Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A structural dissection of protein–RNA interactions based on different RNA base areas of interfaces†
Wen Hu,
Liu Qin,
Menglong Li*,
Xuemei Pu and
Yanzhi Guo *
*
College of Chemistry, Sichuan University, Chengdu 610064, People's Republic of China. E-mail: yzguo@scu.edu.cn; liml@scu.edu.cn; Fax: +86-028-85412290; Tel: +86-028-85412290
First published on 16th March 2018
Abstract
Protein–RNA interactions are very common cellular processes, but the mechanisms of interactions are not fully understood, mainly due to the complicated RNA structures. By the elaborate investigation on RNA structures of protein–RNA complexes, it was firstly found in this paper that RNAs in these complexes could be clearly classified into three classes (high, medium and low) based on the different levels of Pbase (the percentage of base area buried in the RNA interface). In view of the three RNA classes, more detailed analyses on protein–RNA interactions were comprehensively performed from various aspects, including interface area, structure, composition and interaction force, so as to achieve a deeper understanding of the recognition specificity for the three classes of protein–RNA interactions. According to our classification strategy, the three complex classes have significant differences in terms of almost all properties. Complexes in the high class have short and extended RNA structures and behave like protein–ssDNA interactions. Their hydrogen bonds and hydrophobic interactions are strong. For complexes in low class, their RNA structures are mainly double-stranded, like protein–dsDNA interactions, and electrostatic interactions frequently occur. The complexes in medium class have the longest RNA chains and largest average interface area. Meanwhile, they do not show any preference for the interaction force. On average, in terms of composition, secondary structures and intermolecular physicochemical properties, significant feature preferences can be observed in high and low complexes, but no highly specific features are found for medium complexes. We found that our proposed Pbase is an important parameter which can be used as a new determinant to distinguish protein–RNA complexes. For high and low complexes, we can more easily understand the specificity of the recognition process from the interface features than for medium complexes. In the future, medium complexes should be our research focus to further structurally analyze from more feature aspects. Overall, this study may contribute to further understanding of the mechanism of protein–RNA interactions on a more detailed level.
1. Introduction
Nucleic acids, including DNAs (deoxyribonucleic acids) and RNAs (ribonucleic acids), always function through interactions with proteins. Such interactions play crucial roles in a wide variety of biological processes. Protein–DNA interactions (PDIs) are essential for DNA transcription, packaging, replication and repair.1–3 Protein–RNA interactions (PRIs) are indispensable for the regulation of gene expression, protein synthesis, RNA splicing and post-transcriptional control.4–7 It is urgent and quite meaningful to precisely understand the recognition mechanisms of PDIs and PRIs. Since PDIs have been widely reviewed before PRIs, insufficient structure data limit the further development of research on PRIs.8,9 With recent advances in biological technology, the number of available PRI structures are increasing, which provides an opportunity to launch a structure-based analysis on the principles governing the interactions between proteins and RNAs. These research studies on PRIs mainly include the construction of PRI databases,10,11 sequential or structural comparisons between PRIs and PDIs,12,13 prediction of RNA-binding sites,14–17 and structural dissection of protein–RNA interfaces.18–20In recent years, much attention has been paid to examining the general interface properties of protein–RNA complexes.21–28 Bahadur et al.22 analyzed PRIs in terms of interface size, composition, polar interactions and atomic packing and found electrostatic complementation, base recognition and shape complementarity on the interfaces of PRIs. By investigating the preferred RNA structural states in protein-binding regions, Gupta et al.24 observed strong preferences for both RNA bases and RNA structural states in protein–RNA interactions, indicating their mutual importance in protein recognition. The work of Iwakiri et al.26 suggests that nucleotide bases in the RNA loop are flipped out and form hydrogen bonds with the proteins, and different protein surface shapes prefer different RNA base-pairing properties. The most recent report by Barik et al.28 compared the structural, geometric and physicochemical properties of interfaces involved in protein–RNA, protein–DNA and protein–protein interactions. The result indicates that H-bonds, salt bridges and stacking interactions play significant roles in stabilizing PRI interfaces. Despite the great progress in PRI research, the structural mechanism underlying PRIs is still not fully understood, owing to the amazing diversity of RNA structures. Compared to double-stranded DNAs, RNA molecules display a much wider variety of conformations and shapes.29 Moreover, a nucleotide is composed of the negatively charged phosphate, neutral ribose and polar base. As we know, the properties of different RNA interfaces commonly determine the different interacting modes of RNAs with proteins. Therefore, the initial aim of our work is to qualitatively and even quantitatively measure the influence of the structure and composition of RNA interfaces on protein–RNA interactions.
Firstly, the relationship between composition and structure of the RNA interface was explored. We collected a non-redundant dataset of 137 X-ray structures of protein–RNA complexes and analyzed the contents of phosphate, ribose and base on each RNA interface. It was interestingly found that the three RNA groups show obvious composition differences among these 137 complexes, but the most significant difference is observed in terms of base composition. Pbase (the percentage of base buried area in the RNA interface) can be as high as over 80% or lower than 10%. According to the different values of Pbase, the 137 RNA interface structures can be clearly clustered into three classes (high, medium and low). Then, in order to understand the recognition process specificity of the three RNA classes, a comprehensive feature analysis was implemented on interface structures, intermolecular physicochemical properties and interface forces. Systematic comparisons among the three classes of complexes suggest that their interfaces are obviously different in terms of most features, which shows that the classification of RNA interfaces based on Pbase is reasonable. We demonstrate that the interface area contributed by the RNA base group could strongly influence protein recognition and binding, indicating that it can be used as a new determinant to distinguish different types of protein–RNA complexes. Thus, our analysis may contribute to understanding the specificity of the recognition process and the identification of protein–RNA binding sites on a deeper level.
2. Materials and methods
2.1 Dataset of the protein–RNA complexes
Protein–RNA complex structures were obtained from Protein Data Bank (PDB) database30 (Feb 2014) with X-ray structures and resolution better than 3.0 Å as criteria. In the present study, we only extracted those protein–RNA complexes containing proteins with at least 20 amino acid residues and RNA molecules with at least 5 nucleotides (nt). Some PDB chains containing only Cα atoms were excluded from our dataset. Moreover, some ribosomal subunits and viral protein–RNA complexes were also ignored in our dataset because these complexes often contain a large number of amino acid residues on interfaces and most of the RNA interfaces on proteins have not been determined, which could lead to population bias. For each protein–RNA complex, we chose a representative and stable biological assembly using the PDBePISA tool.31 Thus the entire dataset consists of 487 complexes (listed in ESI Table S1†). In order to remove redundancy, we used CDHIT32 to align RNA and protein sequences from the dataset. Sequence identity threshold of 30% for proteins and 90% for RNAs was respectively used. The final non-redundant dataset includes 137 complexes, which are detailed in ESI Table S2.†2.2 Definition of interface
In this paper, the software NACCESS33 was used to calculate the solvent accessible surface (ASA) values. The interface area of a protein–RNA complex was calculated using the web-based tool PRince,34 which uses NACCESS with a probe radius of 1.4 Å and default group radii. The size of a protein–RNA interface area (IA) was estimated by subtracting the ASA of the complex from the sum of the ASAs of the individual subunits, as shown in eqn (1):| IA = ASAprotein + ASARNA − ASAcomplex | (1) |
Here, the interface atoms are referred to as those that lose solvent accessibility and contribute to IA in a complex. In previous studies,14,19,35 a residue with at least one interface atom was always defined as the interface residue. Based on eqn (1), the Pribose, Pphosphate and Pbase were calculated using the following equations:
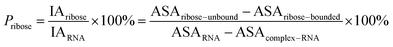 | (2) |
 | (3) |
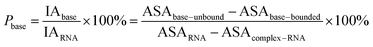 | (4) |
2.3 Definition of RNA structure
We used RNA view36 to identify and classify the types of nucleotide pairs. In our study, paired nucleotides are defined as any of 12 families of base pairs,37 and the remaining nucleotides are considered unpaired. Then, we calculated the Rpair, which indicates the ratio of the number of paired nucleotides to all nucleotides. Rpair represents the degree of pairing of RNA. In a protein–RNA complex, a smaller Rpair indicates more single-stranded regions in the RNA.2.4 Interface properties
Here, six important interface properties were calculated to reveal the structural foundations of different complexes. They are the interface area (IA), the ratio of interface area to surface area (Ri/s), amino acid composition (AAC), amino acid propensity (AAP), secondary structure composition (SSC) and secondary structure propensity (SSP).IA is defined as the total ASA decrease of one protein and one RNA upon interaction, and it reflects the size of the interfaces (eqn (1)). Ri/s is the ratio of the interface area to the rest of the complex surface area (eqn (5)):
 | (5) |
AAC is defined as the occurrence frequencies of the 20 standard amino acids in the interface residue sets, expressed as:
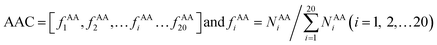 | (6) |
The AAP shows the enrichment or depletion of each type of amino acid in the interface as compared to the entire protein surface.38 The AAP can be calculated as:
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AAP = [PAA1, AAP = [PAA1, ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PAA2, PAA2,![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) … …![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PAAi PAAi![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) … …![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PAA20] PAA20]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) and PAAi = ln(fAAi/ and PAAi = ln(fAAi/![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) fAA, fAA,![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SURFi) SURFi)
| (7) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SURFi is the frequency of the i-th amino acid in the protein surface.
SURFi is the frequency of the i-th amino acid in the protein surface.
The program STRIDE39 was employed to assign the protein secondary structures. Six secondary structure types were considered, including α-helix, β-strand, turn, coil, bridge and 310-helix. Turn, coil, bridge and 310-helix were together deemed as the non-regular (NR) regions. The SSC is defined as follows:
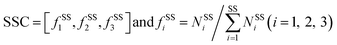 | (8) |
SSP = [PSS1,![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PSS2, PSS2,![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PSS3] and PSS3] and![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PSSi = ln(fSSi/ PSSi = ln(fSSi/![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) fSS,SURFi) fSS,SURFi)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)
| (9) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SURFi is the occurrence frequency of a particular secondary structure type in a protein surface.
SURFi is the occurrence frequency of a particular secondary structure type in a protein surface.
2.5 Interface force
Here, five kinds of noncovalent interactions were considered, including hydrogen bonds, electrostatic forces, van der Waals contacts, hydrophobic interactions and stacking interactions. Hydrogen bonds (H-bonds) at protein–RNA interfaces were calculated using the software HBPLUS,40 and positively charged electrostatic patches on protein surfaces were obtained through BindUP.41 For each protein–RNA complex, we calculated the percent overlap between the largest electrostatic positive patches on protein surfaces and the RNA-binding interfaces of each chain (Pe) and the mean Pe (![[P with combining macron]](https://www.rsc.org/images/entities/i_char_0050_0304.gif) e):42
e):42
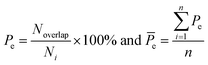 | (10) |
![[P with combining macron]](https://www.rsc.org/images/entities/i_char_0050_0304.gif) e reflects the electrostatic property of the interface. In addition, the exact electrostatic energy of each complex was calculated by the MM/GBSA approach43 using MMPBSA.py tools44 in the Amber16 package.45
e reflects the electrostatic property of the interface. In addition, the exact electrostatic energy of each complex was calculated by the MM/GBSA approach43 using MMPBSA.py tools44 in the Amber16 package.45
van der Waals contacts, hydrophobic interactions and stacking interactions were measured by the program ENTANGLE.46 van der Waals contacts are denoted as the sum of the van der Waals radii of the two atoms plus a maximum distance (defined ≤ 1.0 Å). Stacking interactions are defined as the π–π interactions that can occur between the side chains of Tyr, Trp, Phe, His and the bases. Moreover, we also considered the π–π and π–cation stacking of Arg through its guanidinium moiety onto nucleosides. Hydrophobic interactions are deemed as non-polar atoms that are ≤5.0 Å apart. We calculated the percent overlap between the hydrophobic interface and the RNA-binding interfaces of each chain (Ph) and the average Ph (![[P with combining macron]](https://www.rsc.org/images/entities/i_char_0050_0304.gif) h):
h):
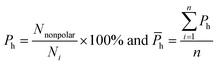 | (11) |
![[P with combining macron]](https://www.rsc.org/images/entities/i_char_0050_0304.gif) h can reflect the hydrophobic property of the interface.
h can reflect the hydrophobic property of the interface.
3. Results and discussion
3.1 Statistical analysis of protein–RNA complex data
Originally, we calculated the content of the ribose, phosphate and base buried in the RNA interface area, designated Pribose, Pphosphate and Pbase, respectively, for the initial dataset (487 complexes, ESI Table S1†), including non-redundant and all remaining redundant complexes. We found that 98% of all complexes have Pphosphate values of <50% and 92%, and of which the Pribose values were lower than 50%. By contrast, the Pbase values show significant differences among all complexes and are widely distributed between 0% and 80%. This can be seen from the violin plot shown in Fig. 1. The ribose and phosphate moieties are the non-specific parts of the RNA molecules, so the differences between RNA molecules are not significant in terms of Pribose and Pphosphate. The Pbase represents the interface area contributed by base groups, which are specific to RNA molecules. Thus, we could consider whether the interface area contributed by RNA base residues can be used as a new standard for distinguishing protein–RNA complexes.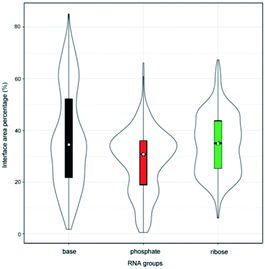 | ||
| Fig. 1 Violin plot combining the box plot and density trace for Pribose, Pphosphate and Pbase in the initial dataset. | ||
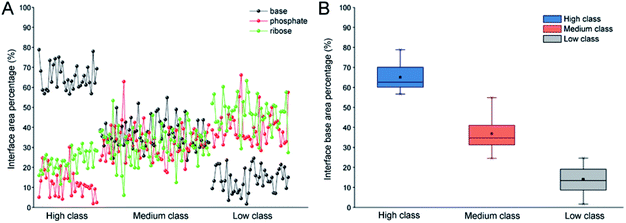
In order to validate the reasonability of this classification, we used the 137 non-redundant complexes for more detailed calculations (ESI Table S2†). Fig. 2A shows the distribution of Pribose, Pphosphate and Pbase in 137 non-redundant protein–RNA complexes. We could easily observe the significant differences between these complexes based on Pbase values and classified them into three classes (high, medium and low). As a result, high includes 33 complexes with the average Pbase value of 65% and standard deviation (SD) of 6.5%. Medium comprises 61 complexes (Pbase = 37% ± 6.2%), and low consists of 43 complexes (Pbase = 14% ± 7.2%, Fig. 2B).
Indeed, previous studies have shown that protein interactions with the RNA ribose-phosphate backbone are more common than interactions with the bases.19,22,24,47,48 So, the number of complexes in high class is lower than that in medium and low. We also counted the numbers of different structures and types of RNAs in the three classes. Fig. 3 shows the distribution of Rpair in different types of protein–RNA interactions. It suggests that most RNA molecules (23/33) in high are single stranded in structure with Rpair = 0, while those in low are double stranded in structure (31/43), with Rpair value greater than 0.8. In medium, RNA structure is more complicated because of the widely distributed Rpair values. For RNA types, we consider the five common types (ssRNA, dsRNA, tRNA, rRNA and mRNA), and other RNAs were deemed as ‘other’ type. The detailed information is listed in ESI Table S3.† The most important RNA types are mRNA, tRNA and dsRNA in high, medium and low, respectively. Therefore, we can conclude that both the RNA structures and types display obvious differences among the complexes in the three classes.
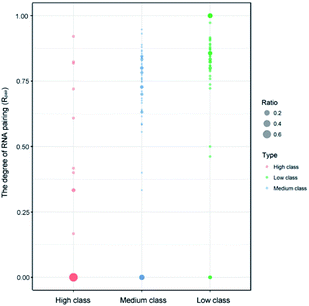 | ||
| Fig. 3 Bubble chart of Rpair in the three classes. The bigger bubble and the deeper color indicate higher frequency of Rpair in each class. | ||
A diagrammatic structure analysis was also performed on the complexes in the three classes, and Fig. S1† gives the 3D structures of three representative samples (PDB ID: 3QJJ, 1F7U and 3VYY). We found that most RNAs are in single-stranded form when interacting with proteins in the high class, which exposes the base groups of RNAs on the interfaces. So, we can explain why Pbase values are high in the high class. Meanwhile, those in low mostly use their stem or double-stranded regions to bind to proteins, so the Pbase values are lower than others. In medium, RNAs can have both single and double-stranded states to interact with proteins, so the Pbase values are medium.
3.2 Interface property analysis of different classes of protein–RNA complexes
Here, we calculated the interface properties for the protein–RNA complexes in three classes. Table 1 gives the average values of different properties.| Interface | Protein–RNAa | |||
|---|---|---|---|---|
| All | High | Medium | Low | |
| a Data are expressed as mean ± standard deviation (SD). | ||||
| Number of complexes | 137 | 33 | 61 | 43 |
| IA (Å2) | 3192 ± 1822 | 2729 | 3808 | 2673 |
| IAprotein (Å2) | 1526 ± 881 | 1255 | 1833 | 1299 |
| IARNA (Å2) | 1666 ± 945 | 1474 | 1975 | 1375 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Number of | ||||
| Amino acids | 54 ± 31 | 46 | 65 | 45 |
| Nucleotides | 20 ± 11 | 12 | 24 | 20 |
| Protein atoms | 178 ± 103 | 154 | 213 | 147 |
| RNA atoms | 172 ± 99 | 147 | 205 | 146 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| IA (Å2) per | ||||
| Amino acid | 28.3 ± 4.8 | 27.1 | 28.3 | 29.3 |
| Nucleotide | 93.4 ± 36.9 | 124.6 | 90.3 | 73.7 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Surface area buried ratio (%) | ||||
| Complex | 14.0 ± 5.9 | 17.5 | 13.1 | 12.5 |
| Protein | 8.8 ± 4.8 | 9.3 | 8.0 | 9.6 |
| Nucleic acid | 28.5 ± 16.3 | 36.7 | 27.1 | 24.2 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 344 Å2 and 11
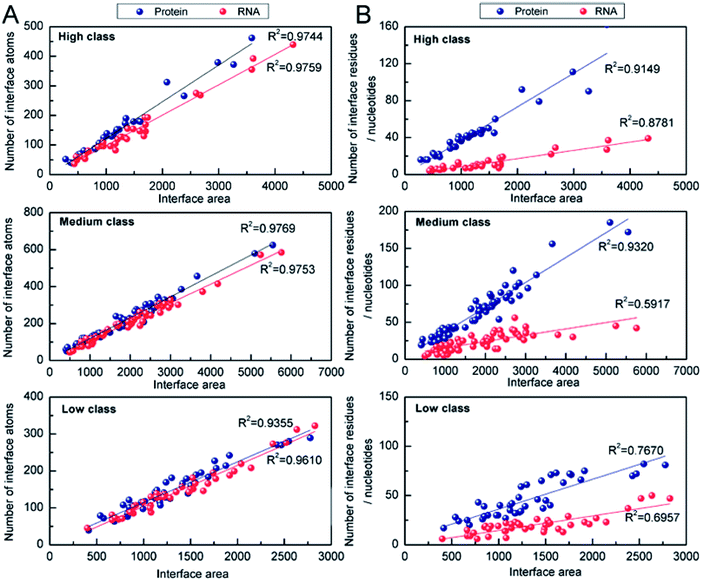
344 Å2 and 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 308 Å2) due to the four or five protein chains on their interfaces. About 50% of medium complexes have IA > 4000 Å2; however, the IA sizes of high and low complexes are from 2000 to 4000 Å2. In high and low class, the distribution of IA has a peak at 2500 Å2. In addition to the same peak at 2500 Å2, the distribution of IA in medium class has another peak at 5000 Å2, which is consistent with the previous report, giving two broad peaks at 2000 Å2 and at 4800 Å2 for whole protein–RNA interfaces.28 This result indicates that the second peak is mainly contributed by the medium class. So, in terms of IA, complexes in high and low are similar to each other, but they are obviously different from those in medium (P < 0.05).
308 Å2) due to the four or five protein chains on their interfaces. About 50% of medium complexes have IA > 4000 Å2; however, the IA sizes of high and low complexes are from 2000 to 4000 Å2. In high and low class, the distribution of IA has a peak at 2500 Å2. In addition to the same peak at 2500 Å2, the distribution of IA in medium class has another peak at 5000 Å2, which is consistent with the previous report, giving two broad peaks at 2000 Å2 and at 4800 Å2 for whole protein–RNA interfaces.28 This result indicates that the second peak is mainly contributed by the medium class. So, in terms of IA, complexes in high and low are similar to each other, but they are obviously different from those in medium (P < 0.05).
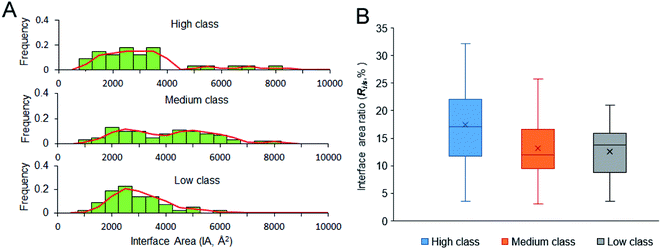 | ||
| Fig. 4 Size of the interfaces and interface area ratio of our dataset. (A) The frequency histogram of interface area size. (B) The box plot for interface area ratio of the three classes. | ||
A stable interface needs not only a large IA but also high Ri/s.51,52 Fig. 4B displays the box plot for Ri/s in different types of protein–RNA interactions. It reveals that complexes in high class have the highest average Ri/s, with the average values of Ri/s declining from high to medium and then to low, showing a different trend from the observation on IA in Fig. 4A. In Fig. 4A, complexes in medium give the largest average IA. This result may indicate that the protein and RNA surfaces in high class are more likely to be involved in the interfaces when they contact with each other to form complexes. Moreover, short and extended RNA structures of high complexes make them more conducive to interact with proteins. The complexes in medium class have both large interface area and large surface area because of the large molecular weight. Lastly, complexes in low may have more unstable interfaces than the two other classes because of the low IA and Ri/s. So, in terms of Ri/s, complexes in high are significantly different from those in medium and low (P < 0.01).
The correlation between the number of interface residues/nucleotides and IA was investigated for each class. We found that the values of R2 in Fig. 5B are always lower than the R2 in Fig. 5A. On the protein side, we obtained a satisfactory R2 of 0.91, 0.93 and 0.76 in high, medium and low class, respectively. On the RNA side, medium class yields the minimum R2 (R2 = 0.59), while R2 is 0.88 and 0.70 in high and low class, respectively. This result may be due to the more complicated RNA structures of medium complexes. Previous studies have reported that the linear correlation between IA and the number of interface nucleotides is low, with R2 of 0.67.22,28 From our results, we can explain that the complexes in medium may be key samples for this mediocre correlation.
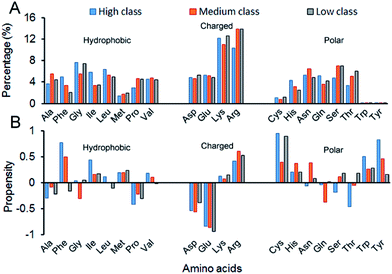 | ||
| Fig. 6 Amino acid composition and propensity of each class. (A) The average percentage of 20 amino acids in the interface of each class. (B) The average propensity of 20 amino acids in each class. | ||
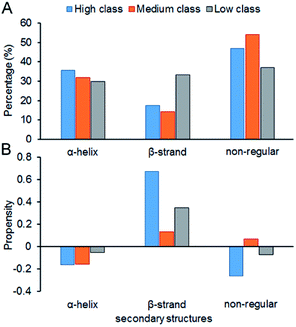
Fig. 7 shows the composition (SSC) and propensity (SSP) of the three types of secondary structures in each class, including α-helix, β-strand and the non-regular regions. Similar to that reported by Gupta and Gribskov,24 the non-regular elements are the primary protein interface structural state (Fig. 7A). Moreover, medium class yields the maximum percentage of non-regular regions. This may be due to the more complicated structures of RNAs in medium, so it is more difficult for them to bind the regular structures of proteins, such as α-helix and β-strand. Therefore, the structures of binding proteins in medium tend to be non-regular. In Fig. 7B, we can easily obtain the same conclusion that β-strands are preferred on protein–RNA interfaces, but α-helix does not show obvious propensity.55,56 In high class, this phenomenon is more obvious. The reason may be that β-strand is less likely to interface with the RNA backbone,24 which gives the RNA base a greater chance to bind with β-strand. The details on the composition and propensity of secondary structures in each class are listed in ESI Table S5.†
3.3 Interaction force analysis on the different classes of protein–RNA complexes
We also counted the frequency of all the chemical components for H-bonds in each class (Table 2). On the protein side, the frequency of main chains increases from high to medium and then to low, probably because in high, the RNA structures are more extended and they more easily interact with protein backbones. The main chain nitrogen has been proven to be more frequently found than the main chain oxygen in protein–RNA H-bonds,28 which can also be obviously observed from the medium- and low-class complexes in our study, but not those in high. The reason is also the influence of different Pbase values for the three classes; the base tends to form hydrogen bonds with protein main chain oxygen atoms, while the phosphate tends to be with nitrogen atoms.22 In the side chain involved in hydrogen bonds, the content of charged groups is nearly twice that of neutral groups in all protein–RNA interactions. However, in protein–DNA complexes, the contents of charged and neutral groups nearly equal each other.47 On the RNA side, the contribution of phosphate and ribose to protein–RNA H-bonds is 61%, which is less than that in protein–DNA H-bonds (76%). The frequencies of different RNA bases involved in H-bonds are also different. U (14%) and G (10%) are more frequently found than A (7%) and C (8%). Interestingly, only in low class is the frequency of G (7%) larger than the frequency of U (2%), and this phenomenon has also been found in protein–DNA H-bonds.27,47 Overall, these results suggest that the complexes in high have the strongest hydrogen bonds, and H-bonds in low are similar to the protein–DNA H-bonds, both on the protein side and the RNA side.
| H-bonds | All | High | Medium | Low |
|---|---|---|---|---|
| a Average number of H-bonds per interface.b Percentage of 2933 protein–RNA H-bonds contributed by the protein or nucleic acid chemical group. | ||||
| Total number | 2933 | 675 | 1529 | 729 |
| Number per interfacea | 22 | 20 | 26 | 17 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Protein chemical group (%)b | ||||
| Main chain O | 12 | 18 | 10 | 9 |
| Main chain N | 15 | 16 | 15 | 14 |
| Side chain groups | ||||
| Charged | 46 | 42 | 49 | 47 |
| Neutral | 27 | 23 | 26 | 31 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Nucleic acid chemical group (%)b | ||||
| Phosphate | 41 | 17 | 50 | 57 |
| Sugar | 20 | 16 | 17 | 26 |
| Base | 39 | 66 | 33 | 17 |
| Guanine | 10 | 15 | 8 | 7 |
| Adenine | 7 | 12 | 7 | 3 |
| Cytosine | 8 | 10 | 8 | 5 |
| Uracil/thymine | 14 | 29 | 11 | 2 |
| High class | Medium class | Low class | All | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Residues | A | G | C | U | A | G | C | U | A | G | C | U | A | G | C | U |
| a Average number of stacking interactions per complex in each class. | ||||||||||||||||
| Phe | 3 | 11 | 1 | 14 | 3 | 3 | 1 | 14 | 0 | 0 | 1 | 0 | 6 | 14 | 3 | 28 |
| Tyr | 9 | 10 | 0 | 29 | 3 | 4 | 0 | 4 | 0 | 0 | 1 | 0 | 12 | 14 | 1 | 33 |
| His | 6 | 9 | 0 | 8 | 3 | 0 | 3 | 3 | 0 | 0 | 0 | 0 | 9 | 9 | 3 | 11 |
| Trp | 3 | 0 | 0 | 0 | 3 | 1 | 1 | 3 | 0 | 1 | 1 | 0 | 6 | 2 | 2 | 2 |
| Arg | 21 | 3 | 6 | 10 | 10 | 5 | 10 | 19 | 2 | 7 | 1 | 0 | 32 | 15 | 17 | 29 |
| Averagea | 4 | 2 | 0 | 2 | ||||||||||||
Overall, our results are consistent with previous studies using the same data set ,25,28,46 and the contribution of residues and bases is similar in each class. Further, our results show significant differences in the strength of stacking interactions among the three classes. The complexes in high have the strongest stacking interactions, while in low complexes, they are very weak and RNAs may interact with the protein by other types of interaction forces.
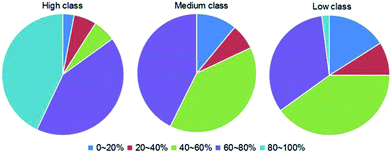
![[P with combining macron]](https://www.rsc.org/images/entities/i_char_0050_0304.gif) h) of each complex. The values are distributed between 0% and 100%, and the details are listed in ESI Table S2.† Then, we divided
h) of each complex. The values are distributed between 0% and 100%, and the details are listed in ESI Table S2.† Then, we divided ![[P with combining macron]](https://www.rsc.org/images/entities/i_char_0050_0304.gif) h into five categories by the values, which are 0–20%, 20–40%, 40–60%, 60–80% and 80–100%, respectively. The distributions of the five categories of
h into five categories by the values, which are 0–20%, 20–40%, 40–60%, 60–80% and 80–100%, respectively. The distributions of the five categories of ![[P with combining macron]](https://www.rsc.org/images/entities/i_char_0050_0304.gif) h in each class are shown in Fig. 9. For almost all complexes of high class (30/33), more than half of the interface residues are involved in hydrophobic interactions. Moreover, 15 complexes in high class have more than 80% overlap between binding interface and hydrophobic interface. However, no complexes were found in medium to have such a high percent overlap, and only 1 was found in low class. These results suggest that hydrophobic interactions play a much more important role in high class complexes than in medium and low, since hydrophobic interactions are defined as non-polar atoms that are ≤5 Å apart in the ENTANGLE package.46,62 In contrast to the phosphates and the bases, due to the presence of the 2′OH, it is harder for the ribose in RNA to form hydrophobic interactions.27
h in each class are shown in Fig. 9. For almost all complexes of high class (30/33), more than half of the interface residues are involved in hydrophobic interactions. Moreover, 15 complexes in high class have more than 80% overlap between binding interface and hydrophobic interface. However, no complexes were found in medium to have such a high percent overlap, and only 1 was found in low class. These results suggest that hydrophobic interactions play a much more important role in high class complexes than in medium and low, since hydrophobic interactions are defined as non-polar atoms that are ≤5 Å apart in the ENTANGLE package.46,62 In contrast to the phosphates and the bases, due to the presence of the 2′OH, it is harder for the ribose in RNA to form hydrophobic interactions.27
4. Conclusions
Protein–RNA interactions play important roles in a wide variety of biological processes.4–7 The different structures or conformations of RNA molecules may influence the binding protein sites.23,28 Moreover, the base is a special part of RNA and is frequently involved in important interactions.22,24,26,48 In the study, to qualitatively and further quantitatively measure the influence of the RNA composition on protein–RNA interactions, we firstly proposed a new standard to distinguish protein–RNA complexes based on the percentage of the base area buried in the RNA interface area. As a result, a dataset of 137 protein–RNA complexes was divided into three classes (high, medium and low). We comprehensive analyzed the properties of protein–RNA interactions, including interface compositions, interface structures, intermolecular physicochemical properties and interface forces, and also analyzed the difference between the three class complexes as well as compared them with protein–DNA interfaces reported in previous research.12,13,47,53 The results are clear: complexes in high class have the shortest RNAs and the RNA structures are mainly single stranded, which facilitates the interaction of the flipped or exposed base group with proteins. These complexes behave like protein–ssDNA interactions. Among the five types of interactions, H-bonding and hydrophobic interaction are strong, while the electrostatic interaction is weak. The complexes in medium have the longest RNAs and the largest interface area; however, the interface ratio is the smallest. The linear correlation between IA and number of interface nucleotides is the worst because of the irregular and more complicated RNA structures. Meanwhile, the interaction forces do not show any preference. In low class, the interface area distribution is similar with that of high class. The RNA structures are mainly double-stranded and behave like protein–dsDNA interactions. The interface propensity of Lys is high. Compared to high class, the electrostatic interaction is strong, while stacking and hydrophobic interactions are very weak. According to our classification strategy, the three classes of complexes have significant differences in terms of almost all properties. Unlike the high or low complexes, we cannot easily understand the specificity of the recognition processes of medium complexes based on the interface features. Therefore, we would pay more attention to the medium complexes in the future. Moreover, it is necessary to develop specific predictors for complexes in different classes, and different classes of protein–RNA complexes should be studied individually. Our study proves that the size of interface area contributed by the RNA base group can highly impact the properties of RNA–binding proteins and may play an important role in understanding the mechanism of protein–RNA interactions.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Funding: this work was funded by the National Natural Science Foundation of China (No. 21675114, 21573151).References
- R. Rohs, S. M. West, A. Sosinsky, P. Liu, R. S. Mann and B. Honig, Nature, 2009, 461, 1248–1253 CrossRef CAS PubMed.
- V. Charoensawan, D. Wilson and S. A. Teichmann, Nucleic Acids Res., 2010, 38, 7364–7377 CrossRef CAS PubMed.
- Y. H. Cai and H. Huang, Amino Acids, 2012, 43, 1141–1146 CrossRef CAS PubMed.
- A. Re, T. Joshi, E. Kulberkyte, Q. Morris and C. T. Workman, Methods Mol. Biol., 2014, 1097, 491–521 CAS.
- J. L. Rinn and J. Ule, Genome Biol., 2014, 15, 401 CrossRef PubMed.
- H. F. Noller, Science, 2005, 309, 1508–1514 CrossRef CAS PubMed.
- D. J. Hogan, D. P. Riordan, A. P. Gerber, D. Herschlag and P. O. Brown, PLoS Biol., 2008, 6, e255 Search PubMed.
- N. M. Luscombe, S. E. Austin, H. M. Berman and J. M. Thornton, Genome Biol., 2000, 1, REVIEWS001 CrossRef CAS PubMed.
- S. Jones, P. van Heyningen, H. M. Berman and J. M. Thornton, J. Mol. Biol., 1999, 287, 877–896 CrossRef CAS PubMed.
- B. A. Lewis, R. R. Walia, M. Terribilini, J. Ferguson, C. Zheng, V. Honavar and D. Dobbs, Nucleic Acids Res., 2011, 39, D277–D282 CrossRef CAS PubMed.
- D. D. Kirsanov, O. N. Zanegina, E. A. Aksianov, S. A. Spirin, A. S. Karyagina and A. V. Alexeevski, Nucleic Acids Res., 2013, 41, D517–D523 CrossRef CAS PubMed.
- J. Yan, S. Friedrich and L. Kurgan, Briefings Bioinf., 2016, 17, 88–105 CrossRef PubMed.
- S. Sonavane and P. Chakrabarti, Nucleic Acids Res., 2009, 37, 4613–4620 CrossRef CAS PubMed.
- Z. Miao and E. Westhof, Nucleic Acids Res., 2015, 43, 5340–5351 CrossRef CAS PubMed.
- J. Luo, L. Liu, S. Venkateswaran, Q. Song and X. Zhou, Sci. Rep., 2017, 7, 614 CrossRef PubMed.
- V. Suresh, L. Liu, D. Adjeroh and X. Zhou, Nucleic Acids Res., 2015, 43, 1370–1379 CrossRef CAS PubMed.
- J. Si, J. Cui, J. Cheng and R. Wu, Int. J. Mol. Sci., 2015, 16, 26303–26317 CrossRef CAS PubMed.
- Z. P. Liu, S. Liu, R. Chen, X. Huang and L. Y. Wu, BMC Bioinf., 2017, 18, 27 CrossRef PubMed.
- S. Jones, D. T. Daley, N. M. Luscombe, H. M. Berman and J. M. Thornton, Nucleic Acids Res., 2001, 29, 943–954 CrossRef CAS PubMed.
- R. Nagarajan, S. P. Chothani, C. Ramakrishnan, M. Sekijima and M. M. Gromiha, Biol. Direct, 2015, 10, 8 CrossRef PubMed.
- Z. Ghaemi, I. Guzman, D. Gnutt, Z. Luthey-Schulten and M. Gruebele, J. Phys. Chem. B, 2017, 121, 8437–8446 CrossRef CAS PubMed.
- R. P. Bahadur, M. Zacharias and J. Janin, Nucleic Acids Res., 2008, 36, 2705–2716 CrossRef CAS PubMed.
- E. Kligun and Y. Mandel-Gutfreund, RNA Biol., 2015, 12, 720–727 CrossRef PubMed.
- A. Gupta and M. Gribskov, J. Mol. Biol., 2011, 409, 574–587 CrossRef CAS PubMed.
- N. Morozova, J. Allers, J. Myers and Y. Shamoo, Bioinformatics, 2006, 22, 2746–2752 CrossRef CAS PubMed.
- J. Iwakiri, H. Tateishi, A. Chakraborty, P. Patil and N. Kenmochi, Nucleic Acids Res., 2012, 40, 3299–3306 CrossRef CAS PubMed.
- A. Barik and R. P. Bahadur, Nucleic Acids Res., 2014, 42, 10148–10160 CrossRef CAS PubMed.
- A. Barik, S. P. Pilla and R. P. Bahadur, J. Biomol. Struct. Dyn., 2015, 33, 2738–2751 CAS.
- E. Jeong, H. Kim, S. W. Lee and K. Han, Mol. Cells, 2003, 16, 161–167 CAS.
- H. M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov and P. E. Bourne, Nucleic Acids Res., 2000, 28, 235–242 CrossRef CAS PubMed.
- E. Krissinel and K. Henrick, J. Mol. Biol., 2007, 372, 774–797 CrossRef CAS PubMed.
- W. Li and A. Godzik, Bioinformatics, 2006, 22, 1658–1659 CrossRef CAS PubMed.
- S. J. Hubbard and J. M. Thornton, NACCESS: Program for calculating accessibilities, Department of Biochemistry and Molecular Biology, University College of London, UK, 1998 Search PubMed.
- A. Barik, A. Mishra and R. P. Bahadur, Nucleic Acids Res., 2012, 40, W440–W444 CrossRef CAS PubMed.
- O. T. Kim, K. Yura and N. Go, Nucleic Acids Res., 2006, 34, 6450–6460 CrossRef CAS PubMed.
- H. Yang, F. Jossinet, N. Leontis, L. Chen, J. Westbrook, H. Berman and E. Westhof, Nucleic Acids Res., 2003, 31, 3450–3460 CrossRef CAS PubMed.
- N. B. Leontis and E. Westhof, RNA, 2001, 7, 499–512 CrossRef CAS PubMed.
- R. P. Bahadur, P. Chakrabarti, F. Rodier and J. Janin, J. Mol. Biol., 2004, 336, 943–955 CrossRef CAS PubMed.
- M. Heinig and D. Frishman, Nucleic Acids Res., 2004, 32, W500–W502 CrossRef CAS PubMed.
- I. K. McDonald and J. M. Thornton, J. Mol. Biol., 1994, 238, 777–793 CrossRef CAS PubMed.
- I. Paz, E. Kligun, B. Bengad and Y. Mandel-Gutfreund, Nucleic Acids Res., 2016, 44, W568–W574 CrossRef CAS PubMed.
- S. Shazman and Y. Mandel-Gutfreund, PLoS Comput. Biol., 2008, 4, e1000146 Search PubMed.
- H. Sun, Y. Li, M. Shen, S. Tian, L. Xu, P. Pan, Y. Guan and T. Hou, Phys. Chem. Chem. Phys., 2014, 16, 22035–22045 RSC.
- B. R. Miller, T. D. McGee, J. M. Swails, N. Homeyer, H. Gohlke and A. E. Roitberg, J. Chem. Theory Comput., 2012, 8, 3314–3321 CrossRef CAS PubMed.
- D. A. Case, D. S. Cerutti, T. E. Cheatham III, T. A. Darden, R. E. Duke, T. J. Giese, H. Gohlke, A. W. Goetz, D. Greene, N. Homeyer, S. Izadi, A. Kovalenko, T. S. Lee, S. LeGrand, P. Li, C. Lin, J. Liu, T. Luchko, R. Luo, D. Mermelstein, K. M. Merz, G. Monard, H. Nguyen, I. Omelyan, A. Onufriev, F. Pan, R. Qi, D. R. Roe, A. Roitberg, C. Sagui, C. L. Simmerling, W. M. Botello-Smith, J. Swails, R. C. Walker, J. Wang, R. M. Wolf, X. Wu, L. Xiao, D. M. York and P. A. Kollman, Amber, University of California, San Francisco, 2017 Search PubMed.
- J. Allers and Y. Shamoo, J. Mol. Biol., 2001, 311, 75–86 CrossRef CAS PubMed.
- D. Lejeune, N. Delsaux, B. Charloteaux, A. Thomas and R. Brasseur, Proteins, 2005, 61, 258–271 CrossRef CAS PubMed.
- S. M. Lima, D. S. Peabody, J. L. Silva and A. C. de Oliveira, Eur. J. Biochem., 2004, 271, 135–145 CrossRef CAS PubMed.
- J. Luo, Y. Guo, Y. Fu, Y. Wang, W. Li and M. Li, Proteins, 2014, 82, 3090–3100 CrossRef CAS PubMed.
- H. Zhu, F. S. Domingues, I. Sommer and T. Lengauer, BMC Bioinf., 2006, 7, 27 CrossRef PubMed.
- J. Luo, Y. Guo, Y. Zhong, D. Ma, W. Li and M. Li, J. Comput.-Aided Mol. Des., 2014, 28, 619–629 CrossRef CAS PubMed.
- R. P. Saha, R. P. Bahadur, A. Pal, S. Mandal and P. Chakrabarti, BMC Struct. Biol., 2006, 6, 11 CrossRef PubMed.
- K. Nadassy, S. J. Wodak and J. Janin, Biochemistry, 1999, 38, 1999–2017 CrossRef CAS PubMed.
- J. Luo, Z. Liu, Y. Guo and M. Li, Sci. Rep., 2015, 5, 14214 CrossRef CAS PubMed.
- J. J. Ellis, M. Broom and S. Jones, Proteins, 2007, 66, 903–911 CrossRef CAS PubMed.
- D. E. Draper, J. Mol. Biol., 1999, 293, 255–270 CrossRef CAS PubMed.
- S. Shazman, G. Celniker, O. Haber, F. Glaser and Y. Mandel-Gutfreund, Nucleic Acids Res., 2007, 35, W526–W530 CrossRef PubMed.
- S. Jones, H. P. Shanahan, H. M. Berman and J. M. Thornton, Nucleic Acids Res., 2003, 31, 7189–7198 CrossRef CAS PubMed.
- N. Bhardwaj, R. E. Langlois, G. Zhao and H. Lu, Nucleic Acids Res., 2005, 33, 6486–6493 CrossRef CAS PubMed.
- B. Honig, K. Sharp and M. Gilson, Prog. Clin. Biol. Res., 1989, 289, 65–74 CAS.
- C. Nilofer, A. Sukhwal, A. Mohanapriya and P. Kangueane, Bioinformation, 2017, 13, 164–173 CrossRef PubMed.
- N. Morozova, J. Allers, J. Myers and Y. Shamoo, Bioinformatics, 2006, 22, 2746–2752 CrossRef CAS PubMed.
- D. Chandler, Nature, 2005, 437, 640–647 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra00598b |
| This journal is © The Royal Society of Chemistry 2018 |