Open Access Article
Open Access ArticleAnalysis of binding properties of pathogens and toxins using multivalent glycan microarrays†
Hyoung Sub Kim,
Ji Young Hyun,
Seong-Hyun Park and
Injae Shin *
*
Center for Biofunctional Molecules, Department of Chemistry, Yonsei University, Seoul 03722, Republic of Korea. E-mail: injae@yonsei.ac.kr
First published on 19th April 2018
Abstract
Pathogens infect hosts often through initial binding of their cell surface lectins to glycans expressed on the exterior of host cells. Thus, methods to evaluate the glycan-binding properties of pathogens are of great importance. Because of the multivalent nature of interactions of pathogens with glycans, the ability to assess the glycan density-dependent binding of pathogens is particularly important. In this study, we developed a facile technique to construct multivalent carbohydrate microarrays through immobilization of unmodified glycans on multivalent hydrazide-derivatized glass surfaces. This immobilization strategy does not require the use of multivalent glycoconjugates, which are typically prepared by using multistep sequences. The results of analysis of microarray images, obtained after incubation of multivalent glycan microarrays with cholera toxin B and pathogens such as uropathogenic E. coli and H. pylori, show that the binding affinities of toxins and pathogens for glycans are highly glycan density-dependent. Specifically, toxins and pathogens bind to glycans more strongly as the valency of the glycans on the microarrays is increased from 1 to 4. It is anticipated that the newly developed immobilization method will be applicable to the preparation of multivalent carbohydrate microarrays that are employed to evaluate multivalent glycan binding properties of a variety of pathogens and toxins.
Introduction
The external surfaces of plasma membranes in mammalian cells are densely covered by diverse glycans in the form of glycoconjugates such as glycoproteins, glycosphingolipids and proteoglycans.1 The patterns of glycosylation on mammalian cell surfaces vary depending on cell and tissue types, and species. Bacterial and viral pathogens frequently use mammalian cell-surface glycans for infection of hosts. In this regard, host cell-surface glycans serve as recognition sites for pathogens that express cell-surface lectins (adhesins or hemagglutinins) which are used for attachment to, and colonization and infection of hosts.2,3 In addition, host cell-surface glycans act as ligands for toxins that are secreted by some pathogens, exemplified by Vibrio cholera.4 Therefore, the ability to assess the interactions of pathogens and toxins with glycans has great importance because it leads to deep insight into glycan-associated infections by pathogens and toxins and to the development of efficacious biomedical agents. In general, lectins bind to monovalent glycans weakly but can associate with multivalent counterparts with high affinities as a result of a cluster effect.5 Thus, methods to analyze glycan density-dependent binding of pathogens and toxins will facilitate our understanding of the nature and consequence of glycan-associated recognition by pathogens and toxins.Carbohydrate microarrays, which consist of various glycans densely and orderly immobilized on solid surfaces, have been widely employed for high-throughput analysis of glycan-associated recognition events.6–9 In addition, this microarray technology has been utilized for assessing substrate specificities of glycosyltransferases, determining protein-glycan interactions quantitatively, and identifying functional glycans.10,11 Furthermore, carbohydrate microarrays have been used to assess binding properties of whole viruses and bacteria.12 Also, these microarrays containing varying glycan densities have been utilized to study the glycan density-dependent binding properties of lectins.13 In spite of this high level of research activity, the microarray-based technology has not been employed to analyze density-dependent binding of bacteria and toxins to glycans. In the study described below, we developed a facile method to construct multivalent carbohydrate microarrays, possessing various glycans that have valencies ranging from 1 to 4. Moreover, these microarrays were employed to demonstrate that binding of bacterial pathogens and toxins is highly glycan density-dependent.
Results and discussion
Several types of carbohydrate microarrays, containing multivalent glycans such as neoglycoproteins/neoglycopeptides, glycodendrimers, peptide nucleic acid-conjugated glycans and DNA-linked glycans with varying glycan densities, have been created previously to evaluate glycan density-dependent binding of lectins.13 However, preparation of the multivalent glycoconjugates used to construct these microarrays require multistep sequences. To avoid the time-consuming and difficult synthesis of multivalent glycoconjugates, we took advantage of an earlier observation showing that unmodified mono-, di-, oligo- and polysaccharides are covalently and site-specifically immobilized on hydrazide-derivatized surfaces.14,15 Accordingly, in this effort we developed a new, facile immobilization protocol for construction of mono- and multivalent carbohydrate microarrays, in which unmodified glycans are linked to mono-, di-, tri- and tetravalent hydrazide-derivatized glass slides (Fig. 1).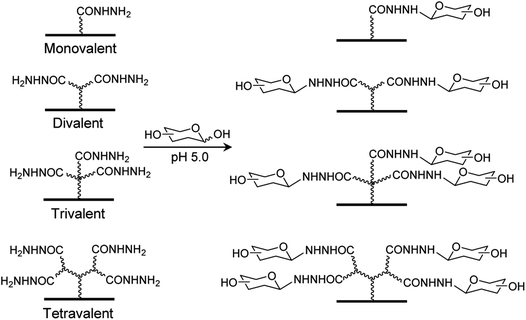 | ||
| Fig. 1 Construction of carbohydrate microarrays containing mono- and multivalent glycans by immobilizing free glycans on surfaces derivatized with mono-, di, tri- and tetravalent hydrazides. | ||
The linkers required for modification of glass slides with multivalent hydrazides were synthesized by using pathways shown in Scheme 1. Briefly, synthesis of di- and tri-t-butyl ester containing linkers 7 and 8, containing primary amine groups required for attachment to glass surfaces, was initiated by reactions of 1 and 2 with t-butyl acrylate under basic conditions, respectively. The adducts 3 and 4 were reacted with 2-(2-(2-(2-azidoethoxy)ethoxy)ethoxy)acetic acid to generate the respective amide coupled products 5 and 6. Reduction of the azide groups in 5 and 6 by catalytic hydrogenation over Pd/C produced the corresponding amines 7 and 8. In addition, the tetra-t-butyl ester containing linker 10 was prepared by sequential removal of t-butyl in 5, the amide coupling reaction of the resulting diacid 5′ with 3 to yield azide 9, and catalytic hydrogenation.
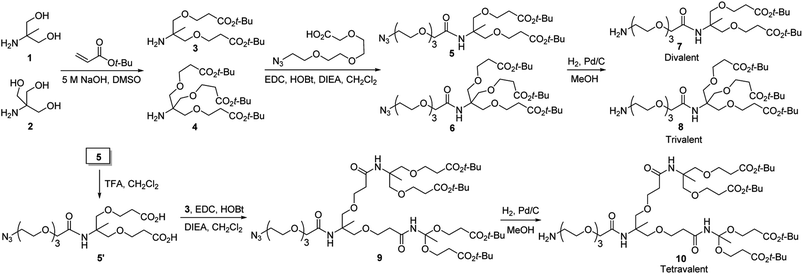 | ||
| Scheme 1 Routes for synthesis of di-, tri- and tetravalent linkers 7, 8 and 10 used for modification of glass slides with multivalent hydrazides. | ||
Next, glass surfaces were modified with mono-, di, tri- and tetravalent hydrazides. For this purpose, epoxide-coated glass slides were converted to N-hydroxysuccinimide (NHS) ester derivatized slides (Fig. 2),16 which were then treated with hydrazine to generate monovalent hydrazide-coated glass slides. In addition, the NHS ester-modified glass slides were individually treated with 7, 8 and 10 followed by removal of t-butyl groups under acidic conditions to generate the respective di-, tri- and tetravalent acid modified slides. Finally, the acid groups on the surfaces of each slide were converted to NHS esters, which were then reacted with hydrazine to generate the target di, tri- and tetravalent hydrazide-modified slides.
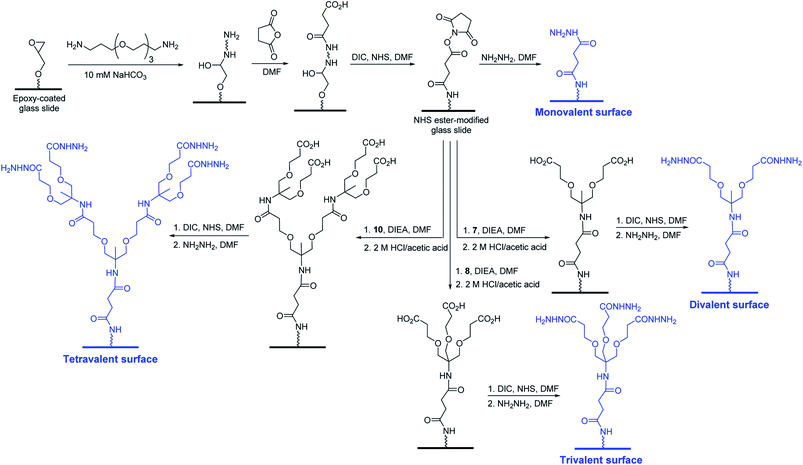
Carbohydrate microarrays containing mono- and multivalent glycans were prepared by printing nineteen unmodified sugars (1.0 nL, 10 and 30 mM of mono- (11–13), di- (14–17) and oligosaccharides (18–29)) on mono-, di-, tri- and tetravalent hydrazide-coated glass slides by using an automatic pin-type microarrayer (Fig. 3).14 To demonstrate that the glycans were successfully immobilized on the mono- and multivalent hydrazide-modified surfaces, the microarrays were incubated with Cy3-labeled plant lectins including Ricinus communis agglutinin I (RCA120, a Gal/GalNAc binding lectin), Concanavalin A (ConA, an α-Man/α-Glc binding lectin) and Aleuria aurantia lectin (AA, a Fuc binding lectin). The results of microarray image analysis showed that the glycans on the microarrays displayed lectin binding profiles that were in agreement with those observed in previous studies (Fig. 4A–C).14,17,18 Specifically, RCA120 recognized glycans containing Gal/GalNAc (binding order: Gal < GalNAc < Galβ1,3GalNAc ≤ asialo-GM1 < NeuNAcα2,6Lac < GM1 < Lac),14,18 ConA bound to Manα1,2Man and Manα1,4Man with similar affinities,14 and AA recognized Fuc containing glycans (binding order: sialyl Lex < H1 < Leb < Lex < Lea < Ley).14,17,18 The observations also showed that lectin binding to glycans becomes stronger as the valency is increased, and that tetravalent glycans immobilized using 10 mM concentrations bind to lectins with similar affinities as do monovalent glycans immobilized using 30 mM concentrations. The findings indicate that glycans are successfully attached to mono- and multivalent hydrazide-coated glass slides.
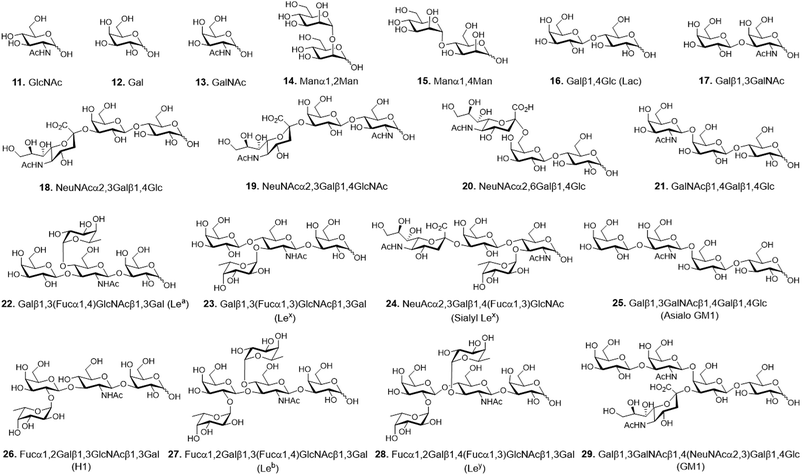 | ||
| Fig. 3 Chemical structures of glycans used for construction of mono- and multivalent carbohydrate microarrays. | ||
Studies were then performed to evaluate binding of cholera toxin B (CT-B) to glycans on the microarray. The Gram-negative Vibrio cholerae colonizes the small intestine and is the main pathogen causing life-threatening acute diarrhoea (cholera). This pathogen secrets cholera toxin (CT), which is composed of two proteins including the monomeric subunit A (CT-A) and the pentameric subunit B (CT-B).19 The released toxin adheres to host cells by binding of CT-B to GM1-ganglioside, which is expressed in various cell types including epithelial cells of the gut and immune cells.20 Thus, studies of the binding properties of CT-B to multivalent glycans is highly significant. To date, binding of CT-B to multivalent glycans has been probed using an SPR technique21 and carbohydrate microarrays.22 However, multivalent interactions between CT-B and glycans have not been assessed thus far. To address this issue, glycan microarrays containing glycans 11–29 with a valency ranging from 1 to 4 were incubated with Cy3-labeled CT-B for 1 h. As shown in Fig. 4D, CT-B bound to GM1-oligosaccharide (Galβ1,3GalNAcβ1,4(NeuNAcα2,3)Galβ1,4Glc) strongly but did not interact with the other glycans.21–23 The results also showed that CT-B bound to glycans more tightly as the glycan valency was increased from 1 to 4 and that recognition of the tetravalent GM1-oligosaccharide immobilized at a concentration of 10 mM by CT-B was comparable to binding of CT-B to a monovalent GM1-oligosaccharide immobilized using a concentration of 30 mM.
Next, the glycan microarrays were applied to study of binding of pathogens, uropathogenic Escherichia coli and Helicobacter pylori, to mono- and multivalent glycans. Uropathogenic E. coli cells adhere to the urinary tract of humans for colonization and infection via a process that is mediated by P, Type 1, S and F1C fimbriae.24 Type 1 fimbriae are the most popular among fimbriae generated by uropathogenic E. coli and are important virulence factors for infection in the urinary tract. FimH, the Type 1 fimbrial adhesin of uropathogenic E. coli, is involved in adhesion to the high-mannosylated glycoprotein receptors in bladder epithelium.25 Based on these observations, we assessed the binding properties of the E. coli ORN178 strain expressing FimH. For this purpose, E. coli ORN178 (cell density: 106 and 107 cells per mL) along with E. coli ORN208 lacking FimH as a negative control were first treated with SYTO 83 for cell staining and then applied to mono- and multivalent glycan microarrays. Bovine serum albumin (BSA, 0.1%) was added to suspensions of the bacterial cells to avoid nonspecific adhesion of pathogens to solid surfaces. As shown in Fig. 5A and S1,† the E. coli ORN178 strain bound to mannobioses 14 (Manα1,2Man) and 15 (Manα1,4Man) with similar affinities, whereas the E. coli ORN208 strain did not bind to these glycans.14 In addition, microarrays incubated with bacteria with cell density of 107 cells per mL exhibited higher fluorescence intensity than those treated with lower cell density (106 cells per mL). Furthermore, the E. coli ORN178 strain bound more strongly to mannobioses 14 and 15 as the glycan valency was increased from 1 to 4.
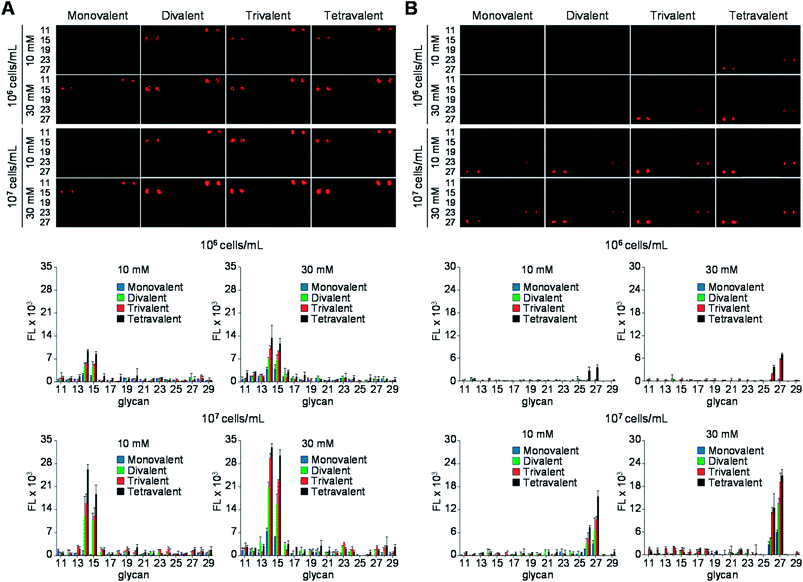 | ||
| Fig. 5 (Upper images) Fluorescence images of mono- and multivalent carbohydrate microarrays treated with SYTO 83-labeled (A) E. coli ORN178 in PBS buffer (pH 7.4) containing 0.1% BSA, 1 mM CaCl2 and 1 mM MnCl2 and (B) H. pylori J99 in TBS buffer (50 mM Tris-Cl, 150 mM NaCl, pH 7.4) containing 0.1% BSA. The distance between centers of adjacent spots is 250 μm. (Bottom graphs) Fluorescence intensities (arbitrary unit) of carbohydrate microarrays (error bar: mean ± s.d., n = 3). The layout of carbohydrate spots on glass slides is shown in Fig. 4. | ||
To obtain additional evidence of binding of E. coli to mannobioses, we prepared the Cy3-labeled BSA-Manα1,2Man conjugate which had been previously successfully utilized to detect the mammalian cell-surface lectin (Fig. 6 and S2†).15 E. coli ORN178 and ORN208 strains were then incubated with the glycoconjugate for 1 h. The results of analysis of cell images obtained using confocal microscopy showed that the BSA-Manα1,2Man conjugate bound to E. coli ORN178 but not to E. coli ORN208, a finding which matches that made in the microarray study (Fig. 7).
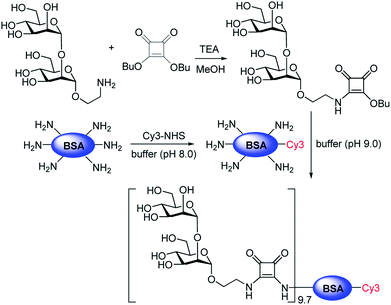 | ||
| Fig. 6 Preparation of the Cy3-BSA-Manα1,2Man conjugate. The ratio of Manα1,2Man conjugation to Cy3-BSA was determined by MS analysis (see ESI† for more details). | ||
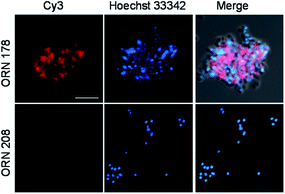
The binding properties of H. pylori expressing BabA to mono- and multivalent glycans on microarrays were evaluated next. H. pylori colonizes the human stomach and is one of the most widespread infectious pathogens that promotes the onset of several gastrointestinal diseases, such as chronic gastritis, peptic ulcer disease and gastric cancer.26 H. pylori frequently expresses cell-surface adhesins that bind to specific host cell glycans, as exemplified by the well-characterized blood group antigen-binding adhesin (BabA). In this case, infection of hosts by H. pylori is initiated by the interaction of BabA with Leb present in the gastric mucosa.26,27
Expression of BabA in the two H. pylori strains, J99 and 26695, was first examined by using reverse transcription-polymerase chain reaction (RT-PCR). The results showed that the H. pylori J99 strain produced a BabA gene but H. pylori 26695 strain did not (Fig. S3†).27 Mono- and multivalent glycan microarrays were then incubated with SYTO 83-labeled H. pylori J99 strain (cell density: 106 and 107 cells per mL) containing 0.1% BSA, along with the H. pylori 26695 strain as a control. The results of microarray image analysis showed that the H. pylori J99 strain adhered to H1 (26, Fucα1,2Galβ1,3GlcNAcβ1,3Gal) and Leb (27, Fucα1,2Galβ1,3(Fucα1,4)GlcNAcβ1,3Gal) with different binding affinities (Fig. 5B). Specifically, the H. pylori J99 strain bound to Leb more strongly than H1, a finding which is consistent with results from studies using purified BabA (Kd = 252 μM for Leb and Kd = 617 μM for H1).28 The pathogen also bound to 26 and 27 in a cell density- and glycan density-dependent manner. However, H. pylori J99 did not recognize the other glycans tested and the H. pylori 26695 strain did not adhere to any of these glycans (Fig. S4†).
Confirmatory evidence for binding of H. pylori J99 to Leb and H1 was gained in a study using fluorescent magnetic nanoparticles (100 μg mL−1) conjugated with Leb and H1 which had been previously successfully employed to detect H. pylori J99.27 The glyconanoparticles were incubated for 1 h with H. pylori J99, which was pretreated for 5 min with 1 μM Hoechst 33342 for bacterial cell staining. As control experiments, H. pylori 26695 strain was incubated with Leb- and H1-conjugated nanoparticles under the same conditions. Analysis of cell images obtained using confocal microscopy showed that Leb- and H1-glycoconjugated nanoparticles bound to BabA expressing J99 but not to BabA lacking 26695 (Fig. 8). The findings are consistent with observations made in the microarray study.
Conclusions
In the investigation described above, we developed a facile and efficient method to prepare multivalent glycan microarrays, in which free glycans are immobilized on multivalent hydrazide-coated surfaces. This protocol does not require multivalent glycoconjugates that are normally prepared by using multistep pathways. By utilizing the glycan microarrays, we observed that the binding affinities of toxins and pathogens for glycans are highly glycan density-dependent. We also found that immobilization concentrations of glycans for tight binding of toxins and pathogens can be reduced from 30 mM to 10 mM when microarrays containing tetravalent glycans are used. To our knowledge, this is the first time that glycan microarray have been applied to analyze the density dependency of glycan binding by pathogens and toxins. We believe that the results of this effort have provided valuable information about the design and preparation of multivalent carbohydrate microarrays and about how they can be utilized for functional studies of pathogens and pathogenic proteins.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by a grant from the National Creative Research Initiative program (2010-0018272) in Korea.References
- S. Park, M. R. Lee and I. Shin, Chem. Soc. Rev., 2008, 37, 1579–1591 Search PubMed.
- A. Imberty and A. Varrot, Curr. Opin. Struct. Biol., 2008, 18, 567–576 Search PubMed.
- (a) J. R. Bishop and P. Gagneux, Glycobiology, 2007, 17, 23r–34r Search PubMed; (b) J. Pettersson, R. Nordfelth, E. Dubinina, T. Bergman, M. Gustafsson, K. E. Magnusson and H. WolfWatz, Science, 1996, 273, 1231–1233 Search PubMed.
- R. N. Pruitt and D. B. Lacy, Front. Cell. Infect. Microbiol., 2012, 2, 28–31 Search PubMed.
- (a) M. Mammen, S. K. Choi and G. M. Whitesides, Angew. Chem., Int. Ed., 1998, 37, 2754–2794 Search PubMed; (b) J. J. Lundquist and E. J. Toone, Chem. Rev., 2002, 102, 555–578 Search PubMed; (c) X. Tian, J. Pai and I. Shin, Chem.–Asian J., 2012, 7, 2052–2060 Search PubMed.
- (a) S. Park and I. Shin, Angew. Chem., Int. Ed., 2002, 41, 3180–3182 Search PubMed; (b) D. N. Wang, S. Y. Liu, B. J. Trummer, C. Deng and A. L. Wang, Nat. Biotechnol., 2002, 20, 275–281 Search PubMed; (c) S. Fukui, T. Feizi, C. Galustian, A. M. Lawson and W. Chai, Nat. Biotechnol., 2002, 20, 1011–1017 Search PubMed.
- (a) J. Y. Hyun, J. Pai and I. Shin, Acc. Chem. Res., 2017, 50, 1069–1078 Search PubMed; (b) C. D. Rillahan and J. C. Paulson, Annu. Rev. Biochem., 2011, 80, 797–823 Search PubMed; (c) S. Park, J. C. Gildersleeve, O. Blixt and I. Shin, Chem. Soc. Rev., 2013, 42, 4310–4326 RSC.
- (a) O. Blixt, S. Head, T. Mondala, C. Scanlan, M. E. Huflejt, R. Alvarez, M. C. Bryan, F. Fazio, D. Calarese, J. Stevens, N. Razi, D. J. Stevens, J. J. Skehel, I. van Die, D. R. Burton, I. A. Wilson, R. Cummings, N. Bovin, C. H. Wong and J. C. Paulson, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 17033–17038 CrossRef CAS PubMed; (b) S. Park, M. R. Lee, S. J. Pyo and I. Shin, J. Am. Chem., Soc., 2004, 126, 4812–4819 CrossRef CAS PubMed; (c) B. Y. Xia, Z. S. Kawar, T. Z. Ju, R. A. Alvarez, G. P. Sachdev and R. D. Cummings, Nat. Methods, 2005, 2, 845–850 CrossRef CAS PubMed; (d) J. C. Manimala, T. A. Roach, Z. T. Li and J. C. Gildersleeve, Angew. Chem. Int. Ed., 2006, 45, 3607–3610 CrossRef CAS PubMed; (e) K. S. Ko, F. A. Jaipuri and N. L. Pohl, J. Am. Chem. Soc., 2005, 127, 13162–13163 CrossRef CAS PubMed; (f) J. L. de Paz, C. Noti and P. H. Seeberger, J. Am. Chem. Soc., 2006, 128, 2766–2767 CrossRef CAS PubMed; (g) S. E. Tully, M. Rawat and L. C. Hsieh-Wilson, J. Am. Chem. Soc., 2006, 128, 7740–7741 CrossRef CAS PubMed; (h) Y. Chevolot, C. Bouillon, S. Vidal, F. Morvan, A. Meyer, J. P. Cloarec, A. Jochum, J. P. Praly, J. J. Vasseur and E. Souteyrand, Angew. Chem., Int. Ed., 2007, 46, 2398–2402 CrossRef CAS PubMed.
- (a) S. Park and I. Shin, Org. Lett., 2007, 9, 1675–1678 CrossRef CAS PubMed; (b) O. Blixt, K. Allin, O. Bohorov, X. F. Liu, H. Andersson-Sand, J. Hoffmann and N. Razi, Glycoconjugate J., 2008, 25, 59–68 CrossRef CAS PubMed.
- (a) Y. F. Wang, C. F. Chang, C. Y. Chi, H. C. Wang, J. R. Wang and I. J. Su, J. Med. Virol., 2012, 84, 679–685 CrossRef CAS PubMed; (b) L. M. Chen, P. Rivailler, J. Hossain, P. Carney, A. Balish, I. Perry, C. T. Davis, R. Garten, B. Shu, X. Xu, A. Klimov, J. C. Paulson, N. J. Cox, S. Swenson, J. Stevens, A. Vincent, M. Gramer and R. O. Donis, Virology, 2011, 412, 401–410 CrossRef CAS PubMed; (c) K. C. Bradley, C. A. Jones, S. M. Tompkins, R. A. Tripp, R. J. Russell, M. R. Gramer, J. Heimburg-Molinaro, D. F. Smith, R. D. Cummings and D. A. Steinhauer, Virology, 2011, 413, 169–182 CrossRef CAS PubMed; (d) M. B. Pearce, A. Jayaraman, C. Pappas, J. A. Belser, H. Zeng, K. M. Gustin, T. R. Maines, X. J. Sun, R. Raman, N. J. Cox, R. Sasisekharan, J. M. Katz and T. M. Tumpey, Proc. Natl. Acad. Sci. U.S.A, 2012, 109, 3944–3949 CrossRef CAS PubMed; (e) H. Yang, L. M. Chen, P. J. Carney, R. O. Donis and J. Stevens, PLoS Pathog., 2010, 6, e1001081 Search PubMed; (f) Z. Wu, E. Miller, M. Agbandje-McKenna and R. J. Samulski, J. Virol., 2006, 80, 9093–9103 CrossRef CAS PubMed; (g) K. C. Nicolaou and H. J. Mitchell, Angew. Chem., Int. Ed., 2001, 40, 1576–1624 CrossRef CAS.
- (a) M.-R. Lee and I. Shin, Org. Lett., 2005, 7, 4269–4272 CrossRef CAS PubMed; (b) A. Walz, S. Odenbreit, J. Mahdavi, T. Borén and S. Ruhl, Glycobiology, 2005, 15, 700–708 CrossRef CAS PubMed; (c) Y. Y. Fei, A. Schmidt, G. Bylund, D. X. Johansson, S. Henriksson, C. Lebrilla, J. V. Solnick, T. Borén and X. D. Zhu, Anal. Chem., 2011, 83, 6336–6341 CrossRef CAS PubMed; (d) C. J. Day, J. Tiralongo, R. D. Hartnell, C.-A. Logue, J. C. Wilson, M. von Itzstein and V. Korolik, PLoS One, 2009, 4, e4927 Search PubMed; (e) J. Y. Hyun, C. W. Park, Y. Liu, D. Kwon, S. H. Park, S. Park, J. Pai and I. Shin, ChemBioChem, 2017, 18, 1077–1082 CrossRef CAS PubMed.
- (a) M. D. Disney and P. H. Seeberger, Chem. Biol., 2004, 11, 1701–1707 CrossRef CAS PubMed; (b) C. L. Bell, L. H. Vandenberghe, P. Bell, M. P. Limberis, G. P. Gao, K. Van Vliet, M. Agbandje-McKenna and J. M. Wilson, J. Clin. Invest., 2011, 121, 2427–2435 CAS; (c) M. A. Campanero-Rhodes, A. Smith, W. G. Chai, S. Sonnino, L. Mauri, R. A. Childs, Y. B. Zhang, H. Ewers, A. Helenius, A. Imberty and T. Feizi, J. Virol., 2007, 81, 12846–12858 CrossRef CAS PubMed; (d) H. J. Nam, B. Gurda-Whitaker, W. Y. Gan, S. Ilaria, R. McKenna, P. Mehta, R. A. Alvarez and M. Agbandje-McKenna, J. Biol. Chem., 2006, 281, 25670–25677 CrossRef CAS PubMed.
- (a) C. Scheibe, A. Bujotzek, J. Dernedde, M. Weber and O. Seitz, Chem. Sci., 2011, 2, 770–775 RSC; (b) K. Gorska, K. T. Huang, O. Chaloin and N. Winssinger, Angew. Chem., Int. Ed., 2009, 48, 7695–7700 CrossRef CAS PubMed; (c) H. M. Branderhorst, R. Ruijtenbeek, R. M. J. Liskamp and R. J. Pieters, ChemBioChem, 2008, 9, 1836–1844 CrossRef CAS PubMed; (d) O. Oyelaran, Q. Li, D. Farnsworth and J. C. Gildersleeve, J. Proteome Res., 2009, 8, 3529–3538 CrossRef CAS PubMed; (e) Y. Chevolot, C. Bouillon, S. Vidal, F. Morvan, A. Meyer, J. P. Cloarec, A. Jochum, J. P. Praly, J. J. Vasseur and E. Souteyrand, Angew. Chem., Int. Ed., 2007, 46, 2398–2402 CrossRef CAS PubMed; (f) L. Moni, G. Pourceau, J. Zhang, A. Meyer, S. Vidal, E. Souteyrand, A. Dondoni, F. Morvan, Y. Chevolot, J. J. Vasseur and A. Marra, ChemBioChem, 2009, 10, 1369–1378 CrossRef CAS PubMed; (g) N. P. Pera, H. M. Branderhorst, R. Kooij, C. Maierhofer, M. van der Kaaden, R. M. J. Liskamp, V. Wittmann, R. Ruijtenbeek and R. J. Pieters, ChemBioChem, 2010, 11, 1896–1904 CrossRef PubMed; (h) Y. Chevolot, J. Zhang, A. Meyer, A. Goudot, S. Rouanet, S. Vidal, G. Pourceau, J. P. Cloarec, J. P. Praly, E. Souteyrand, J. J. Vasseur and F. Morvan, Chem. Commun., 2011, 47, 8826–8828 RSC.
- S. Park, M. R. Lee and I. Shin, Bioconjugate Chem., 2009, 20, 155–162 CrossRef CAS PubMed.
- J. Pai, J. Y. Hyun, J. Jeong, S. Loh, E. H. Cho, Y. S. Kang and I. Shin, Chem. Sci., 2016, 7, 2084–2093 RSC.
- (a) J. Pai, T. Yoon, N. D. Kim, I. S. Lee, J. Yu and I. Shin, J. Am. Chem. Soc., 2012, 134, 19287–19296 CrossRef CAS PubMed; (b) J. Pai, S. Hyun, J. Y. Hyun, S. H. Park, W. J. Kim, S. H. Bae, N. K. Kim, J. Yu and I. Shin, J. Am. Chem. Soc., 2016, 138, 857–867 CrossRef CAS PubMed; (c) S. Park, M. R. Lee and I. Shin, Nat. Protoc., 2007, 2, 2747–2758 CrossRef CAS PubMed; (d) M. R. Lee and I. Shin, Angew. Chem., Int. Ed., 2005, 44, 2881–2884 CrossRef CAS PubMed.
- H. Y. Hsiao, M. L. Chen, H. T. Wu, L. D. Huang, W. T. Chien, C. C. Yu, F. D. Jan, S. Sahabuddin, T. C. Chang and C. C. Lin, Chem. Commun., 2011, 47, 1187–1189 RSC.
- Functional Glycomics website (http://www.functionalglycomics.org).
- P. I. H. Bastiaens, I. V. Majoul, P. J. Verveer, H. D. Soling and T. M. Jovin, EMBO J., 1996, 15, 4246–4253 CAS.
- D. Vanden Broeck, C. Horvath and M. J. S. De Wolf, Int. J. Biochem. Cell Biol., 2007, 39, 1771–1775 CrossRef CAS PubMed.
- G. M. Kuziemko, M. Stroh and R. C. Stevens, Biochem., 1996, 35, 6375–6384 CrossRef CAS PubMed.
- C. S. Kim, J. H. Seo and H. J. Cha, Anal. Chem., 2012, 84, 6884–6890 CrossRef CAS PubMed.
- J. Shi, T. Yang, S. Kataoka, Y. Zhang, A. J. Diaz and P. S. Cremer, J. Am. Chem. Soc., 2007, 129, 5954–5961 CrossRef CAS PubMed.
- B. Foxman, Am. J. Med., 2002, 8, 5S–13S CrossRef.
- (a) M. A. Schembri, K. Kjaergaard, E. V. Sokurenko and P. Klemm, J. Infect. Dis., 2001, 183, S28–S31 CrossRef CAS PubMed; (b) M. M. Sauer, R. P. Jakob, J. Eras, S. Baday, D. Eris, G. Navarra, S. Berneche, B. Ernst, T. Maier and R. Glockshuber, Nat. Commun., 2016, 7, 10738–10750 CrossRef CAS PubMed.
- (a) A. Covacci, J. L. Telford, G. Del Giudice, J. Parsonnet and R. Rappuoli, Science, 1999, 284, 1328–1333 CrossRef CAS PubMed; (b) D. B. Polk and R. M. Peek, Nat. Rev. Cancer, 2010, 10, 593 CrossRef CAS; (c) N. Uemura, S. Okamoto, S. Yamamoto, N. Matsumura, S. Yamaguchi, M. Yamakido, K. Taniyama, N. Sasaki and R. J. Schlemper, N. Engl. J. Med., 2001, 345, 784–789 CrossRef CAS PubMed.
- S. Park, G. H. Kim, S. H. Park, J. Pai, D. Rathwell, J. Y. Park, Y. S. Kang and I. Shin, J. Am. Chem. Soc., 2015, 137, 5961–5968 CrossRef CAS PubMed.
- N. Hage, T. Howard, C. Phillips, C. Brassington, R. Overman, J. Debreczeni, P. Gellert, S. Stolnik, G. S. Winkler and F. H. Falcone, Sci. Adv., 2015, 1, e1500315 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra01285g |
| This journal is © The Royal Society of Chemistry 2018 |