Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Adsorption of tetrakis(4-sulfophenyl)porphyrin onto liposomal surfaces composed of neutral diacylphosphatidylcholine and release by cyclodextrin†
Yuki Tsuchiya,
Toshimi Nakaya,
Tomoyuki Kakigi,
Kouta Sugikawa and
Atsushi Ikeda
and
Atsushi Ikeda *
*
Department of Applied Chemistry, Graduate School of Engineering, Hiroshima University, 1-4-1 Kagamiyama, Higashi-Hiroshima 739-8527, Japan
First published on 27th March 2018
Abstract
Anionic tetrakis(4-sulfophenyl)porphyrin (TPPS) interacts with liposomal surfaces composed of neutral diacylphosphatidylcholine at high lipid concentrations. TPPS interacted with liposomal surfaces through four contact points. The association constant was obtained to be 9.0 × 105 M−4. TPPS was peeled off the liposomal surfaces by the addition of cyclodextrin.
Introduction
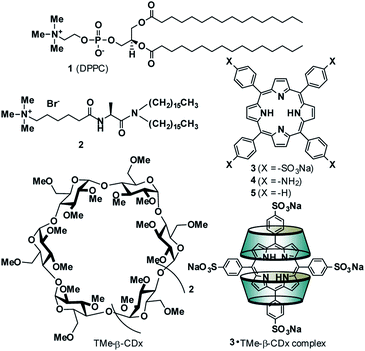
The adsorption of compounds such as polymers and nanoparticles onto the cell surface is important in developing drug carriers1–5 and functional materials.6–12 Recently, several groups reported that polyelectrolytic biomacromolecules, such as DNA or nanoparticles with anionic surfaces, were able to adsorb onto liposomal surfaces composed of neutral diacylphosphatidylcholine (PC).1–12 Although the details of the interactions remain unclear, multipoint interactions might exist between the negative charges of these compounds or materials and the positive charge N+ of the P−–N+ (phosphorous–nitrogen) dipole of PC.13,14 Recently, we showed that 5,10,15,20-tetrakis(4-sulfophenyl)porphyrin (3) (Fig. 1) interacts with the liposomal surface by formation of one-dimensional self-assembled structures (J-aggregates) under acidic conditions.15,16 In contrast, neutralization of the solution deformed the porphyrin J-aggregates, leading to release of 3 from the liposomal surface.15,16 These interactions are important not only for development of novel functional materials by using liposomes, but also for internalization into cells by endocytosis as the first step of intracellular uptake. In this report, 3 was found to interact with liposomal surfaces composed of neutral lipids at high liposome concentrations without the formation of porphyrin J-aggregates (Scheme 1A). Furthermore, 3 was released from the liposomal surfaces by addition of trimethyl-β-cyclodextrin (TMe-β-CDx) (Fig. 1, Scheme 1B).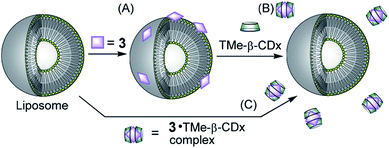 | ||
| Scheme 1 Schematic illustrations of (A) adsorption of 3 on the liposomal surface, (B) exfoliation of 3 by TMe-β-CDx and (C) no interaction between the 3·TMe-β-CDx complex and a liposome. | ||
Results and discussion
Interaction between anionic porphyrin and neutral lipid
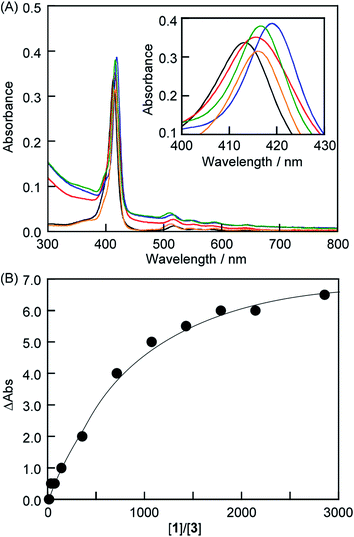
The interactions between anionic porphyrin 3 and the neutral lipid 1 [1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), Fig. 1] were investigated. Concentration dependent UV-vis absorption spectra of 3 were measured by the addition of liposomes composed of 1 (liposomes-1) (Fig. 2) and the red shift in the Soret band was observed at 20 °C (413 → 415.5 nm, ΔAbs = 2.5 nm). The red shift was not because of the formation of one-dimensional self-assembled structures because: (i) 3 cannot form self-aggregates under neutral conditions because protonation of 3 is essential for self-association, and (ii) the shift value is too small for self-aggregates because the Soret band of the porphyrin J-aggregates displayed a significant red shift to 491 nm.15–18 If sulfo groups of 3 interact with ammonio groups of 1 on the liposomal surface, the addition of cationic lipids in the liposome should facilitate the formation of strong interactions between anionic 3 and the liposomal surface via electrostatic interactions.19–21 Therefore, a cationic lipid (2) was mixed with lipid 1 in the liposomes {[1]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [2] = 7
[2] = 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 (mol mol−1)}. As shown in Fig. 2A (blue line), the λmax of 3 (419 nm) was red-shifted by 6 nm when compared with the results obtained by 3 alone in the absence of liposomes [Fig. 2A (black line)]. Therefore, the red shift suggests an interaction between 3 and lipid 1. The shifts of Δλmax in the Soret band of 3 were plotted against the concentration of 1 in Fig. 2B. We were not able to determine the association constant between 3 and lipid 1 from a curve in Fig. 2B. The association constant determined from 1H NMR spectrum is described below. In contrast, we investigated interactions between cationic porphyrin 4 (Fig. 1) and lipid 1. Although the absorbance of 4 increased because of the light scattering of liposomes-1, no shift of Δλmax in the Soret band of 4 was observed (Fig. S1†), indicating that 4 did not interact with the liposomal surface.
3 (mol mol−1)}. As shown in Fig. 2A (blue line), the λmax of 3 (419 nm) was red-shifted by 6 nm when compared with the results obtained by 3 alone in the absence of liposomes [Fig. 2A (black line)]. Therefore, the red shift suggests an interaction between 3 and lipid 1. The shifts of Δλmax in the Soret band of 3 were plotted against the concentration of 1 in Fig. 2B. We were not able to determine the association constant between 3 and lipid 1 from a curve in Fig. 2B. The association constant determined from 1H NMR spectrum is described below. In contrast, we investigated interactions between cationic porphyrin 4 (Fig. 1) and lipid 1. Although the absorbance of 4 increased because of the light scattering of liposomes-1, no shift of Δλmax in the Soret band of 4 was observed (Fig. S1†), indicating that 4 did not interact with the liposomal surface.
Existence of porphyrin on the liposomal surface
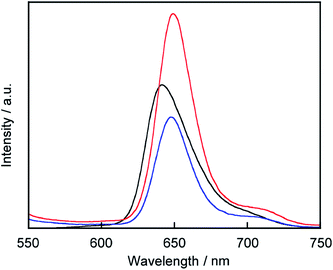
C60 is known to act as a quencher in liposomes.22 To confirm that 3 exists on the liposomal surface, we measured fluorescence quenching of 3 by C60 in lipid-membrane-incorporated C60 (LMIC60, Fig. 3). The presence of C60 led to a fluorescence quenching of 48%, indicating that 3 exists in the neighborhood of C60.Form of porphyrin on the liposomal surface
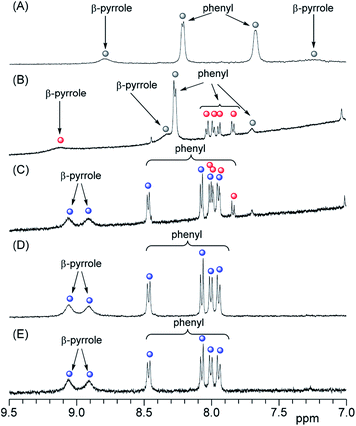
In the 1H NMR spectra, 3 gives rise to two peaks arising from the phenyl protons in the ortho and meta positions because of the D4h symmetry of 3 (Fig. 4A and S2A†). Although the peak assigned to the phenyl protons in the meta position (8.2 ppm) appeared as a sharp doublet, the corresponding peak for the protons in the ortho position (7.7 ppm) was broader and this line-broadening was ascribed to self-aggregation of 3.23,24 In Fig. 4B, the phenyl protons in the ortho positions of 3 were observed as a pronounced broaden peak (grey circle).23 In contrast, when 3 interacted with liposome-1, two pairs of phenyl protons in the ortho and meta positions appeared in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
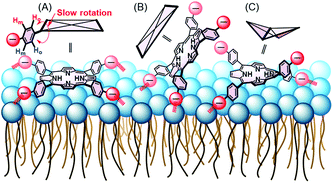
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio [Fig. 4B (red circles) and S2B†]. The result suggests the following three models for the interaction between 1 and 3: C4v, C2v and C2v symmetries, depending on the interaction on the liposomal surface (Scheme 2). In Scheme 2A, 3 interacts with liposomal surfaces by four point interactions. The porphyrin face of 3 is shaped asymmetrical at the upper and lower sides. If the rotation of phenyl units decreases and is slower than the NMR time scale because of steric hindrance by the liposome surface, the two ortho and two meta protons in one phenyl unit are in different chemical environments (see Scheme 2A) and give rise to the four signals in Fig. 4B (red circles). For the other possible models, 3 interacts with the liposomal surface by only two SO3− in the cis-position with C2v (Scheme 2B) or the trans-position with C2v (Scheme 2C), in which 3 interacts with the N+ of P−–N+ in liposome-1. Because 3 has different symmetries in the three models, the β-pyrrole protons should appear as one, four or two sets of peaks in the NMR spectrum for models in Schemes 2A, B and C, respectively. As shown in Fig. 4B (red circle), the β-pyrrole protons appeared as a single broad peak, suggesting that the model in Scheme 2A is correct.
1 ratio [Fig. 4B (red circles) and S2B†]. The result suggests the following three models for the interaction between 1 and 3: C4v, C2v and C2v symmetries, depending on the interaction on the liposomal surface (Scheme 2). In Scheme 2A, 3 interacts with liposomal surfaces by four point interactions. The porphyrin face of 3 is shaped asymmetrical at the upper and lower sides. If the rotation of phenyl units decreases and is slower than the NMR time scale because of steric hindrance by the liposome surface, the two ortho and two meta protons in one phenyl unit are in different chemical environments (see Scheme 2A) and give rise to the four signals in Fig. 4B (red circles). For the other possible models, 3 interacts with the liposomal surface by only two SO3− in the cis-position with C2v (Scheme 2B) or the trans-position with C2v (Scheme 2C), in which 3 interacts with the N+ of P−–N+ in liposome-1. Because 3 has different symmetries in the three models, the β-pyrrole protons should appear as one, four or two sets of peaks in the NMR spectrum for models in Schemes 2A, B and C, respectively. As shown in Fig. 4B (red circle), the β-pyrrole protons appeared as a single broad peak, suggesting that the model in Scheme 2A is correct.
Association constant between anionic porphyrin and neutral lipid
As shown in Scheme 2A, porphyrin 3 interacted with the liposome surface consisted of lipids 1 through four point interactions. Therefore, the equilibrium is defined as (1):| 3 + 4·1 ⇌ 3 − 4·1 | (1) |
The concentrations of free 3 and 3 on the liposomal surface were determined by the peak intensities in Fig. 4B. When these values were substituted into eqn (2), we obtained the association constant (Ka) = 9.0 × 105 M−4.
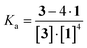 | (2) |
Porphyrin derivatives can form a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 complex with TMe-β-CDx.24–26 Furthermore, all of the tetraphenylporphyrin (5) were released from the TMe-β-CDx cavities and transferred to the lipid membrane after mixing the 5·TMe-β-CDx complex with liposome-1 at 30 °C for 1 h.25 Therefore, we attempted the exchange reaction of 3 from TMe-β-CDx cavities to lipid membranes or the formation of direct interactions between liposome surfaces and the 3·TMe-β-CDx complex. After mixing the 3·TMe-β-CDx complex with liposome-1 under the same conditions reported previously,25 all peaks assignable to TMe-β-CDx in the 3·TMe-β-CDx complex remained in the 1H NMR spectrum (blue circles in Fig. 4D, E, S2D and E†). The result shows that porphyrin 3 did not transfer from the TMe-β-CDx cavities to the liposomes and the porphyrin remained in the TMe-β-CDx cavities (Scheme 1C). This observation suggests that 3 is too hydrophilic to incorporate into the hydrophobic lipid membrane. Furthermore, no chemical shift changes of these peaks indicate that the 3·TMe-β-CDx complex did not interact with the liposome surface by only a single point interaction because of steric hindrance by the two TMe-β-CDxs. Because Ka for the four point interactions was 9.0 × 105 M−4, Ka' per a single point interaction was estimated to be approximately 30 M−1 at most even if entropy and enthalpy gains by multi point interactions were excluded. Therefore, the 3·TMe-β-CDx complex cannot interact with the liposome surface by only a single point interaction. Consequently, the interactions between 3 and 1 need at least four point interactions.
2 complex with TMe-β-CDx.24–26 Furthermore, all of the tetraphenylporphyrin (5) were released from the TMe-β-CDx cavities and transferred to the lipid membrane after mixing the 5·TMe-β-CDx complex with liposome-1 at 30 °C for 1 h.25 Therefore, we attempted the exchange reaction of 3 from TMe-β-CDx cavities to lipid membranes or the formation of direct interactions between liposome surfaces and the 3·TMe-β-CDx complex. After mixing the 3·TMe-β-CDx complex with liposome-1 under the same conditions reported previously,25 all peaks assignable to TMe-β-CDx in the 3·TMe-β-CDx complex remained in the 1H NMR spectrum (blue circles in Fig. 4D, E, S2D and E†). The result shows that porphyrin 3 did not transfer from the TMe-β-CDx cavities to the liposomes and the porphyrin remained in the TMe-β-CDx cavities (Scheme 1C). This observation suggests that 3 is too hydrophilic to incorporate into the hydrophobic lipid membrane. Furthermore, no chemical shift changes of these peaks indicate that the 3·TMe-β-CDx complex did not interact with the liposome surface by only a single point interaction because of steric hindrance by the two TMe-β-CDxs. Because Ka for the four point interactions was 9.0 × 105 M−4, Ka' per a single point interaction was estimated to be approximately 30 M−1 at most even if entropy and enthalpy gains by multi point interactions were excluded. Therefore, the 3·TMe-β-CDx complex cannot interact with the liposome surface by only a single point interaction. Consequently, the interactions between 3 and 1 need at least four point interactions.
Exfoliation of porphyrin from the liposome surface by cyclodextrin
To control the association–dissociation of 3 onto the liposomal surface, TMe-β-CDx was added to the mixture of 3 and liposome-1. After adding TMe-β-CDx, although the λmax of the Soret band of 3 is barely shifted (i.e., 415 to 416 nm), the λmax in the Q-band of 3 is shifted from 517 to 511 nm. These λmax were the same as that observed for the 3·TMe-β-CDx complex (416 and 511 nm) (Fig. 2A, green and orange). Furthermore, peaks were observed in the 1H NMR spectrum for the 3·TMe-β-CDx complex (Fig. 4C and S2C†). The results show that most of 3 peeled off the liposomal surface and had formed a complex with two TMe-β-CDxs (Scheme 1B).Conclusions
In summary, anionic 3 was adsorbed onto liposomal surfaces composed of neutral lipid 1 at high lipid concentrations. The interaction of 3 with 1 was via four contact points. In contrast, the 3·TMe-β-CDx complex cannot be adsorbed onto liposomal surfaces because 3 interacts with 1 by only a single point and is encapsulated by two TMe-β-CDxs. Addition of TMe-β-CDx caused the release of 3 from the liposomal surface and complex formation with two TMe-β-CDxs. Consequently, the association–dissociation of 3 onto the liposomal surfaces can be controlled by the addition of TMe-β-CDx.Experimental
Experimental materials
Trimethyl-β-cyclodextrin (TMe-β-CDx) and 5,10,15,20-tetrakis(4-sulfophenyl)porphyrin (3) were purchased from Wako Pure Chemical Industries Ltd (Tokyo, Japan) and Tokyo Chemical Industries Co., Ltd (Tokyo, Japan), respectively. 1,2-Dimyristoyl-sn-glycero-3-phosphocholine (DMPC, 1) was obtained from Funakoshi Co., Ltd (Tokyo, Japan). Compound 2 was prepared according to the method described previously.27Phosphate buffer
A phosphate buffer was prepared by dissolving a mixture of NaH2PO4·2H2O (390 mg, 2.50 mmol), Na2HPO4·12H2O (181 mg, 0.50 mmol) and K2SO4 (1.70 mg, 9.74 mmol) in pure water or D2O (100 mL), to reach a final pH of 6.5 at 23 °C.Preparation of liposome-1 and liposome-1–2
An appropriate amount of 1 or a mixture of 1 and 2 ([1]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [2] = 7
[2] = 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 mol mol−1) was dissolved in chloroform. The solvent was evaporated under a gentle stream of nitrogen, followed by a period under vacuum to remove any traces of solvent. The resulting thin lipid films were hydrated on the wall of the vial above the phase transition temperature with an appropriate amount of phosphate buffer. The hydrated materials were subjected to eight freeze–thaw cycles (−195 and 50 °C) to give unilamellar vesicles, which were extruded 11 times through 0.05 μm pores using a LiposoFast miniextruder from Avestin (Ottawa, Canada) above the phase transition temperature. The resulting liposomes were uniform in size with a diameter of approximately 80 nm. The final lipid concentration was 3.0 mM.
3 mol mol−1) was dissolved in chloroform. The solvent was evaporated under a gentle stream of nitrogen, followed by a period under vacuum to remove any traces of solvent. The resulting thin lipid films were hydrated on the wall of the vial above the phase transition temperature with an appropriate amount of phosphate buffer. The hydrated materials were subjected to eight freeze–thaw cycles (−195 and 50 °C) to give unilamellar vesicles, which were extruded 11 times through 0.05 μm pores using a LiposoFast miniextruder from Avestin (Ottawa, Canada) above the phase transition temperature. The resulting liposomes were uniform in size with a diameter of approximately 80 nm. The final lipid concentration was 3.0 mM.
Preparation of the 3·TMe-β-CDx complex
Compound 3 (5.00 mg, 5.26 × 10−6 mol) and TMe-β-CDx (15.0 mg, 1.05 × 10−5 mol) were placed in an agate capsule with two agate-mixing balls. The resulting mixture was agitated vigorously at 30 Hz for 20 min using a high-speed vibration mill (MM 200; Retsch Co., Ltd., Haan, Germany). The solid mixture was suspended in either phosphate buffer or D2O–phosphate buffer (1.5 mL) to produce a dark purple emulsion. Subsequent centrifugation (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, 25 °C, 20 min) removed non-dispersed 3 from the solution. The concentration of 3 in the 3·TMe-β-CDx complex was determined to be 0.03 mM by measuring the absorbance of the solution at λmax in water. The molar absorption coefficient for the water-soluble 3·TMe-β-CDx complex is ε416 = 3.79 × 105 dm3 mol−1 cm−1.
000 × g, 25 °C, 20 min) removed non-dispersed 3 from the solution. The concentration of 3 in the 3·TMe-β-CDx complex was determined to be 0.03 mM by measuring the absorbance of the solution at λmax in water. The molar absorption coefficient for the water-soluble 3·TMe-β-CDx complex is ε416 = 3.79 × 105 dm3 mol−1 cm−1.
Spectrophotometric assay
The absorbance spectra were scanned using a UV-2550 spectrophotometer (Shimadzu Corporation, Kyoto, Japan). Fluorescence spectra were obtained using an LS 55 luminescence spectrometer (Perkin-Elmer, Waltham, MA, USA). The excitation and emission wavelengths were 517 and 600–750 nm, respectively.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by a JSPS KAKENHI Grant-in-Aid for Scientific Research (B) (Grant No. JP16H04133) and a Grant-in-Aid for Challenging Exploratory Research (Grant No. JP16K13982). We thank the Edanz Group (http://www.edanzediting.com/ac) for editing a draft of this manuscript.Notes and references
- I. A. Khalil, K. Kogure, H. Akita and H. Harashima, Pharmacol. Rev., 2006, 58, 32–45 CrossRef CAS PubMed.
- F. Zhao, Y. Zhao, Y. Liu, X. Chang, C. Chen and Y. Zhao, Small, 2011, 7, 1322–1337 CrossRef CAS PubMed.
- L. Y. T. Chou, K. Ming and W. C. W. Chan, Chem. Soc. Rev., 2011, 40, 233–245 RSC.
- T. M. Allen and P. R. Cullis, Science, 2004, 303, 1818–1822 CrossRef CAS PubMed.
- S. Mayor and R. E. Pagano, Nat. Rev. Mol. Cell Biol., 2007, 8, 603–612 CrossRef CAS PubMed.
- A. Das, R. Thakur and A. Chakraborty, RSC Adv., 2013, 3, 19572–19581 RSC.
- F. Quemeneur, M. Rinaudo, G. Maret and B. Pepin-Donat, Soft Matter, 2010, 6, 4471–4481 RSC.
- P. Karam, A. A. Hariri, C. F. Calver, X. Zhao, K. S. Schanze and G. Cosa, Langmuir, 2014, 30, 10704–10711 CrossRef CAS PubMed.
- L. Zhang and S. Granick, Nano Lett., 2006, 6, 694–698 CrossRef CAS PubMed.
- R. Michel, T. Plostica, L. Abezgauz, D. Danino and M. Gradzielski, Soft Matter, 2013, 9, 4167–4177 RSC.
- F. Wang, D. E. Curry and J. Liu, Langmuir, 2015, 31, 13271–13274 CrossRef CAS PubMed.
- K. Sugikawa, T. Kadota, K. Yasuhara and A. Ikeda, Angew. Chem., Int. Ed., 2016, 55, 4059–4063 CrossRef CAS PubMed.
- C. Adhikari, A. Das and A. Chakraborty, ChemPhysChem, 2015, 16, 866–871 CrossRef CAS PubMed.
- T. Cha, A. Guo and X.-Y. Zhu, Biophys. J., 2006, 90, 1270–1274 CrossRef CAS PubMed.
- K. Sugikawa, Y. Takamatsu, K. Yasuhara and A. Ikeda, Langmuir, 2017, 33, 1023–1029 CrossRef CAS PubMed.
- K. Sugikawa, Y. Takamatsu, T. Kakigi, K. Yasuhara and A. Ikeda, Chem. Commun., 2017, 53, 10140–10143 RSC.
- O. Ohno, Y. Kaizu and H. Kobayashi, J. Chem. Phys., 1993, 99, 4128–4139 CrossRef CAS.
- D. L. Akins, H. Zhu and C. Guo, J. Phys. Chem., 1994, 98, 3612–3618 CrossRef CAS.
- E. Goormaghtigh, J. Caspers and J. M. Ruysschaert, J. Colloid Interface Sci., 1981, 80, 163–170 CrossRef CAS.
- Z. Wang, K. Yasuhara, H. Ito, M. Mukai and J. Kikuchi, Chem. Lett., 2010, 39, 54–55 CrossRef CAS.
- K. Hall, T.-H. Lee and M.-I. Aguilar, J. Mol. Recognit., 2011, 24, 108–118 CrossRef CAS PubMed.
- A. Ikeda, T. Sue, M. Akiyama, K. Fujioka, T. Shigematsu, Y. Doi, J. Kikuchi, T. Konishi and R. Nakajima, Org. Lett., 2008, 10, 4077–4080 CrossRef CAS PubMed.
- A. Corsini and O. Herrmann, Talanta, 1986, 33, 335–339 CrossRef CAS PubMed.
- K. Kano, R. Nishiyabu, T. Asada and Y. Kuroda, J. Am. Chem. Soc., 2002, 124, 9937–9944 CrossRef CAS PubMed.
- A. Ikeda, S. Hino, T. Mae, Y. Tsuchiya, K. Sugikawa, M. Tsukamoto, K. Yasuhara, H. Shigeto, H. Funabashi, A. Kuroda and M. Akiyama, RSC Adv., 2015, 5, 105279–105287 RSC.
- Y. Tsuchiya, T. Shiraki, T. Matsumoto, K. Sugikawa, K. Sada, A. Yamano and S. Shinkai, Chem.–Eur. J., 2012, 18, 456–465 CrossRef CAS PubMed.
- Y. Murakami, A. Nakano and H. Ikeda, J. Org. Chem., 1982, 47, 2137–2144 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures and UV-vis absorption and 1H NMR spectra. See DOI: 10.1039/c8ra01411f |
| This journal is © The Royal Society of Chemistry 2018 |