscopy images of the immobilized micelles after multiple printing: (a) 1, (b) 5, (c) 10 and (d) 20 times. The insets are the corresponding FFT output images. Scale bars are 20 μm. | ||
To gain a deeper insight in the dependence of micelle density on printing times, AFM measurements were performed on the glass surfaces grafted with micelles. As can be seen in Fig. 2, by increasing the number of printing steps, we could graft more micelles on the glass surface, i.e. achieve higher surface coverage of micelles. The height of a single micelle is around 50 nm (Fig. S3†), which agrees well with our previous study23 and is above the threshold size (35 nm) for contact guidance of fibroblast alignment.24 Fig. 2e illustrates the relationship between printing times and covering area percentage of micelles. The area percentage of micelle coverage raised from around 4.4% to 31.4% as the printing times increased from 1 time to 20 times. A linear relationship was observed, showing that the micelle density on substrate is fully controllable.
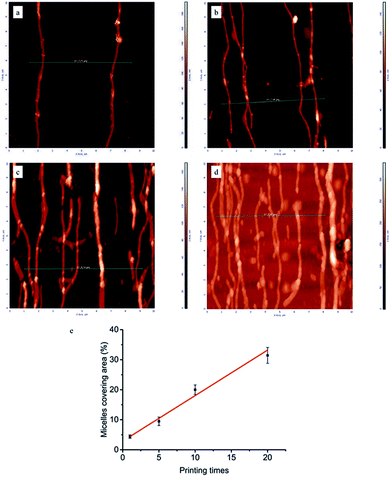 | ||
| Fig. 2 Representative AFM images (10 × 10 μm) of micelles printed and cross-linked on the glass surface: (a) 1, (b) 5, (c) 10, (d) 20 printing times. Line scan profiles corresponding to green lines in AFM images were shown in Fig. S3 of ESI.† (e) Area percentages covered by aligned micelles as a function of printing times. | ||
Unidirectional micelles
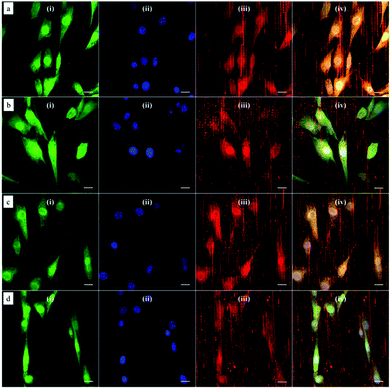
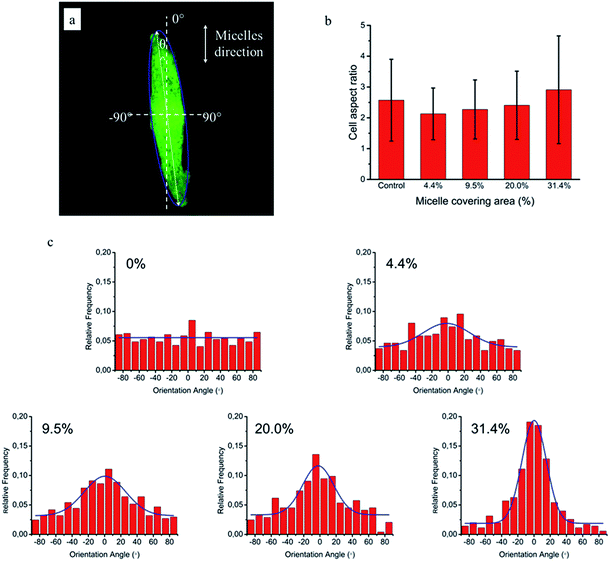
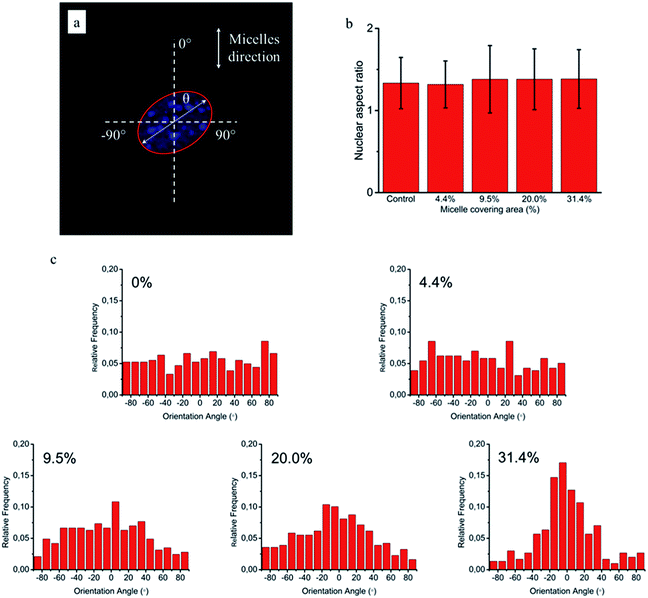
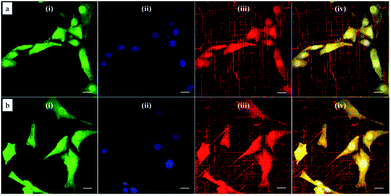
Based on this strategy, we can control the density of micelles in the fibrous platform and study the fundamental understanding of topographic control of cell behaviour. After preparation of the nanofibrous platforms with different degrees of micelles densities, NIH/3T3 fibroblasts were seeded on the substrates and their response was analysed after 24 hours (Fig. 3). We observed that fibroblasts were able to grow on the surface with PS-b-PEO micelles despite the fouling-resistant properties of PEG.25–28 This can be attributed to the presence of methacrylate modified PEO terminal groups in the polymeric fibrous micelles, which may provide the cellular adhesion sites on the biologically inert PEG surface. Moreover, as shown in Fig. 3, the cells seeded on the aligned micelles showed alignment tendency, while cells cultured on flat glass surface without micelles were randomly oriented (Fig. 4). In the platform with the highest density of micelles, corresponding to 20 printing times and an coverage are of 31.4% (Fig. 3d), we observed the highest tendency for cell alignment along the micelle direction and cells also acquired a stretched spindle shape (Fig. 3d). In the substrate with the lower micelle density (1 printing time and 4.4% of coverage area), cells were preferentially oriented along the micelles directions but their morphology was less stretched (Fig. 3a). This characteristic morphological change of cells on these modified substrates strongly suggests that densely aligned copolymer micelles have the ability to guide cell alignment, in consistent with that observed in the aligned electrospun fibers.29To quantitatively analyse cellular behaviour on the substrates with different densities of micelles, the aspect ratio, i.e. ratio between long axis and short axis of ellipse, and orientation angle θ of fibroblasts were measured using ellipse fitting method (Fig. 5a). In the presence of aligned micelles, the aspect ratio (i.e. elongation) of the cells was not considerably different from the control group (Fig. 5b). In contrast to our results, fibroblasts cultured on aligned electro-spun nanofibers were usually shown to elongate along the fiber direction.30 Compared to that work, the density of nanofibrillar micelles used in this research is much lower. This suggests that elongation of fibroblasts may need a higher density of fibers. However, regarding the orientation degree of fibroblasts, we observed that increasing the density of aligned micelles dramatically increases the degree of cell alignment (Fig. 5c). To compare the orientation degrees of cells cultured on substrates with different micelle densities, we used the Gaussian curve to fit each histogram (blue curves) and calculate their full width at half maximum (FWHM). The degree of cell alignment is reflected by the height (apex) and shape (FWHM) of the peaks. As the covering area percentage of the micelles increased, the heights of the peaks increased as well while the shape became sharper, i.e., the FWHM decreased (Fig. 6). It indicates that cells cultured on a platform with higher density of micelles show better orientation profile than the one with lower density of micelles, in agreement with our former observation (Fig. 3 and 4).
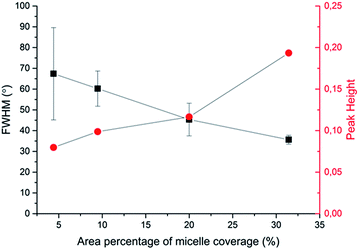 | ||
| Fig. 6 Relationship between area percentage of micelle coverage and characteristic parameters (full width at half maximum (FWHM) and peak height in Gaussian fitting curves) of cellular alignment. | ||
Since cellular alignment and cell differentiation have been demonstrated to correlate with nuclear elongation,31,32 herein we also investigated the nuclear aspect ratio dependence on the covering area percentage of aligned micelles using ellipse fitting method (Fig. 7a). Similar to cellular elongation, no significant nuclear stretching was observed (Fig. 7b). Nevertheless, as the nuclear aspect ratios are around 1.3 in the control group, indicating non-spherical nuclei, the nuclear orientation was still measured to quantitatively evaluate the effect of aligned micelles on the nuclear alignment. As shown in Fig. 7c, the nuclear orientation increased with increasing micelle densities on the glass surface as previously observed in the cellular orientation. In contrast to cellular orientation, poor Gaussian fittings were observed here for platforms with a low micelle densities (curves not shown), limiting the use of the FWHM and apex of the peak to quantitatively reflect the relationship between the extent of nuclei orientation and micelle density.
Based on the above results, we can conclude that coverage area percentage of micelles equal or below 31.4% have no significant effects on cellular and nuclear elongation. On the other hand, the orientation degrees of cells and nuclei increase proportionally to the coverage area of micelles, being maximal at the highest density of micelle coverage (31.4%). This may give a hint that, similarly to orientation, different fiber densities are required for cellular and nuclear stretching.
Multidirectional micelles
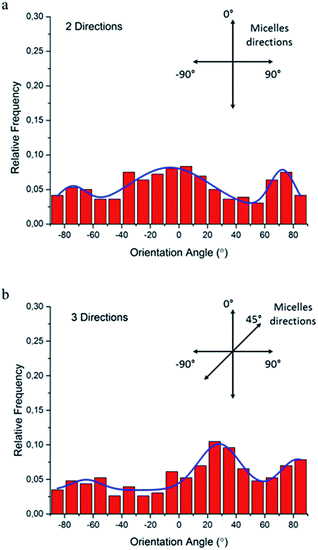
Since the natural extracellular matrix (ECM) is not just one-dimensional topographic pattern, a simple method to fabricate a platform with more complex topographic features is a critical component in tissue engineering. Our method combining self-assembly with soft lithography can provide a solution to build a synthetic platform with multi-direction cues by simply printing micelles in different directions. Based on this concept, we investigated cellular responses to orthogonally and tri-directionally aligned micelles. To ensure that the micelles in each direction have the same possibility to stimulate cells, we printed micelles alternately in different directions at the same density (5 printing times for each direction). Then, NIH/3T3 fibroblasts were cultured on these multi-direction cues for 24 hours. The morphology of cells on multiple aligned micelles is shown in Fig. 8. In the case of orthogonal cues, cells appeared to align in two preferential directions, the same as those of the micelles (Fig. 8a), indicating that micellar contact guidance in these two directions has the same impact. However, when a third direction with further micelle printing was introduced, cellular response became more complex (Fig. 8b), and it is no longer possible to distinguish the three directional alignments.To quantitatively analyze cellular alignment in these two cases, the distribution of orientation angle (θ) was investigated. Regarding the horizontally and vertically aligned micelles, three preferential angles of cell alignment were observed (Fig. 9a), showing cells display a similar contact guidance in both directions. However, it becomes more complicated in the substrate with tri-directionally aligned micelles (Fig. 9b). A larger number of cells oriented towards 30°, indicating that the micelles aligned in the third direction (45°) leaded to more deviation by their mutual contact guidance. It can, therefore, be suggested that cells could integrate multi-direction cues during determining the direction of cell orientation together with mutual interactions, which agrees with former reports.33,34
Finally, it is instructive to discuss the above results in relation to some general properties of the ECM. The ECM is a highly complex 3D network which is composed of many kinds of protein fibers, such as collagen, elastic and reticular fibers, and some non-fiber-forming proteins. It provides various biochemical and biophysical cues to interact with cells. However, until now, the whole interaction process is not fully understood due to the complex and multiple factors involved. The mechanical properties and morphological structures are the two most basic aspects that can be used to mimic the ECM with synthetic nanofibers. According to the literature,18,19 the mean diameter and length of collagen fibrils are around 40–80 nm and 20–200 μm, respectively. The Young's modulus of single collagen fibrils is about 5.4 GPa. The type of micelles used in the present study exhibit diameters of 46 nm, and can easily achieve lengths of ∼200 μm which are comparable to the native ECM collagen fibrils. Their Young's moduli can be tuned between 3–13 GPa (depending on the diameter of their glassy core)20 being of the order of that found for single collagen fibrils.
The electrical properties of native ECM are, of course, different from those of the PS-b-PEO micelles. Certainly, the neutral PEO corona initially excludes effects of surface charging of fibers to induce cell alignment. The difference of cell alignment observed between ECM patterns and the micellar pattern studied above can, however, be attributed to two reasons. On one hand, the ECM pattern formed by the native fibrils have higher density of binding points for cell adhesion on the fibril surface when compared to our micelles. The proteins used for cell adhesion are capable of adsorbing on the whole native fibrils, while in our case only the MA groups provided (non-specific) binding points for cell attachment. On the other hand, the fibril density of ECM pattern35 is usually much higher than the micelle pattern used here, although the present study also shows that increasing the fibril density leads to a higher extent of cell alignment.
Conclusion
In summary, we have demonstrated that ultra-long block copolymer fibrous micelles are capable of regulating cell orientation on a surface. The degree of cell alignment increased with the area density of micelles. When the area percentage of micelle coverage increased to 31.4%, the cellular alignment became more significant even if no significant cell elongation was observed. For high enough micelle surface density, nuclear alignment was also observed although no considerable nuclear elongation was detected. Furthermore, the cells show a competitive response to the micelle networks with multi-directionally alignment. Our unique micellar platform suggests that fibrous micelles of block copolymers can be used to mimic the native fibrous networks surrounding cells. Considering the innumerous possibilities of biochemical modification of the micelle corona end-groups (like terminal methacrylate groups in our work), we believe this work provides a versatile approach to future studies mimicking cell interactions with native fibrous networks containing various chemical functionalities.Materials and methods
Synthesis and preparation of micelles
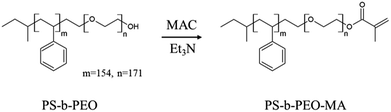
Pristine block copolymer, polystyrene-b-polyethylene oxide (PS-b-PEO), having an –OH terminal group in PEO block was purchased from Polymer Source, Inc. (Canada). Polydispersity index is 1.09 and the molecular weights of PS and PEO blocks are 16.0 kg mol−1 and 7.5 kg mol−1, respectively. In order to fix the micelles onto the substrate after printing, we synthesized PS-b-PEO-MA by reaction of PS-b-PEO with methacryloyl chloride (MAC) as follows (Scheme 2).Briefly, 500 mg of PS-b-PEO (0.02 mmol) was first dissolved in 50 mL of tetrahydrofuran (THF) and then 13 mmol of triethylamine (Et3N) was added under vigorous stirring at room temperature. MAC (13 mmol) was slowly added to the reaction mixture while stirring and resulting mixture was allowed to react for 24 h at 25 °C to substitute terminal alcohol of PS-b-PEO with methacryloyl group. Then, the reaction mixture was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min, after which the supernatant was carefully removed from the precipitated triethylamine hydrochloride salts. This procedure was repeated 2–3 times, after which the remaining solution was concentrated with rotary evaporator to about 5 mL. The resulting solution was slowly added to 600 mL of hexane, and the precipitated PS-b-PEO-MA polymer was isolated by filtration, washed several times with ethanol thoroughly, and dried in a vacuum oven at 50 °C and 50 mbar for 2 days. Following modification of PS-b-PEO, 1H nuclear magnetic resonance (1H-NMR, Agilent-400 MR DD2) spectroscopy was used to verify the structure and substitution efficiency. As shown in Fig. S1,† the methacryloyl functionality was confirmed by 1H-NMR (CDCl3): PS-b-PEO-MA, δ (ppm) 5.6 and 6.2 (CH3–C
000 rpm for 15 min, after which the supernatant was carefully removed from the precipitated triethylamine hydrochloride salts. This procedure was repeated 2–3 times, after which the remaining solution was concentrated with rotary evaporator to about 5 mL. The resulting solution was slowly added to 600 mL of hexane, and the precipitated PS-b-PEO-MA polymer was isolated by filtration, washed several times with ethanol thoroughly, and dried in a vacuum oven at 50 °C and 50 mbar for 2 days. Following modification of PS-b-PEO, 1H nuclear magnetic resonance (1H-NMR, Agilent-400 MR DD2) spectroscopy was used to verify the structure and substitution efficiency. As shown in Fig. S1,† the methacryloyl functionality was confirmed by 1H-NMR (CDCl3): PS-b-PEO-MA, δ (ppm) 5.6 and 6.2 (CH3–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 1.9 (CH3–C
CH2), 1.9 (CH3–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2).
CH2).
Functionalized filamentous micelles (Fig. S2†) were prepared by the evaporation-induced self-assembly method, which has been reported elsewhere.36 Typically, our modified method gives rise to a broad length distribution with a peak around 150–200 μm and a very long tail of quenched ultra-long micelles.22 Instead of using pure PS-b-PEO, stock solutions of 10 mg mL−1 copolymers were prepared with 10% of PS-b-PEO-MA/PS-b-PEO weight fractions. The fluorescent probe 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI) was added to the stock solutions before micelle preparation to facilitate micelle visualization by confocal microscopy.
Fabrication of PDMS stamp
The PDMS stamp with micropillars were produced by conventional soft lithography, which was reported in our previous work.21,22 Briefly, a master silicon template with a pattern (3.5 × 3.5 μm squares with 2 μm gaps) was first produced by deep reactive ion etching technique. Then, the silicon template was treated with a vapour of silanization agent (tridecafluoro-1,1,2,2-tetrahydroocty)trichlorosilane for 4 h, following by curing the mixture of PDMS prepolymer (Sylgard 184, Dow Corning Co. Ltd.) and curing agent (weight ratio: 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with the template at 68 °C for 24 h. After peeling off the formed PDMS stamp from silicon mould, we immersed it into a bath of ethanol for 16 h to remove unreacted compound and dried in the air before using.
1) with the template at 68 °C for 24 h. After peeling off the formed PDMS stamp from silicon mould, we immersed it into a bath of ethanol for 16 h to remove unreacted compound and dried in the air before using.
Alignment of micelles
The micelles were aligned using a dewetting technique reported before.21,22 Briefly, 20 μL of micelle solution was deposited on the edge of stamp pattern and then dragged in one direction by capillary force using lens tissue. As a result of controlled wetting process, micelles were well aligned and deposited on top of the micropillars.Printing and crosslinking micelles onto glass slides
First, methacrylated glass slides (Glass-MA) were prepared by rinsing glass coverslides with ethanol and acetone several times, then cleaned with plasma for 140 s, following by silanization with 3-(trichlorosilyl)propyl methacrylate for 4 h. Second, a PDMS micropillar stamp with aligned micelles was prepared as described above, using 0.1 mg mL−1 of micelle solution containing 5 mg mL−1 of photo initiator, lithium phenyl-2,4,6-trimethyl-benzoyphosphinate (LAP), which was synthesized as previously described.37 Then, the PDMS micro-pillar stamp with the aligned micelles on top was brought into contact with Glass-MA slides under gentle force (around 40 g) for 30 s and then slowly peeled off at low speed (around 1 mm s−1) to transfer print the aligned micelle array. Finally, the Glass-MA with micelles was irradiated for 20 min to crosslink the micelles onto glass slides under UV light. For preparing substrates with different micelle coverages, the number of times for printing micelles were adjusted before UV irradiation. Note, to prevent degradation of LAP, aluminium foil was used to cover all glass vials and Petri dishes during the whole aligning and printing process.Cell culture
The cell line used in this study (NIH/3T3 mouse embryonic fibroblasts) was obtained from the American Type Culture Collection (ATCC, USA) and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% newborn calf serum (NCS) and 0.5% (v/v) penicillin–streptomycin. Cell cultures were prepared from deep-frozen stock vials, seeded in 75 cm2 culture bottles (Cellstar, Greigner Bio-One), incubated under standard cell culture conditions (37 °C, 5% CO2 atmosphere and water-saturated 95% air) and maintained until sub-confluence was reached (70–80%). For the experimental studies, cells were trypsinized and seeded at a density of 2 × 104 cells per cm2 in the modified nanofibrous glass substrates, which were placed in 6-well plates. Cells were allowed to interact with the substrates overnight. After this incubation period, the adherent cells were labelled with a CellTracker™ Green 5-chloromethylfluorescein diacetate solution (Molecular Probes, C7025) at a concentration of 10 μM in serum-free medium for 45 minutes at 37 °C. Cells were then washed twice with Dulbecco's Phosphate-Buffered Saline (DPBS, Gibco) and fixed with pre-warmed 3.7% formaldehyde in PBS for 15 min. Cells were washed again thrice with DPBS and mounted with DAPI Vectashield (Vector Laboratories) for confocal microscopy. Images were captured with a Carl Zeiss LSM710 microscope and were superimposed to determine the localization of the CellTracker dye, the DAPI nuclear dye and the DiI micellar dye. Experiments were performed in duplicate.Characterization
The micelles and NIH/3T3 fibroblasts were visualized using a laser scanning confocal microscopy (LSM 710, Carl Zeiss Microscopy GmbH, Germany) with a Fluar 40×/1.30 oil M27 objective lens. The nuclei were stained with DAPI for fluorescence imaging. Atomic force microscopy (AFM) was performed using an NT-MDT (NTEGRA) microscope and HA_NA tips with resonance frequency around 240 kHz. All topography scans were obtained in tapping mode in the air and at room temperature. Then, the grain threshold method38 was utilized to quantitatively analyse AFM results and investigate the coverage area of micelles.Micelle alignment analysis
Micelle alignment was investigated by Fast Fourier Transform (FFT) image analysis as a function of printing times.29,39,40 The FFT function converts information present in the original confocal image from “real” space into mathematically defined “frequency” space. The resulting FFT output image contains grayscale pixels that are distributed in a pattern that reflects the degree of micelle alignment present in the original confocal image. The confocal images with 1024 × 1024 pixels were first converted to 8 bit grayscale TIF files and then processed with ImageJ software (version 1.48v, National Institutes of Health, USA, http://imagej.nih.gov/ij).Cellular alignment analysis
To quantify orientation and elongation of cells and nuclei on substrates, an in-house program was developed with Matlab® using the image processing toolbox. Briefly, cell or nucleus was fitted with an ellipse shape using the moments algorithm41,42 and various elliptical parameters were obtained. The angle between the major axis of fitting ellipse and aligned direction of micelles was defined as orientation angle (θ) and used to quantify cellular alignment, while the aspect ratio between the length of major axis and minor axis was applied to evaluate the elongation behavior. Around 300 data points were taken from confocal images for analysis of each sample.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank the China Scholarship Council (CSC) for the financial support.References
- R. Langer and J. P. Vacanti, Science, 1993, 260, 920–926 Search PubMed.
- R. P. Mecham, Current protocols in cell biology, 2012, ch. 10, Unit 10.11 Search PubMed.
- J. K. Mouw, G. Q. Ou and V. M. Weaver, Nat. Rev. Mol. Cell Biol., 2014, 15, 771–785 CrossRef PubMed.
- C. Frantz, K. M. Stewart and V. M. Weaver, J. Cell Sci., 2010, 123, 4195–4200 CrossRef PubMed.
- E. Engel, A. Michiardi, M. Navarro, D. Lacroix and J. A. Planell, Trends Biotechnol., 2008, 26, 39–47 CrossRef PubMed.
- P. X. Ma, Adv. Drug Delivery Rev., 2008, 60, 184–198 CrossRef PubMed.
- T. J. Sill and H. A. von Recum, Biomaterials, 2008, 29, 1989–2006 CrossRef PubMed.
- X. F. Wang, B. Ding and B. Y. Li, Mater. Today, 2013, 16, 229–241 CrossRef PubMed.
- X. H. Liu and P. X. Ma, Biomaterials, 2009, 30, 4094–4103 CrossRef PubMed.
- F. Yang, R. Murugan, S. Ramakrishna, X. Wang, Y. X. Ma and S. Wang, Biomaterials, 2004, 25, 1891–1900 CrossRef PubMed.
- Y. S. Nam and T. G. Park, J. Biomed. Mater. Res., 1999, 47, 8–17 CrossRef PubMed.
- G. M. Whitesides and B. Grzybowski, Science, 2002, 295, 2418–2421 CrossRef PubMed.
- L. E. R. O'Leary, J. A. Fallas, E. L. Bakota, M. K. Kang and J. D. Hartgerink, Nat. Chem., 2011, 3, 821–828 CrossRef PubMed.
- A. C. Mendes, E. T. Baran, R. L. Reis and H. S. Azevedo, Wires Nanomed. Nanobi., 2013, 5, 582–612 CrossRef PubMed.
- A. Harada and K. Kataoka, Prog. Polym. Sci., 2006, 31, 949–982 CrossRef.
- K. Kataoka, A. Harada and Y. Nagasaki, Adv. Drug Delivery Rev., 2001, 47, 113–131 CrossRef PubMed.
- M. H. Park, B. G. Choi and B. Jeong, Adv. Funct. Mater., 2012, 22, 5118–5125 CrossRef.
- T. Ushiki, Arch. Histol. Cytol., 2002, 65, 109–126 CrossRef PubMed.
- L. Yang, K. O. van der Werf, B. F. J. M. Koopman, V. Subramaniam, M. L. Bennink, P. J. Dijkstra and J. Feijen, J. Biomed. Mater. Res., Part A, 2007, 82A, 160–168 CrossRef PubMed.
- L. Jennings, P. Glazer, A. C. Laan, R. M. de Kruijff, G. Waton, F. Schosseler and E. Mendes, Soft Matter, 2016, 12, 7324–7329 RSC.
- P. J. Glazer, L. Bergen, L. Jennings, A. J. Houtepen, E. Mendes and P. E. Boukany, Small, 2014, 10, 1729–1734 CrossRef PubMed.
- K. Zhang, P. J. Glazer, L. Jennings, S. Vedaraman, S. Oldenhof, Y. Wang, F. Schosseler, J. H. van Esch and E. Mendes, Chem. Commun., 2016, 52, 12360–12363 RSC.
- E. Meijering, M. Jacob, J. C. F. Sarria, P. Steiner, H. Hirling and M. Unser, Cytometry, Part A, 2004, 58a, 167–176 CrossRef PubMed.
- W. A. Loesberg, J. te Riet, F. C. M. J. M. van Delft, P. Schon, C. G. Figdor, S. Speller, J. J. W. A. van Loon, X. F. Walboomers and J. A. Jansen, Biomaterials, 2007, 28, 3944–3951 CrossRef PubMed.
- W. R. Gombotz, G. H. Wang, T. A. Horbett and A. S. Hoffman, J. Biomed. Mater. Res., 1991, 25, 1547–1562 CrossRef PubMed.
- S. J. Sofia, V. V. Premnath and E. W. Merrill, Macromolecules, 1998, 31, 5059–5070 CrossRef PubMed.
- T. McPherson, A. Kidane, I. Szleifer and K. Park, Langmuir, 1998, 14, 176–186 CrossRef.
- S. I. Jeon, J. H. Lee, J. D. Andrade and P. G. Degennes, J. Colloid Interface Sci., 1991, 142, 149–158 CrossRef.
- Y. Orlova, N. Magome, L. Liu, Y. Chen and K. Agladze, Biomaterials, 2011, 32, 5615–5624 CrossRef PubMed.
- T. Fee, S. Surianarayanan, C. Downs, Y. Zhou and J. Berry, PLoS One, 2016, 11, e0154806 Search PubMed.
- K. S. Brammer, S. Oh, C. J. Cobb, L. M. Bjursten, H. van der Heyde and S. Jin, Acta Biomater., 2009, 5, 3215–3223 CrossRef PubMed.
- A. J. Maniotis, C. S. Chen and D. E. Ingber, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 849–854 CrossRef.
- X. T. Zhou, J. Hu, J. J. Li, J. Shi and Y. Chen, ACS Appl. Mater. Interfaces, 2012, 4, 3888–3892 Search PubMed.
- H. Y. Mi, M. R. Salick, X. Jing, W. C. Crone, X. F. Peng and L. S. Turng, J. Biomed. Mater. Res., Part A, 2015, 103, 593–603 CrossRef PubMed.
- C. Chaubaroux, F. Perrin-Schmitt, B. Senger, L. Vidal, J. C. Voegel, P. Schaaf, Y. Haikel, F. Boulmedais, P. Lavalle and J. Hemmerle, Tissue Eng., Part C, 2015, 21, 881–888 CrossRef PubMed.
- J. T. Zhu and R. C. Hayward, J. Am. Chem. Soc., 2008, 130, 7496–7502 CrossRef PubMed.
- B. D. Fairbanks, M. P. Schwartz, C. N. Bowman and K. S. Anseth, Biomaterials, 2009, 30, 6702–6707 CrossRef PubMed.
- Q. B. Guan, B. Norder, L. Y. Chug, N. A. M. Besseling, S. J. Picken and T. J. Dingemans, Macromolecules, 2016, 49, 8549–8562 CrossRef.
- C. Ayres, G. L. Bowlin, S. C. Henderson, L. Taylor, J. Shultz, J. Alexander, T. A. Telemeco and D. G. Simpson, Biomaterials, 2006, 27, 5524–5534 CrossRef PubMed.
- T. A. M. Valente, D. M. Silva, P. S. Gomes, M. H. Fernandes, J. D. Santos and V. Sencadas, ACS Appl. Mater. Interfaces, 2016, 8, 3241–3249 Search PubMed.
- K. Voss and H. Suesse, IEEE Trans. Pattern Anal. Machine Intell., 1997, 19, 80–84 CrossRef.
- S. Lenhert, M. B. Meier, U. Meyer, L. F. Chi and H. P. Wiesmann, Biomaterials, 2005, 26, 563–570 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra03527j |
| This journal is © The Royal Society of Chemistry 2018 |