Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Effect of surfactant on Pseudomonas aeruginosa colonization of polymer microparticles and flat films†
Amanda Hüsler abc,
Simon Haasb,
Luke Parry
abc,
Simon Haasb,
Luke Parry b,
Manuel Romero
b,
Manuel Romero d,
Takasi Nisisako
d,
Takasi Nisisako c,
Paul Williams
c,
Paul Williams d,
Ricky D. Wildman
d,
Ricky D. Wildman b and
Morgan R. Alexander
b and
Morgan R. Alexander *a
*a
aAdvanced Materials and Healthcare Technologies Division, School of Pharmacy, University of Nottingham, Nottingham, NG7 2RD, UK. E-mail: morgan.alexander@nottingham.ac.uk
bCentre for Additive Manufacturing, Faculty of Engineering, University of Nottingham, Nottingham, NG7 2RD, UK
cInstitute of Innovative Research, Tokyo Institute of Technology, Yokohama, Japan
dCentre for Biomolecular Sciences, School of Life Sciences, University of Nottingham, Nottingham, NG7 2RD, UK
First published on 24th April 2018
Abstract
Micro- and nanoparticles are of great interest because of their potential for trafficking into the body for applications such as low-fouling coatings on medical devices, drug delivery in pharmaceutics and cell carriers in regenerative medicine strategies. Particle production often relies on the use of surfactants to promote stable droplet formation. However, the presence of residual surfactant has been shown to complicate the surface chemistry and resultant properties. When forming particles from polymerizable monomer droplets, these polymeric surfactant chains can become physically entangled in the particle surface. Due to the key role of the outermost layers of the surface in biomaterial interactions, the surface chemistry and its influence on cells needs to be characterized. This is the first study to assess surfactant retention on microfluidic produced particles and its effect on bacterial attachment; surfactant contaminated microparticles are compared with flat films which are surfactant-free. Polymeric microparticles with an average diameter of 76 ± 1.7 μm were produced by using a T-junction microfluidic system to form monomer droplets which were subsequently photopolymerized. Acrylate based monomer solutions were found to require 2 wt% PVA to stabilize droplet formation. ToF-SIMS was employed to assess the surface chemistry revealing the presence of PVA in a discontinuous layer on the surface of microparticles which was reduced but not removed by solvent washing. The effect of PVA on bacterial (Pseudomonas aeruginosa) attachment was quantified and showed reduction as a function of the amount of PVA retained at the surface. The insights gained in this study help define the structure–function relationships of the particulate biomaterial architecture, supporting materials design with biofilm control.
Introduction
Over the last decade, materials discovery using high throughput screening has been applied to identify novel materials with unique properties that can be tailored for specific applications, such as maintaining stem cell pluripotency for tissue engineering applications or reducing bacterial attachment and biofilm formation for medical devices.1–3 Polymer microarrays allow screening of many hundreds of materials in parallel to identify a desired biological response with the ability to discover novel materials without the need to fully understand the cell–material interactions.4 However, this approach has only been applied to flat, or two-dimensional (2D) materials, and it is well recognized that cells can behave differently on three-dimensional (3D) or topographically patterned structures, for example by contact guidance.5 Consequently, the materials discovery procedures need to be expanded into the third dimension; from flat microarrays to spheres, possibly the simplest 3D shape. Particles have wide applicability for use in a range of areas including drug delivery,6,7 tissue engineering,8,9 diagnostics10 and photonics.11 They can be produced by many techniques, with the most commonly used for polymers being precipitation,12 emulsion polymerization13 and most recently, microfluidic based methods.14,15 Droplet formation in microfluidic devices is capable of producing highly uniform droplets ranging from the nano- to the microscale from a variety of materials.16,17 Particle control has been demonstrated by Kim et al. to be feasible with microfluidics, ranging from control of shape to compartmentalization of different components.14 This technique has been shown to be suitable for manufacturing large quantities of particles,18 and has the potential to generate particles of diverse compositions when coupled with automated fluid handling and a library of UV polymerizable monomers. A drawback that is shared among many droplet generation methods is that surfactants are often required in order to stabilize the fluid–fluid interfaces of the droplets.19,20 Salman et al. have recently reported that poly(vinyl alcohol-co-vinyl acetate) (PVA) and a mixture of PVA with a biosurfactant reduces the formation of Staphylococcus aureus and Pseudomonas aeruginosa biofilms by up to 98% on glass and plastic plates, depending on the molecular weight of PVA used.21Novel materials that can control bacterial attachment and biofilm formation are in demand to reduce device-centered infections in the fight against antimicrobial resistance (AMR).22 Moreover, the adhesion of marine fouling organisms to surfaces such as ships' hulls poses a worldwide problem and is subject to on-going research.23,24 Microorganisms attach to surfaces and form biofilms i.e. populations of microbial cells enmeshed in a matrix consisting of extracellular polymers such as polysaccharides, proteins and nucleic acids. In such bacterial communities, bacteria are up to 1000 times more tolerant to antimicrobials and host defenses than individual planktonic bacterial cells.25 Addressing this at the earliest possible stage ideally requires that surfaces are engineered in such a way that they can prevent bacterial attachment and subsequent biofilm formation.
In this paper, the production of monodisperse microparticles with an average diameter of 76 ± 1.7 μm was demonstrated using a simple microfluidic T-junction configuration by photopolymerizing acrylate based monomer solutions. First, we show the need for a surfactant, in this case 2 wt% PVA, for successful droplet formation. Then, the surface chemistry of the fabricated particles was assessed using Time-of-Flight secondary ion mass spectrometry (ToF-SIMS). The effect of PVA on the attachment of P. aeruginosa was investigated on acrylate-based microparticles using washing cycles to control the amount of PVA.
Results and discussion
Formation of microparticles in a T-junction
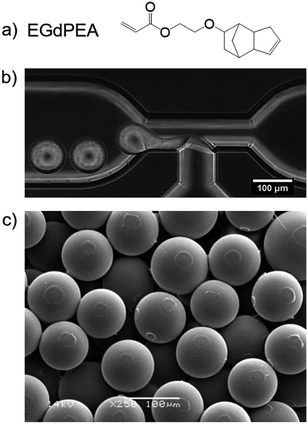
Microparticles were successfully prepared from the monomer ethylene glycol dicyclopentenyl ether acrylate (EGdPEA) in a microfluidic approach using a glass T-junction chip with a feature size of 100 μm. EGdPEA was selected since it has been identified as a ‘hit’ homopolymer exhibiting low surface area coverage when incubated with P. aeruginosa.2 The underlying mechanism by which the polyEGdPEA resists biofilm formation has still not been fully elucidated and is part of ongoing research. The chemical structure of the EGdPEA monomer is depicted in Fig. 1(a). The formation of monomer droplets in the glass T-junction is shown in Fig. 1(b). The continuous phase emerging from the right channel is an aqueous poly(vinyl alcohol-co-vinyl acetate) (PVA) solution (2 wt%), while the dispersed phase coming from the bottom is the respective monomer solution, in this case comprising of EGdPEA and 1 wt% of 2,2-dimethoxy-2-phenylacetophenone (DMPA) as photo-initiator for the subsequent polymerization of the monomer droplets to microparticles.PVA is able to stabilize the monomer solution droplets within the deionized water due to its amphiphilicity, hence the surfactant was required for successful droplet formation. Without a surfactant in the continuous phase, a constant stream of monomer solution was observed in the microfluidic channel. Moreover, employing a PVA concentration below 2 wt% failed to produce droplets or led to unstable droplet formation which resulted in a lack of both control and particle homogeneity. No data was gathered for a PVA concentration above 2 wt% because the scope was to reduce the amount of surfactant as much as possible.
Using the methodology outlined, homogeneous microparticles with a coefficient of variation of 2.2% were obtained. Fig. 1(c) shows a micrograph of polyEGdPEA microparticles with an average diameter of 76 μm imaged by SEM. The circular surface features arise from particle to particle contact prior to full hardening in the collection vial.
Assessment of surface chemistry using ToF-SIMS
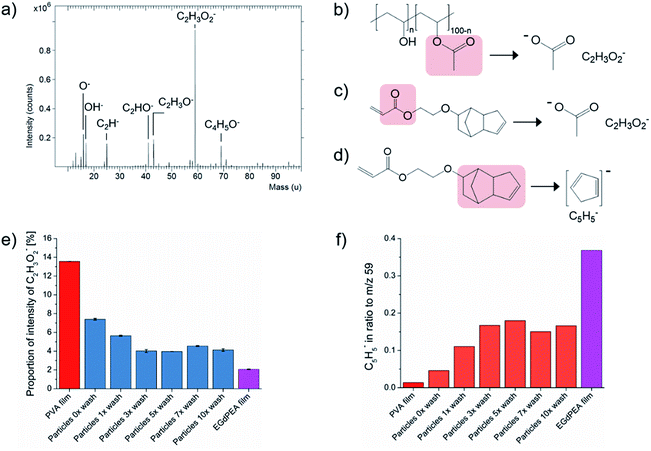
The surface chemistry of polyEGdPEA particles produced via microfluidics was assessed using ToF-SIMS. This surface mass spectral technique is not inherently quantitative, but is good at providing a relative measure of the amount of a particular species in a similar sample down to sub-monolayer sensitivities by virtue of its 3 nm depth of analysis.26 To study the presence of PVA surfactant on the polymer particles, aliquots of particles were washed with varying the numbers of wash cycles. A wash cycle involved sonicating the particles in isopropanol for 5 min followed by another sonication in distilled water for 5 min. SIMS spectra of PVA powder as-received (Fig. 2(a)) and a flat thin PVA-free film made of polyEGdPEA were acquired as references for PVA and the particle polymer itself. The secondary ions that are most dominant for detecting PVA arose from the unhydrolyzed vinyl acetate component within the 88% hydrolyzed PVA (structure in Fig. 2(b)) used as particle surfactant and control in this study. The peak detected at m/z = 59 in the negative SIMS spectrum represents the acetate anion [CH3COO]−. Fragmentation of the PVA into the acetate anion is illustrated in Fig. 2(b). In the positive ion spectrum, the base peak was found at m/z = 43 corresponding to the acetyl cation [CH3CO]+ which had previously been reported by Scholes et al. as a diagnostic peak for PVA.27 Both peaks, however, cannot only be attributed to ion fragments from the PVA surfactant but also to secondary ions derived from the polymer itself. This was confirmed by the flat polyEGdPEA film revealing peaks at the same m/z in both secondary ion polarities as shown to be representative for the PVA surfactant (Fig. 2(c)). The proportion of total counts of the acetate anion are plotted for the polyEGdPEA microparticles against the wash cycles and its corresponding flat thin film in Fig. 2(e).A reduction in intensity of the acetate anion was observed with increased washing as shown in Fig. 2(e). A gradual decrease of 45.7% was quantified from particles as-produced (particles 0× wash) to particles analyzed after the third wash cycle. The normalized counts (%) of the PVA characteristic secondary ion remained reasonably constant without any significant changes from the 3rd to the 10th wash cycle. Increased values, however, were obtained on microparticles after the 7th wash. One of the possible reasons is that particles, which were initially agglomerated during washing, became separated in the sample preparation for the ToF-SIMS analysis. Consequently, not all particles were washed with the same efficiency leaving a higher amount of PVA surfactant behind. The same observations were made in the positive ion SIMS spectra when considering the acetyl cation.
A secondary ion representative of the EGdPEA polymer, C5H5−, was selected to verify the increase in the polymer concentration with decreased amount of PVA surfactant at the surfaces. The fragmentation of the EGdPEA into the cyclopentadienyl anion (C5H5−) is presented in Fig. 2(d). An increase in the ratio of the cyclopentadienyl anion to the acetate anion of 264.2% was observed from particles as-produced (particles 0× wash) to particles after the 3rd wash cycle (Fig. 2(f)). The increase of the anion intensity characteristic for the EGdPEA fragment was inversely proportional to the decrease in intensity found for the PVA compound, confirming that the PVA surfactant was removed from the particle surfaces. However, the intensity of the cyclopentadienyl anion after extensive washing did not reach the same intensity that was observed in a prepared film known to be high purity EGdPEA, suggesting that not all the PVA was removed by washing. Consequently, it was hypothesized that most of the PVA surfactant was physically entangled in the polymeric particle surfaces preventing its complete removal.
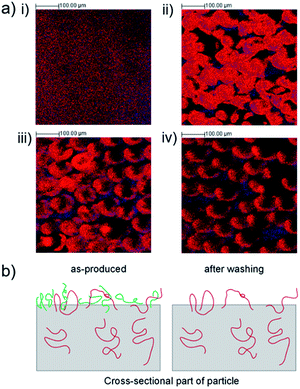
Applying the characteristic ions, C2H3O2− and C5H5−, the ToF-SIMS spectra were reconstructed as images to examine the distribution of PVA surfactant and EGdPEA on the surfaces. The overlay of PVA surfactant (red) and EGdPEA (blue) is shown for the thin film (i), particles as-produced (ii), after the first wash (iii) and after 10 washes (iv) in Fig. 3(a). The particle shape became distorted in the SIMS imaging process as a result of the particle shape and primary ion beam configuration.28 The flat thin film revealed a homogeneous distribution of the two diagnostic ions across the surface. In contrast, the untreated EGdPEA particles (no washing) were almost fully covered by the PVA surfactant such that the proportion of the characteristic ion C5H5− to the PVA was only 0.05. PolyEGdPEA appeared only on a few spots in the image (Fig. 3(a)(ii)) indicating that the PVA surfactant might cover the surfaces as a discontinuous layer as previously reported by Rafati et al.19 These images, however, cannot be resolved in such a detail to confirm this possibility. Comparing the ToF-SIMS images of the microparticles as-produced (ii) with the image of the extensively washed particles (iv), a significant reduction in PVA was observed. Therefore, the reduction in PVA surfactant at the particle surfaces resulted in an increased detection of the cyclopentadienyl anion derived from the EGdPEA polymer (Fig. S4(b)†).
The distribution of PVA surfactant was further analyzed on microparticles after the first wash because microfluidic produced particles were washed once with phosphate buffer saline (PBS) before being tested for bacterial attachment. As shown in Fig. 3(a)(iii), the concentration of PVA surfactant was not uniform within a sample aliquot.
A model proposing the PVA chain entanglement is illustrated in Fig. 3(b). Microparticles as-produced reveal both polymeric PVA chains physically entangled (red) and loosely bound (green) to the particle surface. Washing the particles excessively leads to removal of the loosely attached PVA chains leaving the physically entangled PVA chains behind.
Effect of PVA on bacterial attachment on washed microparticles
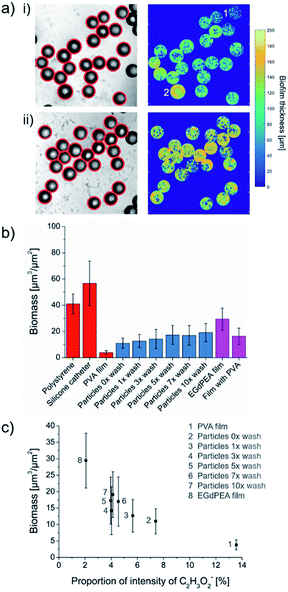
To study the effect of residual PVA surfactant on the ability of the particle to resist bacterial attachment, 5 mg each of microparticle sample varying in the number of wash cycles were cultured with P. aeruginosa PAO1-N expressing the mCherry fluorescent protein for 24 h in 48-well microtitre plates with agitation (shaking at 60 rpm) and imaged using confocal microscopy. The number of bacterial cells attached was quantified using the mCherry fluorescence, exclusively on the surfaces of microparticles and flat films by using a custom MATLAB algorithm (details in ESI 1.7†). Thin films, made by pipetting the EGdPEA monomer into microtitre well plates, were photopolymerized in the presence of a 2 wt% aqueous PVA solution. Curing the acrylate in the presence of PVA was conducted to mimic the microfluidic process of crosslinking PVA surfactant into particle surfaces as confirmed by ToF-SIMS analysis. The bacterial attachment quantification method allowed assessment of the variance of biomass within a sample. The successful particle detection on optical brightfield images and the captured biomass, resulting from applying the mask throughout the volumetric images, is depicted in Fig. 4(a) for microfluidic produced polyEGdPEA particles as-produced (i) and after 10 wash cycles (ii); wash cycles were performed before incubation in medium inoculated with PAO1-N mCherry. The biofilm thickness detected across the particle surfaces varied which is best seen in Fig. 4(a)(i). The particle numbered 2 exhibited considerably more bacterial cells attached compared to the particle numbered 1. These differences in biomass volume may have developed due to variations in PVA surface coverage (Fig. 3(a)) or due to heterogeneity of colonization across microparticles within a test aliquot. Moreover, alterations in surface topography were proposed by Feng et al.29 to lead to different levels of bacterial attachment. This study revealed that bacterial attachment was reduced on anodic alumina surfaces with cylindrical nanopores with diameters of 15 and 25 nm as compared with surfaces with larger pores. The bacteria-repelling effect of the topographies was observed across several bacterial species and attributed to densely distributed pores exerting increased repulsive forces on the bacteria found in the proximity of the surfaces.29 In contrast to Feng et al., the microfluidic produced polyEGdPEA particles exhibited mostly detachment marks and only few surface pores on some particles, as shown in the Fig. S5.†The overall biomass for each sample is shown in Fig. 4(b) including comparison samples of high bacterial attachment surfaces: non-tissue culture polystyrene (bottom of well) and sections of a silicone catheter. A significantly lower bacterial attachment (90.7% reduction) was quantified on PVA compared with polystyrene. The PVA sample consisted of 5 mg of loose PVA powder in a microtitre plate well that dissolved during sample washing and testing. This observation revealed that the low-fouling properties of the well surfaces covered with dissolved bulk PVA were similar to that reported by Salman et al. on PVA films (5 wt%) cast on glass and plastic plates.21 For the microfluidic produced polyEGdPEA microparticles, a gradual increase in the bacterial cells attached was observed from zero up to three particle pre-wash cycles. However, the level of bacterial attachment remained unaltered after performing five wash cycles. The number of bacterial cells attached was inversely proportional to the amount of PVA surfactant determined on the particle surfaces using ToF-SIMS (shown in Fig. 2(c)). Consequently, the reduction in PA01-N mCherry attachment observed with higher PVA surfactant concentration at the particle surfaces confirmed an anti-attachment effect of the surfactant. Furthermore, these findings were substantiated when considering the reduction in bacterial attachment of 44.6% on the PVA embedded polyEGdPEA films compared to the control films without PVA.
The relationship between bacterial attachment and PVA concentration is shown by plotting the biomass captured on PVA, polyEGdPEA particles and films against the proportion of total counts of the ToF-SIMS acetate ion representative for PVA (Fig. 4(c)). The graph demonstrates that having more PVA on the surface resulted in less biomass. The number of bacterial cells quantified on polyEGdPEA particles and films was higher than the amount determined on bulk PVA. This could be explained by two possible reasons: first, differences in the low-attachment mechanism for PVA bulk samples and PVA surfactant physically entrapped into polymerized surfaces; second, differences in the surface area of PVA exposed to bacterial cells. It was postulated that PVA in the bulk works by dissolution of the PVA followed by sloughing off the bacterial attachment from the weak interfacial layer. However, in the case of the rinsed samples exhibiting PVA surfactant which cannot be fully removed from the particle surfaces, this proposed mechanism of action for the bulk PVA would not be applicable. PVA physically entangled into the surface (surfactant) is considered as a water-soluble polymer that can induce surface hydration via hydrogen bonding. A tightly bound layer of water molecules near the surface is known to form a physical and energetic barrier able to prevent stable bacterial attachment.30 It is believed that PVA coatings to prevent bacterial attachment would be a short-lived strategy since it is likely to be depleted over time.
Conclusions
In this study, polyEGdPEA microparticles were successfully produced with average sizes of 76 ± 1.7 μm by combining microfluidics with photopolymerization of an acrylate monomer. It was demonstrated that surfactant was needed to achieve stable droplet formation, in this work 2 wt% aqueous PVA solution. ToF-SIMS analysis revealed the presence of PVA surfactant on the particle surfaces, which was partially removed through excessive washing (45.7% reduction). A chain entanglement model was proposed to explain the residual PVA. A bulk PVA sample had 91% less P. aeruginosa cell coverage compared to polystyrene. By testing microparticles varying in repeats of wash cycles, a trend was observed which revealed greater reduction in bacterial attachment for particles with increased amount of retained PVA surfactant at particle surfaces. The finding of this work emphasizes the role of surfactants in controlling cellular response to materials in this system, and the importance of carrying out chemical surface analysis to detect and rationalize the role of such confounding species. This insight will help improve the understanding of the structure–function relationships and hence, facilitate to move towards rational materials design with biofilm control in the future.Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by the Japan Society for the Promotion of Science London [grant number PE15777]; and the Engineering and Physical Sciences Research Council [grant number EP/N006615/1]. The authors kindly acknowledge Associate Professor Takasi Nisisako and Professor Takeshi Hatsuzawa for enabling collaboration with the Tokyo Institute of Technology and the University of Nottingham from which the expertise in microfluidics methodology originated. Furthermore, the authors would like to acknowledge David Scurr for his assistance with ToF-SIMS measurements. Martyn Davies kindly highlighted the importance to us of surface based PVA.References
- A. D. Celiz, J. G. W. Smith, A. K. Patel, R. Langer, D. G. Anderson, D. A. Barrett, L. E. Young, M. C. Davies, C. Denning and M. R. Alexander, Biomater. Sci., 2014, 2, 1604–1611 RSC.
- A. L. Hook, C.-Y. Chang, J. Yang, J. Luckett, A. Cockayne, S. Atkinson, Y. Mei, R. Bayston, D. J. Irvine, R. Langer, D. G. Anderson, P. Williams, M. C. Davies and M. R. Alexander, Nat. Biotechnol., 2012, 30, 868–875 CrossRef CAS PubMed.
- S. Brocchini, K. James, V. Tangpasuthadol and J. Kohn, J. Biomed. Mater. Res., 1998, 42, 66–75 CrossRef CAS PubMed.
- M. C. Davies, M. R. Alexander, A. L. Hook, J. Yang, Y. Mei, M. Taylor, A. J. Urquhart, R. Langer and D. G. Anderson, J. Drug Targeting, 2010, 18, 741–751 CrossRef CAS PubMed.
- A. Curtis and C. Wilkinson, Biomaterials, 1997, 18, 1573–1583 CrossRef CAS PubMed.
- S. Iwanaga, N. Saito, H. Sanae and M. Nakamura, Colloids Surf., B, 2013, 109, 301–306 CrossRef CAS PubMed.
- D. J. Siegwart, K. a Whitehead, L. Nuhn, G. Sahay, H. Cheng, S. Jiang, M. Ma, A. Lytton-Jean, A. Vegas, P. Fenton, C. G. Levins, K. T. Love, H. Lee, C. Cortez, S. P. Collins, Y. F. Li, J. Jang, W. Querbes, C. Zurenko, T. Novobrantseva, R. Langer and D. G. Anderson, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 12996–13001 CrossRef CAS PubMed.
- M. Ma, A. Chiu, G. Sahay, J. C. Doloff, N. Dholakia, R. Thakrar, J. Cohen, A. Vegas, D. Chen, K. M. Bratlie, T. Dang, R. L. York, J. Hollister-Lock, G. C. Weir and D. G. Anderson, Adv. Healthcare Mater., 2013, 2, 667–672 CrossRef CAS PubMed.
- D. Y. S. Chau, K. Agashi and K. M. Shakesheff, Mater. Sci. Technol., 2008, 24, 1031–1044 CrossRef CAS.
- K. Ferrara, R. Pollard and M. Borden, Annu. Rev. Biomed. Eng., 2007, 9, 415–447 CrossRef CAS PubMed.
- D. Dendukuri and P. S. Doyle, Adv. Mater., 2009, 21, 4071–4086 CrossRef CAS.
- G. L. Li, H. Möhwald and D. G. Shchukin, Chem. Soc. Rev., 2013, 42, 3628–3646 RSC.
- V. R. Sinha and A. Trehan, J. Controlled Release, 2003, 90, 261–280 CrossRef CAS PubMed.
- J. H. Kim, T. Y. Jeon, T. M. Choi, T. S. Shim, S.-H. Kim and S.-M. Yang, Langmuir, 2014, 30, 1473–1488 CrossRef CAS PubMed.
- T. Nisisako and T. Hatsuzawa, Microfluid. Nanofluid., 2010, 9, 427–437 CrossRef.
- D. Baah and T. Floyd-Smith, Microfluid. Nanofluid., 2014, 17, 431–455 CrossRef CAS.
- R. Vasiliauskas, D. Liu, S. Cito, H. Zhang, M.-A. Shahbazi, T. Sikanen, L. Mazutis and H. a. Santos, ACS Appl. Mater. Interfaces, 2015, 7, 14822–14832 CAS.
- T. Nisisako and T. Torii, Lab Chip, 2008, 8, 287–293 RSC.
- A. Rafati, A. Boussahel, K. M. Shakesheff, A. G. Shard, C. J. Roberts, X. Chen, D. J. Scurr, S. Rigby-singleton, P. Whiteside, M. R. Alexander and M. C. Davies, J. Controlled Release, 2012, 162, 321–329 CrossRef CAS PubMed.
- S. L. Anna, N. Bontoux and H. A. Stone, Appl. Phys. Lett., 2003, 82, 364–366 CrossRef CAS.
- J. A. S. Salman, M. F. H. Kadhemy, M. S. Jaleel and A. K. Abdal, Int. J. Curr. Microbiol. Appl. Sci., 2014, 3, 301–309 CAS.
- J. O'Neill, Comm. by UK Prime Minist.
- M. E. Callow and J. A. Callow, Biologist, 2002, 49, 10–14 Search PubMed.
- S. Cao, J. Wang, H. Chen and D. Chen, Chin. Sci. Bull., 2011, 56, 598–612 CrossRef CAS.
- R. D. Monds and G. A. O'Toole, Trends Microbiol., 2009, 17, 73–87 CrossRef CAS PubMed.
- R. Ogaki, F. M. Green, I. S. Gilmore, A. G. Shard, S. Luk, M. R. Alexander and M. C. Davies, Surf. Interface Anal., 2007, 39, 852–859 CrossRef CAS.
- P. D. Scholes, A. G. A. Coombes, L. Illum, S. S. Davis, J. F. Watts, C. Ustariz, M. Vert and M. C. Davies, J. Controlled Release, 1999, 59, 261–278 CrossRef CAS PubMed.
- J. C. Vickerman and D. Briggs, TOF-SIMS: Materials Analysis by Mass Spectrometry, IM Publications, 2nd edn, 2013 Search PubMed.
- G. Feng, Y. Cheng, S.-Y. Wang, D. A. Borca-Tasciuc, R. W. Worobo and C. I. Moraru, NPJ Biofilms Microbiomes, 2015, 1 DOI:10.1038/npjbiofilms.2015.22.
- S. Chen, L. Li, C. Zhao and J. Zheng, Polymer (Guildf), 2010, 51, 5283–5293 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: The Supporting information include the experimental details and some additional results. In the experimental details, the following subjects are discussed: preparation of polymer microparticles, flat thin film formation, scanning electron microscopy (SEM), ToF-SIMS, bacterial attachment studies, confocal microscope image acquisition and confocal image processing. Movies are uploaded for the successful, using 2 wt% PVA solution (mp4, Fig. 1(b)), and for the unsuccessful droplet formation without the addition of PVA (mp4). See DOI: 10.1039/c8ra01491d |
| This journal is © The Royal Society of Chemistry 2018 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) with a hydrolysis degree of 88% was used as particle surfactant and control.
000) with a hydrolysis degree of 88% was used as particle surfactant and control.