Open Access Article
Open Access ArticleExpression of phosphotransacetylase in Rhodosporidium toruloides leading to improved cell growth and lipid production†
Xiaobing Yang *ab,
Wenyi Suna,
Hongwei Shenac,
Sufang Zhangac,
Xiang Jiaoa and
Zongbao K. Zhao
*ab,
Wenyi Suna,
Hongwei Shenac,
Sufang Zhangac,
Xiang Jiaoa and
Zongbao K. Zhao ac
ac
aDivision of Biotechnology, Dalian Institute of Chemical Physics, CAS, Dalian 116023, PR China
bCollege of Enology, Northwest A&F University, Yangling, Shaanxi 712100, PR China. E-mail: yangxb@nwafu.edu.cn; Tel: +86-187-2952-0029
cDalian National Laboratory for Clean Energy, Dalian Institute of Chemical Physics, CAS, Dalian 116023, PR China
First published on 10th July 2018
Abstract
Microbial lipids (MLs) are potential alternatives to vegetable oils and animal fats for production of biofuels and oleochemicals. It remains critical to improve ML production efficiency and costs for further commercial development. In the present study, we overexpressed a gene encoding phosphotransacetylase (Pta) in the oleaginous yeast Rhodosporidium toruloides for enhanced cell growth and lipid production. Compared with the parental strain R. toruloides NP11, the engineered strain NP-Pta-15 showed significant improvement in glucose consumption, cell growth and lipid accumulation when cultivated under nitrogen limited conditions in an Erlenmeyer flask as well as a stirred tank bioreactor. The introduction of Pta may establish an additional acetyl-CoA formation route by utilization of acetylphosphate. Our results should inspire more engineering efforts to facilitate the economic viability of ML technology.
1. Introduction
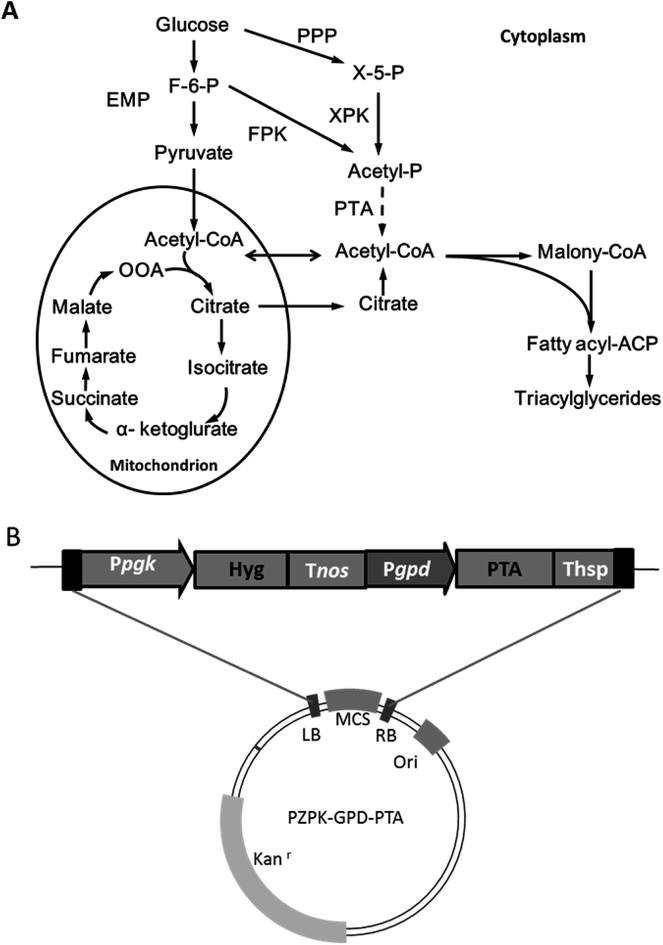
Oleo-chemicals are attractive alternatives to traditional petrochemicals and usually derived from vegetable oils and animal fats, which have limited availability.1 MLs prepared from oleaginous microorganisms have been demonstrated to have comparable properties to typical plant oils and a number of advantages, such as a shorter production and distribution cycle, and without competing with food supply and land use.2 However, it remains challenging to produce MLs at a large scale because of high costs.3There are several strategies to improve the economic feasibility of MLs: first of all, exploration of low cost substrates such as lignocellulosic biomass and various organic wastes;4,5 second, developing more efficient culture strategies and product separation methods to reduce process costs;6 in another scenario, manufacturing valuable (co-)products such as unsaturated fatty acids and carotenoids through metabolic engineering of oleaginous species.7,8 However, the theoretical yield of per mole of hexose is limited to two moles acetyl-CoA, since the glycolysis loses a carbon equivalent for each C3 unit upon pyruvate decarboxylation.9 Ideally, cell growth and lipid production may be further improved by devising more efficient acetyl-CoA generating pathways, but such efforts remain limited in oleaginous yeast. Recently, phosphoketolase (Pk) and phosphotransacetylase (Pta) were successfully expressed in the yeast Yarrowia lipolytica, and established an alternative cytosolic acetyl-CoA formation pathway, leading to improved lipid production performance.10 Pk can convert fructose-6-phosphate (F6P) or xylose-5-phosphate (X5P) into acetylphosphate without ATP consumption, followed by Pta activity to acetyl-CoA (Fig. 1). Thus, acetyl-CoA production was realized independent of the ATP-citrate lyase mediated process being utilized by most oleaginous yeasts for lipid accumulation under nitrogen limited conditions.9
The oleaginous yeast R. toruloides is a robust lipid producer.11,12 We have obtained multi-omic information of R. toruloides and established reliable genetic manipulation strategies.13,14 In the present study, the Pta gene originated from Bacillus subtilis was sequence-optimized, de novo synthesized, and integrated into the genome of R. toruloides NP11 through Agrobacterium mediated transformation (ATMT). The engineered strain showed significant improvement in glucose consumption, cell mass and lipid production when cultivated in Erlenmeyer flask as well as in stirred tank bioreactor. Our results should inspire more engineering efforts to facilitate the economic viability of MLs.
2. Results and discussion
2.1 Expression of Pta in R. toruloides NP11
The codon redressed Pta was introduced into the R. toruloides genome by using the ATMT strategy and confirmed by colony PCR analysis.14 Further genome walking results revealed that 3 of 4 insertion events happened at the same chromosome and that the left-border of two transformants, namely, NP-Pta-4 and NP-Pta-20, was inserted at an identical site (ESI†). However, no sequence homology was found among those inserted T-DNA flanking sequences and the insertion positions. The results suggested that T-DNA integrations occurred at random positions in the R. toruloides genome via a non-homologous recombination process as reported previously.16,17 RT-PCR results also confirmed successful expression of Pta in those transformants (ESI†).2.2 Lipid production by engineered R. toruloides strains
A number of colonies were first cultured in 50 mL test tubes in 5.0 medium to screen those with improved cell growth profile. This was because colonies obtained by the Agrobacterium-mediated transformation usually had the expression cassette randomly integrated into the genome, and thus could have varied phenotypic profiles.17 Indeed, many R. toruloides transformants were tested to find good fatty alcohol producer in a recent study.7 Our preliminary results showed that 4 out of 24 transformants, namely, NP-Pta-4, NP-Pta-6, NP-Pta-15 and NP-Pta-20, grew much faster than the wild strain NP11. Next, these four strains were grown on N-limited medium with an initial glucose concentration of 50 g L−1 under shake-flask culture conditions. It was found that the wild strain NP11 utilized glucose in 10 d, while NP-Pta-4, NP-Pta-6 and NP-Pta-20 did by 9 d, and NP-Pta-15, by 7 d (Fig. 2), clearly indicating an improved sugar consumption profile by those engineered strains. The cell mass of NP-Pta-4, NP-Pta-6 and NP-Pta-20 were slightly higher than that of NP11. However, NP-Pta-15 produced 12.8 g L−1 cell mass, about 8.5% more than that of NP11. The engineered strains produced 9.6% to 15.1% more lipids than the parental strain NP11 did. More importantly, lipid productivity of the engineered strain NP-Pta-15 reached 1.20 g L−1 d−1, about 64.4% higher than that of the NP11. These data clearly demonstrated the beneficial effects of Pta expression in R. toruloides for lipid production.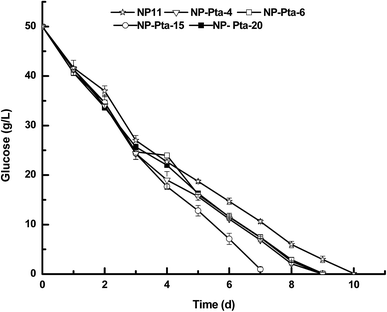 | ||
| Fig. 2 Glucose depletion curves of various R. toruloides strains under shake-flask culture condition. | ||
Therefore, we further compared lipid production performance between NP11 and NP-Pta-15 on 3 L bioreactor (Fig. 3). NP-Pta-15 consumed 200 g L−1 glucose within 54 h while it took 72 h for NP11, indicating that the engineered strain had 33.3% faster substrate consumption than the parental strain. NP-Pta-15 produced cell mass and lipid of 57.5 g L−1 and 15.7 g L−1, respectively, about 27.1% and 7.6% more than NP11 did. However, cellular lipid content of 28.7% was about 10.3% lower. The fact that NP-Pta-15 cells had a lower lipid content upon consumption of 200 g L−1 glucose suggested that more cells were produced and thus might be considered as an advantage for high cell density process by feeding more substrates.
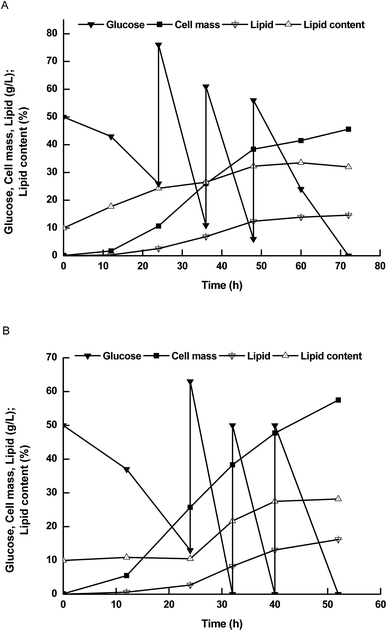 | ||
| Fig. 3 The time courses of lipid production by R. toruloides strains in 3 L bioreactor. (A) Results with NP11; (B) results with NP-Pta-15. | ||
The results above indicated that the presence of Pta in R. toruloides could facilitate cell growth and lipid production. It should be noted that our genome annotation study had suggested a Pk gene RHTO_04463 (ESI†) for potential formation of acetylphosphate from F6P or X5P by R. toruloides NP11.13 Therefore, the introduction of Pta may establish an alternative acetyl-CoA supplement by utilization of acetylphosphate from the non-oxidative glycolysis pathway (Fig. 1). Recently, improved higher alcohols and fatty acid derivatives production was achieved in a number of studies, in which a spare acetyl-CoA supplying channel was constructed besides the native acetyl-CoA generating pathway through the combination of Pk and Pta.10,18 However, more efforts should be conducted to verify the function of RHTO_04463 in R. toruloides NP11.
2.3 Fatty acid compositional profile
Table 2 showed the fatty acid compositional profiles of neutral lipids produced by R. toruloides strains. It was found that distributions of major fatty acids were 47.5–55.2% oleic acid (C18:1), 26.1–33.6% palmitic acid (C16:0) and 7.8–11.3% stearic acid (C18:0). Other minor fatty acids included 0.82–1.38% myristic acid (C14:0), 1.7–2.9% linoleic acid (C18:2) and 1.5–3.5% linolenic acid (C18:3). Such fatty acid compositional profiles were in line with the previous study with R. toruloides and similar to those of vegetable oils.11 The contents of linoleic acid were below the specified limit of 12% (w/w) for biodiesel production according to the European Standard EN 14214. The fact that no difference was found in terms of fatty acid species among those strains indicated that the Pta enzyme had little direct effects on the enzymes of fatty acid biosynthetic pathway, but might facilitate the fatty acid precursor acetyl-CoA supply. The MLs from engineered strains can be used as alternative feedstock for oleochemicals production (Table 1).| Strains or plasmids | Relevant characteristics | Source or resources |
|---|---|---|
| Strains | ||
| R. toruloides NP11 GCMCC A 2.224 | MAT A | Zhu et al., (2012) |
| R. toruloides NP-Pta-4 | PZPK-Ppgk-HYG-Tnos-Pgpd-PTA-Thsp, HYGR | This study |
| R. toruloides NP-Pta-6 | PZPK-Ppgk-HYG-Tnos-Pgpd-PTA-Thsp, HYGR | This study |
| R. toruloides NP-Pta-15 | PZPK-Ppgk-HYG-Tnos-Pgpd-PTA-Thsp, HYGR | This study |
| R. toruloides NP-Pta-20 | PZPK-Ppgk-HYG-Tnos-Pgpd-PTA-Thsp, HYGR | This study |
| Agrobacterium tumefaciens AGL1-HYG-Pta | PZPK-Ppgk-HYG-Tnos-Pgpd-PTA-Thsp | This study |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| Plasmids | ||
| PZPK-Pgpd-MCS-Thsp | PZPK-Ppgk-HYG-Tnos-Pgpd-MCS-Thsp in PZPK | This study |
| PZPK-Pgpd-PTA-Thsp | PZPK-Ppgk-HYG-Tnos-Pgpd-PTA-Thsp in PZPK | This study |
3. Experimental
3.1 Strain, media and reagents
R. toruloides NP11, a haploid derivative of R. toruloides AS 2.1389 from China General Microbiological Culture Collection Center, was maintained at 4 °C on YEPD agar slant containing (g L−1): glucose 20, yeast extract 10, peptone 10 and agar powder 20, pH 6. All the strains and plasmids are listed in Table 2.| Strains | Cell mass (g L−1) | Lipid content (%) | Lipid yield (g g−1) | Lipid productivity (g L−1 d−1) | Relative fatty acid content (%) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| C14:0 | C16:0 | C18:0 | C18:1 | C18:2 | C18:3 | |||||
| a Experiments with shale flasks.b Experiments with 3 L stirred-tank bioreactor. | ||||||||||
| NP11a | 11.8 ± 0.5 | 62.1 ± 2.4 | 0.146 ± 0.002 | 0.73 ± 0.01 | 1.38 ± 0.22 | 33.6 ± 0.4 | 9.6 ± 0.8 | 47.5 ± 1.5 | 2.7 ± 0.3 | 3.3 ± 0.6 |
| NP-Pta-4a | 12.5 ± 0.2 | 66.0 ± 4.7 | 0.166 ± 0.009 | 0.92 ± 0.05 | 1.02 ± 0.03 | 31.9 ± 0.1 | 7.8 ± 0.1 | 52.1 ± 0.4 | 1.9 ± 0.1 | 3.5 ± 0.2 |
| NP-Pta-6a | 12.3 ± 0.3 | 65.8 ± 1.7 | 0.160 ± 0.008 | 0.89 ± 0.03 | 0.87 ± 0.04 | 30.8 ± 0.1 | 8.5 ± 0.6 | 52.1 ± 0.9 | 2.2 ± 0.5 | 3.1 ± 0.3 |
| NP-Pta-15a | 12.8 ± 0.5 | 65.6 ± 2.4 | 0.168 ± 0.008 | 1.20 ± 0.06 | 0.82 ± 0.02 | 26.1 ± 0.2 | 11.3 ± 0.8 | 55.2 ± 1.8 | 1.7 ± 0.9 | 3.0 ± 1.0 |
| NP-Pta-20a | 12.2 ± 0.2 | 68.0 ± 1.0 | 0.166 ± 0.002 | 0.92 ± 0.01 | 1.14 ± 0.24 | 31.7 ± 1.2 | 7.8 ± 0.7 | 53.1 ± 2.1 | 2.9 ± 1.9 | 1.5 ± 0.5 |
| NP11b | 45.6 | 32.0 | 0.073 | 2.43 | 2.1 | 30.5 | 9.5 | 36.5 | 13.3 | 4.8 |
| NP-Pta-15b | 57.5 | 28.2 | 0.081 | 3.75 | 1.5 | 31.2 | 12.0 | 43.0 | 7.8 | 2.0 |
Lipid production media for flask cultures contained (g L−1): glucose 50, yeast extract 0.75, (NH4)2SO4 0.1, Mg2SO4·7H2O 1.5 and KH2PO4 0.4, pH 6.0. The lipid production media for bioreactor experiments contained (g L−1): glucose 50, (NH4)2SO4 10.0, yeast extract 10.0, KH2PO4 1.0, MgSO4·7H2O 1.0 and trace element stock solution (1%, v/v), pH 6.0. For feeding experiments, 1000 g L−1 glucose solution was used. All media were sterilized by autoclaving at 121 °C for 18 min. Trace element solution was added to all media after sterilization to a final concentration (mg L−1): CaCl2·2H2O 40, FeSO4·7H2O 5.5, citric acid·H2O 5.2, ZnSO4·7H2O 1.0 and MnSO4·H2O 0.76.
DNA polymerases and restriction enzymes were supplied by TaKaRa (Dalian, China). Acetosyringone, kanamycin, hygromycin and primers were from Dingguo Biotechnology Co. Ltd (Beijing, China). All other chemicals were of analytical grade and bought locally.
3.2 Construction of plasmids
The binary vector was constructed based on the pZPK plasmid with kanamycin resistance coding cassette.14 A B. subtilis originated phosphoacetyltransferase gene (Pta) was redressed by referring to the R. toruloides codon preference (ESI†) and synthesized (Synbio Technologies, Suzhou, China). The promoter of glyceraldehyde-3-phosphate dehydrogenase (Pgpd) was cloned from R. toruloides genome with the primers GPD-Xbal I-F and GPD-EcoR V-R. The PTA gene was then cloned with the primer pair Pta-EcoR V-F/Pta-Spe I-R (ESI†). Both the plasmid PZPK-Pgpd-MCS-Thsp and Pta gene were cleaved by restriction enzyme EcoR I and Spe I, and ligated to generate the plasmid PZPK-Ppgk-HYG-Tnos-Pgpd-PTA-Thsp for further engineering of R. toruloides.3.3 Genetic transformation and characterization
The Agrobacterium-mediated transformation protocol was based on a published method.14 R. toruloides transformants were picked and grew for five successive generations on YEPD plate containing 50 mg L−1 hygromycin for stability test. For reverse-transcriptional PCR analysis, total RNA and cDNA samples were prepared from R. toruloides transformants using the PrimeScript™ RT reagent Kit gDNA Eraser (Takara, Dalian, China). The Pta expression was detected with the primer pair Pta-RtPCR-F/Pta-RtPCR-R (ESI†).For genome walking experiments, total genomic DNA samples were extracted from R. toruloides transformants according to a former study.14 The thermal asymmetrical interlaced-polymerase chain reaction (TAIL-PCR) was used to identify the flanking sequence of T-DNA as described.15 Four arbitrary degenerated primers (LAD-1, LAD-2, LAD-3, LAD-4) and three specific right border (RB) primers (Pta-RB764, Pta-RB826, Pta-RB974) were used (ESI†). The four arbitrary degenerate primers and the Pta-RB764 were used in pre-amplification. The secondary TAIL-PCR cooperated with Pta-RB826 and LAD-1. Pta-RB973 and LAD-1 were applied for the last round genome walking. Only the most specific product over 700 bp was purified with San Prep Column DNA Gel Extraction Kit of Sangon Biotech (Shanghai, China), ligated into pMD™ 18-T Vector and transformed into Escherichia coli DH5α. Transformants were picked out by colony PCR and the sequences between the T-cloning sites were determined by Sangon Biotech. T-DNA location was identified by comparing the sequences with published R. toruloides genome using the BLAST program.13
3.4 Lipid production procedures
The inocula were prepared in YEPD liquid medium. A loop of NP11 or the engineered NP11 was inoculated into the YEPD medium and cultivated at 30 °C, 200 rpm for 24 h. Then the seed was transferred into lipid production medium for lipid production. The initial OD600 was 0.8 and the culture was sampled at intervals of 24 h for cell mass, lipid and glucose consumption analysis. All the experiments in flasks were done with a working volume of 50 mL and performed in triplicate. The experiments on 3 L bioreactor were initiated with a working volume of 1.8 L and an initial OD600 of 0.8. The air flow rate was 2 L min−1 and the dissolved oxygen was maintained above 40% by auto-controlled stirring. The cell mass, lipid and glucose consumption analysis were conducted at time intervals of 24 h and the glucose stock solution was added when needed.3.5 Analytical methods
Glucose was quantified by a glucose analyzer (SBA-50B; Shandong Academy of Sciences, Jinan, China). Cell mass was expressed as dry cell weight (DCW). Cells from 30 mL of culture broth were harvested by centrifugation at 8000 × g for 5 min, washed twice with distilled water, and dried at 105 °C to a constant weight. Lipids were extracted and the fatty acid composition of lipid samples was profiled by using a 7890F gas chromatography instrument after transesterification.11 Lipid content was expressed as g lipid per g DCW multiplied by 100%. The cell mass yield was defined as g cell mass per g sugar consumed. The lipid yield was defined as g lipid per g sugar consumed. The lipid productivity was defined as g lipid per litre per day. All values for flask cultures were the average of three independent experiments.4. Conclusions
In summary, this study demonstrated that overexpression of the Pta gene in the yeast R. toruloides NP11 can lead to strains with improved performance in terms of glucose consumption, cell growth and lipid production. The presence of Pta may establish an additional acetyl-CoA formation route by utilization of acetylphosphate. Our results should inspire more engineering efforts to facilitate the economic viability of large-scale ML production.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We sincerely thank Ms Xueying Wang for her comments on the manuscript. This study was supported by National Natural Science Foundation of China (21602218; 21325627; 51761145014) and China Postdoctoral Science Foundation (2016M591469).Notes and references
- U. Biermann, U. Bornscheuer, M. A. R. Meier, J. O. Metzger and H. J. Schäfer, Oils and fats as renewable raw materials in chemistry, Angew. Chem., Int. Ed., 2011, 50(17), 3854–3871 CrossRef PubMed.
- M. J. Jin, P. J. Slininger, B. S. Dien, S. Waghmodeet, B. Moser, A. Orjuela, L. C. Sousa and V. Balan, Microbial lipid-based lignocellulosic biorefinery: feasibility and challenges, Trends Biotechnol., 2015, 33(1), 43–54 CrossRef PubMed.
- A. A. Koutinas, A. Chatzifragkou, N. Kopsahelis, S. Papanikolaou and I. K. Kookoset, Design and techno-economic evaluation of microbial oil production as a renewable resource for biodiesel and oleochemical production, Fuel, 2014, 116, 566–577 CrossRef.
- C. Huang, X. F. Chen, L. Xiong, L. Xiong, X. D. Chen, L. L. Ma and Y. Chen, Single cell oil production from low-cost substrates: the possibility and potential of its industrialization, Biotechnol. Adv., 2013, 31(2), 129–139 CrossRef PubMed.
- Z. W. Gong, H. W. Shen, X. B. Yang, Q. Wang, H. B. Xie and Z. B. K. Zhao, Lipid production from corn stover by the oleaginous yeast Cryptococcus curvatus, Biotechnol. Biofuels, 2014, 7, 158 CrossRef PubMed.
- G. J. Jin, F. Yang, C. M. Hu, H. W. Shen and Z. B. K. Zhao, Enzyme-assisted extraction of lipids directly from the culture of the oleaginous yeast Rhodosporidium toruloides, Bioresour. Technol., 2012, 111, 378–382 CrossRef PubMed.
- S. Fillet and J. L. Adrio, Microbial production of fatty alcohols, World J. Microbiol. Biotechnol., 2016, 32, 152 CrossRef PubMed.
- J. J. L. Lee, L. W. Chen, B. Cao and W. N. Chen, Engineering Rhodosporidium toruloides with a membrane transporter facilitates production and separation of carotenoids and lipids in a bi-phasic culture, Appl. Microbiol. Biotechnol., 2016, 100(2), 869–877 CrossRef PubMed.
- S. Papanikolaou and G. Aggelis, Lipids of oleaginous yeasts. Part I: Biochemistry of single cell oil production, Eur. J. Lipid Sci. Technol., 2011, 113(8), 1031–1051 CrossRef.
- P. Xu, K. Qiao, W. S. Ahn and G. Stephanopoulos, Engineering Yarrowia lipolytica as a platform for synthesis of drop-in transportation fuels and oleochemicals, Proc. Natl. Acad. Sci. U. S. A., 2016, 113(39), 10848–10853 CrossRef PubMed.
- Y. H. Li, Z. B. Zhao and F. W. Bai, High-density cultivation of oleaginous yeast Rhodosporidium toruloides Y4 in fed-batch culture, Enzyme Microb. Technol., 2007, 41(3), 312–317 CrossRef.
- J. Yaegashi, J. Kirby, M. Ito, J. Sun, T. Dutta, M. Mirsiaghi, E. R. Sundstrom, A. Rodriguez, E. Baidoo, D. Tanjore, T. Pray, K. Sale1, S. Singh, J. D. Keasling, B. A. Simmons, S. W. Singer, J. K. Magnuson, A. P. Arkin, J. M. Skerker and J. M. Gladden, Rhodosporidium toruloides: a new platform organism for conversion of lignocellulose into terpene biofuels and bioproducts, Biotechnol. Biofuels, 2017, 10, 241 CrossRef PubMed.
- Z. W. Zhu, S. F. Zhang, H. W. Liu, H. W. Shen, X. P. Lin, F. Yang, Y. J. J. Zhou, G. J. Jin, M. L. Ye, H. F. Zou and Z. K. Zhao, A multi-omic map of the lipid-producing yeast Rhodosporidium toruloides, Nat. Commun., 2012, 3, 1112 CrossRef PubMed.
- X. Lin, Y. Wang, S. Zhang, Z. W. Zhu, Y. J. J. Zhou, F. Yang, W. Y. Sun, X. Y. Wang and Z. B. K. Zhao, Functional integration of multiple genes into the genome of the oleaginous yeast Rhodosporidium toruloides, FEMS Yeast Res., 2014, 14(4), 547–555 CrossRef PubMed.
- Y. G. Liu and Y. Chen, High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences, Biotechniques, 2007, 43(5), 649–656 CrossRef PubMed.
- Y. B. Liu, C. M. J. Koh, L. H. Sun, L. H. Sun, M. M. Hlaing, M. G. Du, N. Peng and L. H. Ji, Characterization of glyceraldehyde-3-phosphate dehydrogenase gene RtGPD1 and development of genetic transformation method by dominant selection in oleaginous yeast Rhodosporidium toruloides, Appl. Microbiol. Biotechnol., 2013, 97(2), 719–729 CrossRef PubMed.
- X. Lin, N. Gao, S. Liu, S. F. Zhang, S. Song, C. F. Ji, X. P. Dong, Y. C. Su, Z. B. K. Zhao and B. W. Zhu, Characterization the carotenoid productions and profiles of three Rhodosporidium toruloides mutants from Agrobacterium tumefaciens-mediated transformation, Yeast, 2017, 34(8), 335–342 CrossRef PubMed.
- I. W. Bogorad, C. T. Chen, M. K. Theisen, T. Y. Wu, A. R. Schlenz, A. T. Lam and J. C. Liao, Building carbon–carbon bonds using a biocatalytic methanol condensation cycle, Proc. Natl. Acad. Sci. U. S. A., 2014, 111(45), 15928–15933 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: 2 ESI tables and 3 ESI figures are provided. See DOI: 10.1039/c8ra03028f |
| This journal is © The Royal Society of Chemistry 2018 |