Open Access Article
Open Access ArticleSynergy of carbon and nitrogen removal of a co-culture of two aerobic denitrifying bacterial strains, Acinetobacter sp. GA and Pseudomonas sp. GP†
Yan Guoab,
Ruili Yangab,
Zhaoji Zhang *a,
Xiaojun Wanga,
Xin Yea and
Shaohua Chen
*a,
Xiaojun Wanga,
Xin Yea and
Shaohua Chen *a
*a
aCAS Key Laboratory of Urban Pollutant Conversion, Institute of Urban Environment, Chinese Academy of Sciences, 1799 Jimei Road, Xiamen 361021, People's Republic of China. E-mail: shchen@iue.ac.cn; Fax: +86 592 6190977; Tel: +86 592 6190995
bUniversity of Chinese Academy of Sciences, Beijing 100049, People's Republic of China
First published on 13th June 2018
Abstract
Two newly isolated aerobic denitrifying bacterial strains (Acinetobacter sp. GA and Pseudomonas sp. GP) were co-cultured to investigate the synergy of carbon and nitrogen removal of different functional bacteria. The co-culture showed higher efficiency for removing total organic carbon (TOC) and total dissolved nitrogen (TDN) than strain GA or GP cultured separately. The inoculation ratio of 50 (strain GA/GP) was advantageous for the TOC and TDN removal efficiencies of the co-culture. The sequential co-culture tests showed that strain GP being inoculated after strain GA was inoculated for 36 h increased the TDN removal efficiency from 53.3% to 86.8%. This finding indicated that the activity of strain GA was important in the co-culture system, and the sequential co-culture could be advantageous to the synergistic effect of strains GA and GP. The co-culture of different functional bacteria can be an alternative method for improving the performance of aerobic denitrifying microorganisms for pollutant removal.
1. Introduction
Biological treatment processes are widely used in municipal wastewater treatment plants because of their high efficiency and low cost.1 Typical biological treatment processes are often constructed by combining several aerobic, anaerobic, and anoxic reactors to remove organic matter and nutrients because of the diverse and complex metabolism of microorganisms.2 For example, the nitrogen removal process in wastewater treatment plants is often composed of autotrophic aerobic nitrification and heterotrophic anaerobic denitrification.3 However, various habitat requirements of different microorganisms can adversely affect the efficiency of pollutant removal. Moreover, traditional biological treatment processes present limitations in terms of large floor space and fluctuant efficiency in extreme environments.4 Hence, new biological treatment processes must be developed for designing model wastewater treatment plants.Aerobic denitrification is a new biological nitrogen removal process that has been widely investigated in the last decade. Several functional bacterial strains have also been isolated in recent years. These bacterial strains belong to various genera, such as Acinetobacter,5,6 Pseudomonas,7,8 Bacillus,9 and Paracoccus.10 In contrast to traditional nitrification–denitrification, aerobic denitrifying bacteria can remove organic matter and nitrogen compounds simultaneously and efficiently in one reactor.11 To date, most studies on aerobic denitrification focused on isolation and characterization of functional bacterial strains.5,7,10 Different strains show varied carbon and nitrogen metabolic performances. The optimal C/N, pH, and other culture conditions of different functional bacterial strains have been extensively studied.12,13 However, few scholars have focused on mixed aerobic denitrifying bacterial consortia. Compared with the pure culture of functional strains, mixed bacterial consortia might be more resistant to stress factors due to their composition variability and good adaptability.14 The slow growth rate and complicated regulation limit the development of mixed aerobic denitrifying bacterial consortia.15 The co-culture of two different bacterial strains might be an alternative for fundamental research of bacterial consortia, which exhibit rapid growth rate and primary interaction of different bacteria. Thus far, no study has investigated yet the co-culture of two different aerobic denitrification bacterial strains.
The co-culture technology of different microorganisms has been used widely in environmental or bioengineering fields. For example, the co-culture of anammox bacteria and ammonium oxidizing bacteria can be used for complete removal of autotrophic nitrogen removal over nitrite, which is known as the CANON process.16 In laboratory-scale studies, Mujtaba et al. co-cultured the bacterium Pseudomonas putida and microalga Chlorella vulgaris to treat simulated wastewater.17 The mixed consortium showed high efficiency for removal of organic matter and ammonia. Zhao et al. researched the dehalogenation of trihalomethanes by a co-culture containing Dehalobacter sp., which was responsible for the degradation of trihalomethanes, and Desulfovibrio sp., which provided a stepwise reductive environment for the metabolism of the former.18 Furthermore, the co-culture of different microorganisms has been broadly investigated during the production of surfactin,19 methane,20 and H2.21 Results showed that the co-culture of different microorganisms can cause synergy on pollutant degradation or resource production. The co-culture of complementary bacteria can be more efficient than the pure culture of a sole bacterial strain.
In this study, two aerobic denitrifying bacterial strains were isolated. The carbon and nitrogen removal performance of the two strains was characterized with different carbon and nitrogen sources. The main objective of this study was trying to provide a method to improve the capacity of biodegradation of substrates for aerobic denitrifying bacterial strains through investigating the co-culture performance of the two strains in terms of degradation of substrates and change in bacterial abundance. The influence of inoculation ratio and time was also studied to explore the optimal incubation condition for the co-culture.
2. Materials and methods
2.1 Culture media
Table 1 shows the carbon and nitrogen sources used in different media (media M1 to M7). The concentrations of total organic carbon (TOC) and total dissolved nitrogen (TDN) was 1000 and 100 mg L−1, respectively, in each medium. The other medium components were identical (Table 2). Bromothymol blue (BTB) medium used for bacterial isolation was modified from medium M3 by adding 5.0 mL L−1 1% alcoholic solution of BTB and 1.5% agar. The preculture before each shaking experiment was conducted in medium M3. The harvested bacterial solution was washed twice by sterile water before use.| Medium | Carbon source (g L−1) | Nitrogen source (g L−1) | ||
|---|---|---|---|---|
| M1 | Sodium acetate | 3.45 | NH4Cl | 0.38 |
| M2 | Sodium acetate | 3.45 | NaNO2 | 0.50 |
| M3 | Sodium acetate | 3.45 | KNO3 | 0.72 |
| M4 | Sodium acetate | 2.84 | Acetamide | 0.42 |
| M5 | Sodium succinate hexahydrate | 5.62 | KNO3 | 0.72 |
| M6 | Sodium citrate dihydrate | 4.08 | KNO3 | 0.72 |
| M7 | Sodium citrate dihydrate | 3.39 | Acetamide | 0.42 |
| Inorganic components (g L−1) | Trace element (mg L−1) | ||||
|---|---|---|---|---|---|
| KH2PO4 | 1.50 | EDTA | 20.00 | MnCl2·4H2O | 10.00 |
| Na2HPO4·12H2O | 7.90 | ZnSO4 | 7.80 | CaCl2·2H2O | 14.60 |
| MgSO4·7H2O | 0.10 | FeSO4·7H2O | 10.00 | CoCl2·6H2O | 3.22 |
| CuSO4·5H2O | 3.14 | (NH4)6Mo7O2·4H2O | 2.20 | ||
2.2 Isolation of aerobic denitrifying bacterial strains
Two bacterial strains were isolated from an aerobic denitrifying reactor.11 In brief, diluted activated sludge was inoculated on plates with BTB medium. After incubation for several days, the blue colonies were selected and inoculated into fresh BTB plates through streak plate method. The last step was repeated three times to purify the bacterial strains. The candidate bacterial strains were cultured in medium M3 to assess their nitrate removal efficiency. Bacterial strains with high nitrate removal efficiency (>50%) were identified by 16S rRNA gene amplification and sequencing. Phylogenetic trees were constructed by MEGA 6.06 program through neighbor-joining method.2.3 Nitrogen removal tests
Media M1–M6 were used to determine the nitrogen removal capacity of the two selected bacterial strains (named as strains GA and GP). In brief, 6 mL of the pre-cultured bacteria solution was inoculated into 100 mL of medium, which was then incubated at 25 °C under shaking at 150 rpm. All experiments were conducted in triplicate. The concentrations of ammonium nitrogen (NH4+–N), nitrite nitrogen (NO2−–N), nitrate nitrogen (NO3−–N), TDN, TOC, and optical density at 600 nm (OD600) were detected periodically during the incubation. Measurements were performed according to the standard methods.222.4 Co-culture of two bacterial strains
DNA was extracted from the co-culture periodically during the incubation by using FastDNA SPIN Kit (MP Biomedicals, Illkirch, France) according to the manufacturer's instructions. The combined V3–V4 hypervariable region of the bacterial 16S rRNA gene was amplified using oligonucleotide barcode tags containing the 341F and 806R primers. Polymerase chain reaction amplification, amplicon library preparation, Illumina MiSeq sequencing, and data analysis were conducted according to our previously described protocols.23 The sequences were aligned with the Greengenes database (version 13.5).24
3. Results and discussion
3.1 Isolation and identification of bacterial strains
Two bacterial strains were isolated and selected for subsequent experiments. Analysis of the phylogenetic trees showed that strain GA belongs to Acinetobacter, and strain GP belongs to Pseudomonas. These bacterial genera are commonly discussed in published papers about aerobic denitrification.5–8 In the source bioreactor, the abundance of these bacterial genera was higher than that of other reported aerobic denitrifying bacterial genera. The sequence data about 16S rRNA of two bacterial strains have been submitted to the NCBI Sequence Read Archive database (strain GA: SRR6073705; strain GP: SRR3073704).3.2 Nitrogen removal performance of two bacterial strains
Fig. 1 shows the OD600 and nitrogen removal performance of the two bacterial strains in six media. M1–M4 media were used to investigate the effect of nitrogen source on the growths and nitrogen removal performance of strain GA and GP. In these media, the carbon source was acetate, and the nitrogen source was ammonium, nitrite, nitrate, and acetamide, respectively. The results showed that both strain GA and GP grew rapidly in the medium with inorganic nitrogen as nitrogen source (Fig. 1a–c). When the nitrogen source was acetamide, only strain GA grew (Fig. 1d). Strain GA showed efficient nitrogen removal performance in all the four media, and the nitrogen removal efficiency reached 90% after incubation for 24 h. For strain GP, the nitrogen removal efficiency in the ammonium medium is higher than that in the medium with nitrite or nitrate as nitrogen source. This result was also obtained in other bacterial species, such as Paracoccus versutus LYM10 and Aeromonas sp. HN-02.11 Ammonium could be the most suitable nitrogen source for most aerobic denitrifying bacteria. However, some bacterial species, such as Pseudomonas stutzeri YZN-001, showed high nitrogen removal rate when nitrate was used as the nitrogen source.8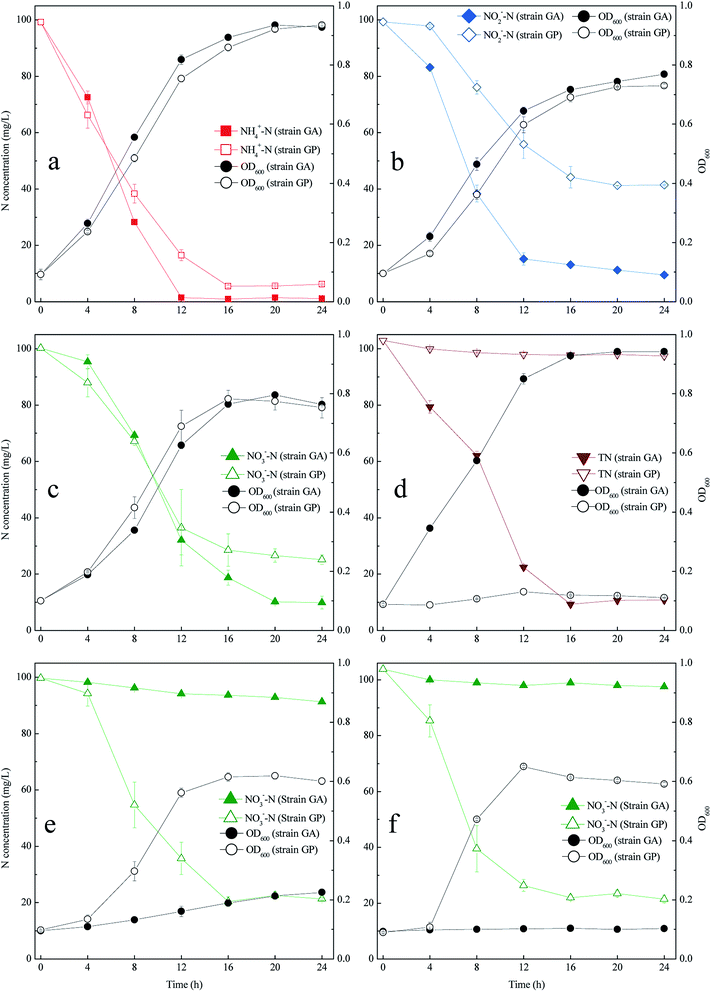 | ||
| Fig. 1 OD600 and nitrogen removal performance of strains GA and GP in the following media: M1 (a); M2 (b); M3 (c); M4 (d); M5 (e); and M6 (f). | ||
Media M3, M5, and M6 were used to investigate the influence of carbon source on the growth and nitrogen removal efficiency of the two bacterial strains (Fig. 1c, e and f). In these media, the nitrogen source was nitrate, and the carbon source was acetate, succinate, and citrate, respectively. Strain GA only grew in the acetate medium, while strain GP could utilize all these three carbon sources. The nitrate removal efficiency of strain GP in different carbon sources reached 75%. However, the OD600 in the succinate or citrate medium is lower than that in the acetate medium. Based on these results, micromolecular organic matter was suitable for bacterial growth because of its simple molecular structure. Several studies reported acetate or succinate as the optimal carbon source for many species of Acinetobacter and Pseudomonas, such as Acinetobacter junii YB,26 Acinetobacter sp. HA2,6 Pseudomonas sp. T13,27 and Pseudomonas tolaasii Y-11.28 However, some aerobic denitrifying bacterial species, such as Chryseobacterium sp. R31,29 and Bacillus methylotrophicus L7,30 cannot utilize acetate. In addition, several aerobic denitrifying bacteria cannot use citrate or sucrose as carbon source.30,31 However, Agrobacterium sp. LAD9 can utilize citrate or sucrose as carbon source.32
These results indicated that aerobic denitrifying bacteria from different species displayed varied performance for carbon and nitrogen metabolism. However, it also implied that using only one kind of aerobic denitrifying bacteria is insufficient for treating complex wastewater. The reasonable co-culture of different bacterial strains may improve the capacity of biodegradation of substrates for aerobic denitrifying bacterial strains. It might be an alternative solution to improve the pollutant removal efficiency.
3.3 Co-culture of two bacterial strains
Fig. 1 shows that in the medium with appropriate carbon source or nitrogen source, strain GA utilized acetamide but did not degrade citrate; meanwhile, strain GP utilized citrate but did not consume acetamide. Therefore, when the carbon source was citrate and the nitrogen source was acetamide (medium M7), the growth rate of separately cultured strain GA or GP was low (Fig. 2a and b). The TDN and TOC removal efficiencies of strain GA were 30.18% and 25.17%. For strain GP, the TDN and TOC removal efficiencies were 7.13% and 11.76%.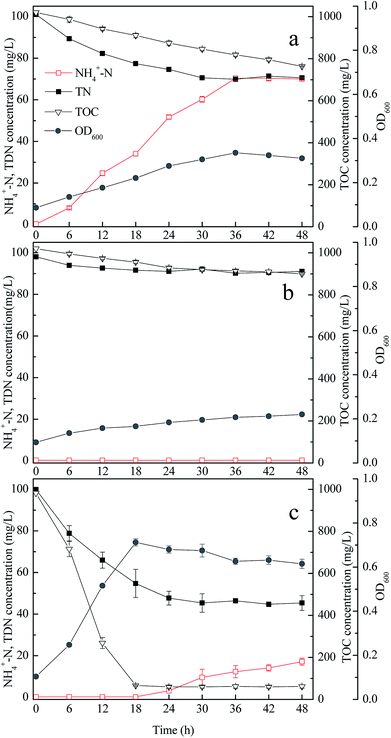 | ||
| Fig. 2 Performance of strains GA and GP in medium M7; separate culture of strain GA (a); separate culture of strain GP (b); and co-culture of strains GA and GP (c). | ||
When strain GA was incubated solely in medium M7, the concentration of ammonium increased gradually during the entire process (Fig. 2a). It was the result of the biodegradation of acetamide by the strain GA. In medium M7, strain GA utilized acetamide as both carbon and nitrogen sources. Given that acetamide is a nitrogenous organic matter with low C/N, the excess nitrogen was discharged from bacterial cells in the form of ammonium.33
When strains GA and GP were co-cultured in medium M7, the TDN and TOC removal efficiencies were higher than those when the strains were cultured separately (Fig. 2c). Hence, the co-culture of strains GA and GP exerted synergy on the carbon and nitrogen removal performance. After 48 h, the TDN and TOC removal efficiencies of the co-culture reached 54.61% and 93.67%, respectively. In the first 18 h, the TOC and TDN concentrations decreased quickly. Ammonium was not detected because the released ammonium by strain GA might be utilized by strain GP. With citrate as the carbon source, and ammonium as the nitrogen source, strain GP could grow and remove TOC and TDN efficiently. After 18 h, the TOC concentration was below 100 mg L−1, and the TDN concentration remained approximately 50 mg L−1. The ammonium concentration increased slowly in the later period of the co-culture, indicating that the carbon source for strain GP was insufficient in subsequent culture. Therefore, ammonia released by strain GA accumulated.
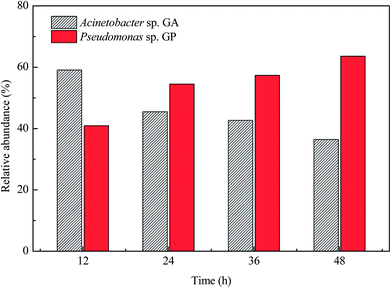
Fig. 3 shows the changes in the relative abundance of strains GA and GP during the co-culture. In the first 12 h, the relative abundance of strain GA was higher than that of strain GP. However, the relative abundance of strain GP increased and exceeded that of strain GA after 24 h. In the beginning of the co-culture, strain GA was found to be dominant in the mixed system because it grew by utilizing acetamide, whereas strain GP did not grow because of the lack of nitrogen source. As the culture proceeded, citrate and ammonium released by strain GA constructed an advantageous habitat for strain GP. Therefore, the relative abundance of strain GP became preponderant in the later hours of incubation.
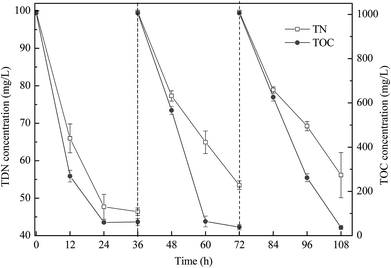
The co-culture was conducted for three cycles to investigate its stability (Fig. 4). After culture for 36 h in each cycle, the TOC removal efficiency in the three cycles was similar, whereas the TDN removal efficiency slightly decreased. Hence, the co-culture system exhibited good repeatability, especially for TOC removal.
3.4 Effect of inoculum ratio of the two strains on the performance of co-culture
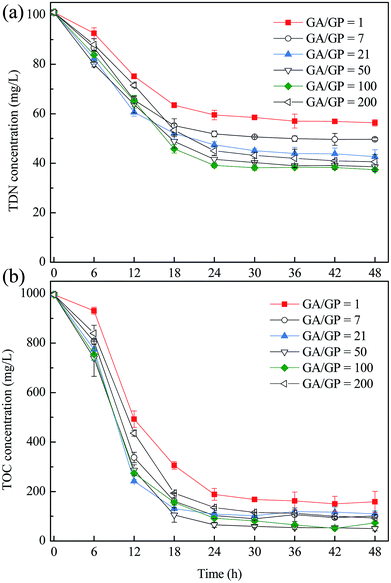
Fig. 5 shows the TDN and TOC removal performance of the co-culture with different inoculum ratios. When the cell number ratio of strain GA to GP was increased from 1 to 50, the TDN and TOC removal efficiency were both increased. When the ratio was higher than 50, the TDN and TOC removal efficiency were keeping stable and then showed a little decline at the ratio of 200. It indicated that the cell number ratio of the inoculum has an obvious influence on the performance of the co-culture. When the cell number ratio was lower than 50, the activity of strain GA seems to be the restrictive factor, since the growth of strain GP was dependent on the metabolism of strain GA. A higher abundance of strain GA could release the ammonium faster and then promote the TDN and TOC removal rate of the co-culture. However, when the ratio of strain GA was higher than 100, the excess strain GA and the deficient strain GP also brought down the TDN and TOC efficiency of the co-culture. This result showed that the abundance of strain GA and strain GP affect the performance of the co-culture jointly, and the optimal cell number ratio was about 50 to 100.In Fig. 3, the abundance of strain GP was higher than that of GA after co-culture for 24 h. When the co-culture system was re-cultured in the fresh medium, the ratio of GA/GP would be less than the former batch culture. Correspondingly, the TDN and TOC removal rates were lower than those in the former batch culture (Fig. 4). This result was consistent with that shown in Fig. 5, that is, a low ratio of GA/GP would lead to decreased TOC and TDN removal rates. Hence, the activity of strain GA was found to be the decisive factor in the co-culture system.
For different co-culture systems, the inoculum ratio influenced the co-culture efficiency to some extent. Mujtaba and Lee reported that in the co-culture system of C. vulgaris and activated sludge,34 the optimal ratio of activated sludge to microalgae was 0.5. A high proportion of sludge would decrease the efficiency for removing ammonium and phosphate. Zhi et al. indicated that different inoculation ratios of two Bacillus amyloliquefaciens strains adversely influenced the total cell number and surfactin production.19 The ratio of 1/5 was considered advantageous to cell growth, and the ratio of 1/0.2 was the best for resource production. Hence, inoculation ratio would decide the initial relative abundance of different microorganisms and influence the interaction of the co-cultured microorganisms. The optimal inoculation ratio can be confirmed by combining the mechanisms of the co-culture system and the research targets of researchers.
3.5 Effect of inoculation time of strain GP on the performance of the co-culture
Fig. 6 shows that the inoculation time of GP influenced the performance of the co-culture, particularly in terms of OD600 increase and TDN removal. As the inoculation time of strain GP increased from 0 to 36 h, the OD600 and TDN removal efficiency of the co-culture increased. When strain GP was inoculated at 36 h, the TDN removal efficiency of the co-culture reached 91.97%, which considerably increased compared with the batch culture when strain GP was inoculated at 0 h (Fig. 6c). Fig. 6b shows the changes in the ammonium concentration during the co-culture. Prolonged culture of strain GA led to high amounts of ammonium that accumulated in the medium. When strain GP was inoculated into the medium, the concentrations of ammonium and TDN rapidly decreased. Moreover, the TOC concentration decreased quickly after strain GP was inoculated in all batch tests. Different ammonium concentrations did not influence the TOC removal efficiency. This result indicates that strain GP can grow in an appropriate medium with a wide range of C/N, like many other published aerobic denitrifying bacterial strains.30,31,35 In conclusion, strain GP inoculated later can improve the TDN removal efficiency but would not affect the TOC removal efficiency.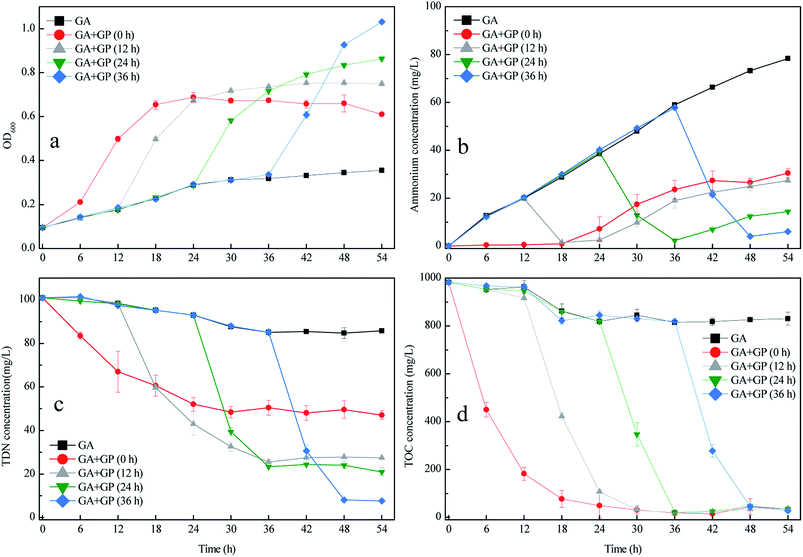 | ||
| Fig. 6 Changes in OD600 (a) and in the concentrations of NH4+–N (b); TDN (c); and TOC (d) during co-culture at different inoculation times. | ||
Sequential co-culture is often used in biological fermentation. Wen et al. fermented acetone–butanol–ethanol (ABE) through the sequential co-culture of Clostridium thermocellum and Clostridium beijerinckii;15 that is, the former was cultured for 100 h before the latter was inoculated. This method could efficiently produce ABE from lignocellulose in one step. Liu et al. conducted sequential co-culture to ferment Solaris white wine.36 In this study, four non-Saccharomyces yeasts were inoculated and cultured for 3 days before Saccharomyces cerevisiae was inoculated. Based on the results, the three Metschnikowia strains exhibited potential for winemaking from Solaris grapes. The greatest advantage of sequential co-culture is that it retained the synergy and coordinated the discrepancy in the growth rate of different microorganisms. In the present study, strain GP inoculated later can provide sufficient time for strain GA to transform acetamide to ammonium. The high amount of ammonium that accumulated in the medium before the inoculation of strain GP led to high TDN removal efficiency. Therefore, the flexible and reasonable use of sequential co-culture could improve the performance of mixed cultivation.
4. Conclusions
Two newly isolated aerobic denitrifying bacteria, namely, Acinetobacter sp. GA and Pseudomonas sp. GP, were co-cultured in one medium. The TOC and TDN removal efficiencies of the co-culture are higher than those of strains GA and GP cultured separately. Strain GA was abundant in the first 12 h, whereas strain GP was dominant after 24 h. The optimal inoculation ratio of strain GA to strain GP was about 50. The sequential co-culture tests showed that strain GP inoculated later increased the TDN removal efficiency but did not affect the TOC removal efficiency.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the China-Japanese Research Cooperative Program (No. 2016YFE0118000) and the National Science and Technology Major Project of China under Grant No. 2016ZX05060-022. We wish to thank the anonymous reviewers for their valuable suggestions on revising the work.References
- A. A. Khardenavis, A. Kapley and H. J. Purohit, Appl. Microbiol. Biotechnol., 2007, 77, 403–409 CrossRef PubMed.
- T. Alvarino, S. Suarez, J. Lema and F. Omil, Sci. Total Environ., 2018, 615, 297–306 CrossRef PubMed.
- K. Bernat and I. Wojnowska-Baryła, Biochem. Eng. J., 2007, 36, 116–122 CrossRef.
- M. X. Chen, W. C. Wang, Y. Feng, X. H. Zhu, H. Z. Zhou, Z. L. Tan and X. D. Li, Bioresour. Technol., 2014, 167, 456–461 CrossRef PubMed.
- J. F. Su, J. X. Shi, T. L. Huang and F. Ma, Mar. Pollut. Bull., 2016, 109, 87–94 CrossRef PubMed.
- S. Yao, J. R. Ni, T. Ma and C. Li, Bioresour. Technol., 2013, 139, 80–86 CrossRef PubMed.
- Y. Xu, T. X. He, Z. L. Li, Q. Ye, Y. L. Chen, E. Y. Xie and X. Zhang, BioMed Res. Int., 2017, 1429018 Search PubMed.
- J. B. Zhang, P. X. Wu, B. Hao and Z. N. Yu, Bioresour. Technol., 2011, 102, 9866–9869 CrossRef PubMed.
- Z. F. Song, J. An, G. H. Fu and X. L. Yang, Aquaculture, 2011, 319, 188–193 CrossRef.
- Z. Shi, Y. Zhang, J. T. Zhou, M. X. Chen and X. J. Wang, Bioresour. Technol., 2013, 148, 144–148 CrossRef PubMed.
- B. Ji, K. Yang, L. Zhu, Y. Jiang, H. Y. Wang, J. Zhou and H. N. Zhang, Biotechnol. Bioprocess Eng., 2015, 20, 643–651 CrossRef.
- P. Z. Chen, J. Li, Q. X. Li, Y. C. Wang, S. P. Li, T. Z. Ren and L. G. Wang, Bioresour. Technol., 2012, 116, 266–270 CrossRef PubMed.
- Y. X. Liu, T. T. Hu, Y. J. Song, H. P. Chen and Y. K. Lv, J. Biosci. Bioeng., 2015, 120, 549–554 CrossRef PubMed.
- L. Vacková, M. Srb, R. Stloukal and J. Wanner, Bioresour. Technol., 2011, 102, 4661–4666 CrossRef PubMed.
- Z. Q. Wen, M. B. Wu, Y. J. Lin, L. R. Yang, J. P. Lin and P. L. Cen, Process Biochem., 2014, 49, 1941–1949 CrossRef.
- Y. F. Deng, X. L. Zhang, Y. Miao and B. Hu, Water Sci. Technol., 2016, 73, 534–542 Search PubMed.
- G. Mujtaba, M. Rizwan and K. Lee, J. Ind. Eng. Chem., 2017, 49, 145–151 CrossRef.
- S. Y. Zhao, M. J. Rogers and J. Z. He, Appl. Microbiol. Biotechnol., 2017, 101, 5481–5492 CrossRef PubMed.
- Y. Zhi, Q. Wu and Y. Xu, Bioresour. Technol., 2017, 235, 96–103 CrossRef PubMed.
- T. Yeung, M. Kwan, L. Adler, T. J. Mills, B. A. Neilan, G. Conibeer and R. Patterson, Bioresour. Technol., 2017, 234, 686–692 CrossRef PubMed.
- P. Sharma and U. Melkania, Bioresour. Technol., 2017, 239, 49–56 CrossRef PubMed.
- Chinese NEPA, Water and wastewater monitoring methods, Environ. Sci. Pub. House, Beijing, 4th edn., 2002 Search PubMed.
- Y. Y. Wang, Z. J. Zhang, L. Qiu, Y. Guo, X. J. Wang, X. J. Xiong and S. H. Chen, Biochem. Eng. J., 2015, 101, 200–208 CrossRef.
- T. Z. DeSantis, P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu and G. L. Andersen, Appl. Environ. Microbiol., 2006, 72, 5069–5072 CrossRef PubMed.
- P. Foladori, S. Tamburini and L. Bruni, Water Res., 2010, 44, 4888–4899 CrossRef PubMed.
- L. Yang, Y. X. Ren, X. Liang, S. Q. Zhao, J. P. Wang and Z. H. Xia, Bioresour. Technol., 2015, 193, 227–233 CrossRef PubMed.
- Y. L. Sun, A. Li, X. N. Zhang and F. Ma, Appl. Microbiol. Biotechnol., 2015, 99, 3243–3248 CrossRef PubMed.
- T. X. He, Z. L. Li, Q. Sun, Y. Xu and Q. Ye, Bioresour. Technol., 2016, 200, 493–499 CrossRef PubMed.
- P. Kundu, A. Pramanik, A. Dasgupta, S. Mukherjee and J. Mukherjee, BioMed Res. Int., 2014 DOI:10.1155/2014/436056.
- Q. L. Zhang, Y. Liu, G. M. Ai, L. L. Miao, H. Y. Zheng and A. P. Liu, Bioresour. Technol., 2012, 108, 35–44 CrossRef PubMed.
- J. Chen, J. Zheng, Y. Li, H. H. Hao and J. M. Chen, Appl. Microbiol. Biotechnol., 2015, 99, 10695–10702 CrossRef PubMed.
- Q. Chen and J. R. Ni, J. Biosci. Bioeng., 2012, 113, 619–623 CrossRef PubMed.
- B. H. M. Schmidt, K. Kalbitz, S. Braun, R. Fuß, W. H. McDowelle and E. Marzner, Soil Biol. Biochem., 2011, 43, 1742–1745 CrossRef.
- G. Mujtaba and K. Lee, Water Res., 2017, 120, 174–184 CrossRef PubMed.
- Y. Zhang, Z. Shi, M. X. Chen, X. Y. Dong and J. T. Zhou, Bioresour. Technol., 2015, 175, 602–605 CrossRef PubMed.
- J. Liu, N. Arneborg, T. B. Toldam-Andersen, S. J. Zhang, M. A. Petersen and W. L. P. Bredie, Eur. Food Res. Technol., 2017, 243, 437–445 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra02721h |
| This journal is © The Royal Society of Chemistry 2018 |