Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Prediction of the targets of the main components in blood after oral administration of Xanthii Fructus: a network pharmacology study†
Yanshuang Zhuang‡
 a,
Kunming Qin‡bcd,
Bing Yang‡a,
Xiao Liu‡a,
Baochang Cai‡*ab and
Hao Cai‡*a
a,
Kunming Qin‡bcd,
Bing Yang‡a,
Xiao Liu‡a,
Baochang Cai‡*ab and
Hao Cai‡*a
aEngineering Center of State Ministry of Education for Chinese Medicine Processing, Nanjing University of Chinese Medicine, Nanjing 210023, China. E-mail: haocai_98@126.com; 295331981@qq.com; bingbingyang_2012@163.com; 1498223254@qq.com; Fax: +86-25-68193500; Tel: +86 13770500190 Tel: +86 13585148874
bNanjing Haichang Chinese Medicine Group Co., Ltd., Nanjing 210061, China. E-mail: qinkm123@126.com; baochangcai_2012@126.com
cNanjing Haiyuan Prepared Slices of Chinese Crude Drugs Co., Ltd., Nanjing 210061, China
dHuaihai Institute of Technology, Lianyu Gang 222005, China
First published on 27th February 2018
Abstract
Xanthii Fructus (XF), a famous traditional Chinese medicine (TCM), has been widely used in the treatment of rhinitis and other diseases. However, the targets of the main XF components found in the blood after oral administration of XF extract are still unclear. In the current study, a feasible systems pharmacology method was developed to predict these targets. In accordance with our previous research, XF components were selected including cleomiscosin A, myristic acid, succinic acid, xanthosine, sitostenone, emodin, apigenin, and chrysophanol. Three components, namely emodin, apigenin, and chrysophanol, failed to be detected with target proteins, thus the other five components, namely cleomiscosin A, myristic acid, succinic acid, xanthosine and sitostenone, were eventually chosen for further systematic analysis. Ninety-nine target proteins and fifty-two pathways were found after a series of analyses. The frequency of some target proteins was much higher than that of others; high frequencies were obtained for P15086, P07360, P07195, MAOM_HUMAN (P23368), P35558, P35520, ACE_HUMAN (P12821), C1S_HUMAN (P09871), PH4H_HUMAN (P00439), FPPS_HUMAN (P14324), P50613, P12724, IMPA1_HUMAN (P29218), HXK1_HUMAN (P19367), P14061, and MCR_HUMAN (P08235). The frequency of eight pathways was also high, including Generic Transcription Pathway, RNA Polymerase II Transcription, Metabolism, Metabolism of steroids, Gene expression (Transcription), Cellular responses to stress, Platelet activation, signaling and aggregation, Signaling by Receptor Tyrosine Kinases, and Cellular Senescence. This study identified a common pathway – the Metabolism pathway – for all five XF components. We successfully developed a network pharmacology method to predict the potential targets of the main XF components absorbed in serum after oral administration of XF extract.
1. Introduction
Over thousands of years, abundant clinical experience has accumulated in the use of traditional Chinese medicine (TCM). TCM has exerted synergistic effects in the treatment of complex diseases with its multi-component properties and multi-target functioning, creating a difficult challenge for its modernization. Recently, network pharmacology has risen rapidly in the research field. It explores drug targets by finding the overall correlation between drugs and diseases when combined with systems biology, multidirectional pharmacology and multidisciplinary technology, such as in network analysis, computational biology and disease-gene–drug network construction. It could therefore provide a new approach for overcoming barricades in the way of TCM modernization.Network pharmacology, based on the network of “disease-gene-target-drug” interactions, is a way of revealing the synergistic effects of complex drugs on the human system and finding efficient and low toxicity multi-target new drugs at the network level by observing the intervention of drugs and their impact on disease. With information databases such as gene network libraries, protein network libraries, disease network libraries, and drug network libraries, and systematic spectrogram data analysis, network pharmacology is able to reveal mysterious disease–disease, disease phenotype-target protein, target protein–drug and drug–drug linkages.1–7
Uncovering the material basis of TCM is the key and precondition for TCM quality control, which puts it at the core of TCM modernization. In a network pharmacology study, drug–drug networks can be constructed based on the similarities in the structures and efficacies of different drugs. In the process of TCM modernization, some researchers have achieved good initial results in exploring the essential properties of TCMs and revealing their comprehensive overall effects on multi-pathways, multi-targets and multi-components via the research ideas of network pharmacology.8–11
Xanthii Fructus (XF) is the ripe fruit of Xanthium sibiricum Patr. XF is used for the treatment of cramping, numbness of the limbs, ulcers, sinusitis, catarrhs, and pruritus, for its function in smoothing nasal orifices and eliminating wind-dampness.12 In modern clinic application, XF is commonly used for the treatment of rhinitis. Particularly when combined with Magnoliae flos, mint and other Chinese medicines, XF has enhanced effects in curing chronic rhinitis, allergic rhinitis and other rhinitis.13
2. Materials and methods
2.1 Screening active ingredients
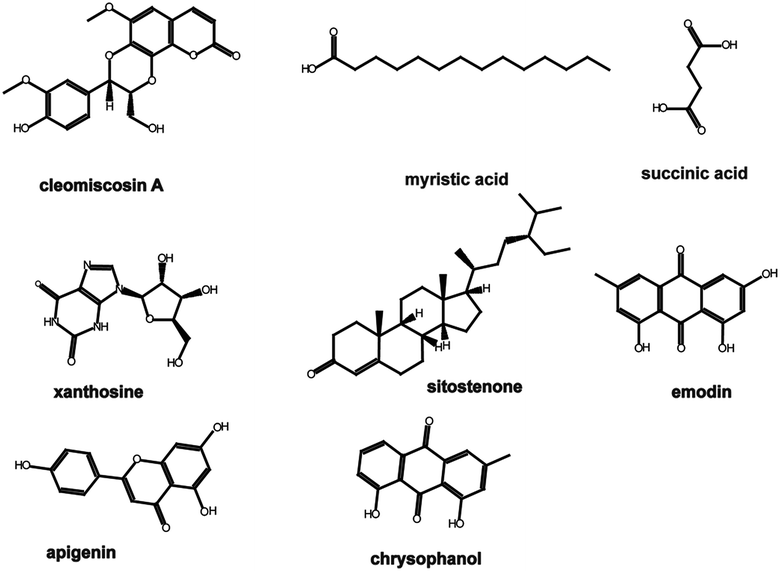
In our previous study (unpublished), components such as myristic acid, succinic acid, xanthosine, emodin, apigenin, and chrysophanol were identified from serum samples after oral administration of XF extracts. Components such as cleomiscosin A and sitostenone were filtered using the traditional Chinese medicine systems pharmacology (TcmSP™) database, and the parameters were set as follows: oral bioavailability (OB) ≥ 30%, drug-likeness (DL) ≥ 0.18. The structures of the components mentioned above are shown in Fig. 1.2.2 Prediction of active component targets
Firstly, the MDL SD (*.sdf) type files of the above active ingredients were searched using the PubMed database. Secondly, targets, including information like the target name, matching value, target protein abbreviation, function, disease and applicable results related to the modified compound, could be predicted by importing each component file in *sdf format into the PharmMapper database. The top 20 high-matching targets, by value, were used as the TCM target proteins related to the components. The targets were then searched for in the UniProt database to identify human-related target codes.2.3 Pathway comments and analysis
The retrieved target protein information was analyzed using the Reactome database to obtain the result of the related pathway “pathwayIdexByPathway_kegg”. A pathway was selected as reliable when its P value was less than 0.01.2.4 Drug-target-pathway relationship
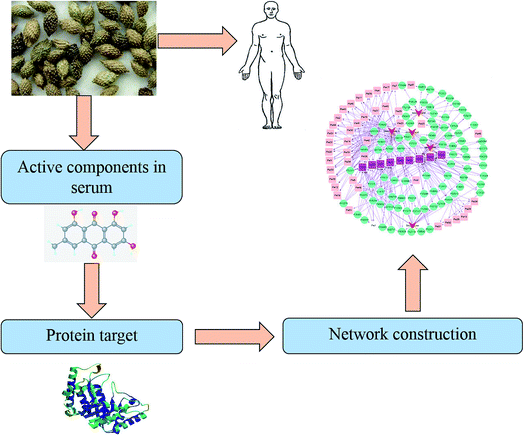
The predicted targets of five chemical components of XF, namely cleomiscosin A, myristic acid, succinic acid, xanthosine and sitostenone, were recorded in excel tables titled as ‘component-protein’ and ‘protein-pathway’. The tables were imported into Cytoscape software to construct the main effect components of the XF-target-pathway network. The network was mainly composed of three types of nodes: effect component, protein and pathway. The effect components and their related target proteins, and the proteins and their related pathways were all side-linked. When the target protein of the effect component was the same as the target protein of the pathway, the effect component was side-linked to the pathway. A complete network diagram was built by the establishment of connections including effect component-protein-pathway, effect component-protein-effect component, pathway-protein-pathway, protein-effect component-protein and other four kinds of connection. The whole framework, based on the active component strategy of system pharmacology, is shown in Fig. 2.3. Results
3.1 Potential target information for five components in XF
Eight components in XF were initially selected to uncover potential target proteins. Of these, five components, namely cleomiscosin A, myristic acid, succinic acid, xanthosine, and sitostenone, were successfully analyzed. A total of 99 target proteins were related to these five XF components as shown in Table 1. The frequency of some target proteins was much higher than that of others; high frequencies were obtained for P15086, P07360, P07195, MAOM_HUMAN (P23368), P35558, P35520, ACE_HUMAN (P12821), C1S_HUMAN (P09871), PH4H_HUMAN (P00439), FPPS_HUMAN (P14324), P50613, P12724, IMPA1_HUMAN (P29218), HXK1_HUMAN (P19367), P14061, and MCR_HUMAN (P08235).| No. | Compound | Protein code | Protein name | Frequency |
|---|---|---|---|---|
| 1 | Cleomiscosin A | P06276 | CHLE_HUMAN | 3 |
| 2 | Cleomiscosin A | P23141 | EST1_HUMAN | 3 |
| 3 | Cleomiscosin A | P62937 | P62937 | 3 |
| 4 | Cleomiscosin A | P00918 | CAH2_HUMAN | 3 |
| 5 | Cleomiscosin A | P24941 | P24941 | 3 |
| 6 | Cleomiscosin A | P07339![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
CATD_HUMAN | 3 |
| 7 | Cleomiscosin A | P03372 | ESR1_HUMAN | 3 |
| 8 | Cleomiscosin A | Q15078 | CD5R1_HUMAN | 3 |
| 9 | Cleomiscosin A | P00915 | CAH1_HUMAN | 3 |
| 10 | Cleomiscosin A | P04062 | GLCM_HUMAN | 3 |
| 11 | Cleomiscosin A | P11309 | PIM1_HUMAN | 3 |
| 12 | Cleomiscosin A | P00491 | PNPH_HUMAN | 3 |
| 13 | Cleomiscosin A | Q9NP99 | Q9NP99 | 3 |
| 14 | Cleomiscosin A | O14965 | STK6_HUMAN | 3 |
| 15 | Cleomiscosin A | Q16539 | Q16539 | 4 |
| 16 | Cleomiscosin A | Q92731 | ESR2_HUMAN | 3 |
| 17 | Cleomiscosin A | Q07343 | PDE4B_HUMAN | 4 |
| 18 | Cleomiscosin A | O14757 | CHK1_HUMAN | 4 |
| 19 | Cleomiscosin A | P45983 | MK08_HUMAN | 4 |
| 20 | Cleomiscosin A | P08758 | ANXA5_HUMAN | 4 |
| 21 | Myristic acid | P12643 | BMP2_HUMAN | 3 |
| 22 | Myristic acid | P28482 | MK01_HUMAN | 3 |
| 23 | Myristic acid | P09211 | GSTP1_HUMAN | 3 |
| 24 | Myristic acid | P15121 | ALDR_HUMAN | 3 |
| 25 | Myristic acid | P49137 | P49137 | 3 |
| 26 | Myristic acid | P10828 | P10828 | 3 |
| 27 | Myristic acid | P11309 | P11309 | 3 |
| 28 | Myristic acid | P27338 | AOFB_HUMAN | 3 |
| 29 | Myristic acid | P62937 | P62937 | 3 |
| 30 | Myristic acid | P02774 | VTDB_HUMAN | 4 |
| 31 | Myristic acid | P02768 | ALBU_HUMAN | 3 |
| 32 | Myristic acid | P52732 | KIF11_HUMAN | 4 |
| 33 | Myristic acid | P02652 | APOA2_HUMAN | 3 |
| 34 | Myristic acid | P00918 | CAH2_HUMAN | 3 |
| 35 | Myristic acid | P08842 | STS_HUMAN | 3 |
| 36 | Myristic acid | P02766 | TTHY_HUMAN | 3 |
| 37 | Myristic acid | Q14994![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
NR1I3_HUMAN | 3 |
| 38 | Myristic acid | P37231 | PPARG_HUMAN | 3 |
| 39 | Myristic acid | P30044![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
PRDX5_HUMAN | 3 |
| 40 | Succinic acid | P09012 | P09012 | 3 |
| 41 | Succinic acid | P02743 | P02743 | 4 |
| 42 | Succinic acid | P12931 | SRC_HUMAN | 4 |
| 43 | Succinic acid | O15382 | O15382 | 4 |
| 44 | Succinic acid | P18031 | PTN1_HUMAN | 4 |
| 45 | Succinic acid | P15086 | P15086 | 5 |
| 46 | Succinic acid | P07360 | P07360 | 5 |
| 47 | Succinic acid | P02788 | TRFL_HUMAN | 4 |
| 48 | Succinic acid | P03950 | ANGI_HUMAN | 4 |
| 49 | Succinic acid | P07195 | P07195 | 5 |
| 50 | Succinic acid | P23368 | MAOM_HUMAN | 5 |
| 51 | Succinic acid | Q9P2W7 | B3GA1_HUMAN | 4 |
| 52 | Succinic acid | P35558 | P35558 | 6 |
| 53 | Succinic acid | P35520 | P35520 | 7 |
| 54 | Succinic acid | P12821 | ACE_HUMAN | 7 |
| 55 | Succinic acid | P09871 | C1S_HUMAN | 6 |
| 56 | Succinic acid | P00439![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
PH4H_HUMAN | 5 |
| 57 | Succinic acid | P14324 | FPPS_HUMAN | 8 |
| 58 | Succinic acid | P50613 | P50613 | 8 |
| 59 | Succinic acid | P12724 | P12724 | 6 |
| 60 | Xanthosine | Q9BW91 | Q9BW91 | 3 |
| 61 | Xanthosine | P37173 | TGFR2_HUMAN | 3 |
| 62 | Xanthosine | P04062 | GLCM_HUMAN | 3 |
| 63 | Xanthosine | O14965![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
STK6_HUMAN | 3 |
| 64 | Xanthosine | Q13126 | Q13126 | 3 |
| 65 | Xanthosine | P00533![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
EGFR_HUMAN | 3 |
| 66 | Xanthosine | P24941 | P24941 | 4 |
| 67 | Xanthosine | Q07343 | PDE4B_HUMAN | 3 |
| 68 | Xanthosine | P00915 | CAH1_HUMAN | 3 |
| 69 | Xanthosine | Q12884 | SEPR_HUMAN | 3 |
| 70 | Xanthosine | O14757 | CHK1_HUMAN | 3 |
| 71 | Xanthosine | Q05315 | LPPL_HUMAN | 3 |
| 72 | Xanthosine | P04745 | P04745 | 3 |
| 73 | Xanthosine | P18075 | BMP7_HUMAN | 4 |
| 74 | Xanthosine | P03950 | ANGI_HUMAN | 4 |
| 75 | Xanthosine | P00491 | PNPH_HUMAN | 4 |
| 76 | Xanthosine | P29218 | IMPA1_HUMAN | 5 |
| 77 | Xanthosine | Q99933![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
BAG1_HUMAN | 4 |
| 78 | Xanthosine | P19367 | HXK1_HUMAN | 5 |
| 79 | Xanthosine | P17707 | DCAM_HUMAN | 4 |
| 80 | Sitostenone | P52895 | AK1C2_HUMAN | 3 |
| 81 | Sitostenone | P49137 | P49137 | 3 |
| 82 | Sitostenone | P55210 | CASP7_HUMAN | 3 |
| 83 | Sitostenone | P12643 | BMP2_HUMAN | 3 |
| 84 | Sitostenone | P08842 | STS_HUMAN | 3 |
| 85 | Sitostenone | P27338 | AOFB_HUMAN | 3 |
| 86 | Sitostenone | P02774![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
VTDB_HUMAN | 4 |
| 87 | Sitostenone | P11309 | P11309 | 3 |
| 88 | Sitostenone | P02768 | ALBU_HUMAN | 3 |
| 89 | Sitostenone | P28482 | MK01_HUMAN | 3 |
| 90 | Sitostenone | P45452 | MMP13_HUMAN | 4 |
| 91 | Sitostenone | P10828 | P10828 | 3 |
| 92 | Sitostenone | P52732 | KIF11_HUMAN | 3 |
| 93 | Sitostenone | P00918 | CAH2_HUMAN | 3 |
| 94 | Sitostenone | P14061 | P14061 | 5 |
| 95 | Sitostenone | P02652 | APOA2_HUMAN | 3 |
| 96 | Sitostenone | P08235 | MCR_HUMAN | 5 |
| 97 | Sitostenone | P06401 | PRGR_HUMAN | 4 |
| 98 | Sitostenone | P10275 | ANDR_HUMAN | 3 |
| 99 | Sitostenone | P02766 | TTHY_HUMAN | 3 |
3.2 Pathway analysis of potential target proteins
The potential pathway information for the five effect components in XF is shown in Table 2.| No. of pathway | Pathway name | Frequency |
|---|---|---|
| Pw1 | Nuclear receptor transcription pathway | 1 |
| Pw2 | Activation of the AP-1 family of transcription factors | 3 |
| Pw3 | MAPK targets/nuclear events mediated by MAP kinases | 10 |
| Pw4 | p38MAPK events | 3 |
| Pw5 | Generic Transcription Pathway | 42 |
| Pw6 | Transcriptional regulation by RUNX2 | 10 |
| Pw7 | Signalling to RAS | 3 |
| Pw8 | RNA polymerase II transcription | 73 |
| Pw9 | Regulation of TP53 Activity through phosphorylation | 5 |
| Pw10 | Metabolism | 68 |
| Pw11 | Nuclear events (kinase and transcription factor activation) | 7 |
| Pw12 | RUNX2 regulates osteoblast differentiation | 5 |
| Pw13 | Metabolism of steroids | 23 |
| Pw14 | MAP kinase activation in TLR cascade | 15 |
| Pw15 | Erythrocytes take up oxygen and release carbon dioxide | 1 |
| Pw16 | RUNX2 regulates bone development | 5 |
| Pw17 | Signalling to ERKs | 3 |
| Pw18 | Gene expression (transcription) | 80 |
| Pw19 | Interleukin-17 signaling | 15 |
| Pw20 | Digestion of dietary carbohydrate | 2 |
| Pw21 | Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation | 3 |
| Pw22 | DSCAM interactions | 2 |
| Pw23 | NGF signalling via TRKA from the plasma membrane | 10 |
| Pw24 | Reversible hydration of carbon dioxide | 2 |
| Pw25 | O2/CO2 exchange in erythrocytes | 2 |
| Pw26 | Erythrocytes take up carbon dioxide and release oxygen | 1 |
| Pw27 | Cellular responses to stress | 27 |
| Pw28 | MyD88 cascade initiated on plasma membrane | 15 |
| Pw29 | Toll like receptor 10 (TLR10) cascade | 15 |
| Pw30 | Toll like receptor 5 (TLR5) cascade | 15 |
| Pw31 | TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation | 15 |
| Pw32 | Platelet activation, signaling and aggregation | 32 |
| Pw33 | Oxidative stress induced senescence | 12 |
| Pw34 | MyD88 dependent cascade initiated on endosome | 15 |
| Pw35 | Toll like receptor 7/8 (TLR7/8) cascade | 15 |
| Pw36 | MyD88:Mal cascade initiated on plasma membrane | 15 |
| Pw37 | Toll like receptor TLR6:TLR2 cascade | 15 |
| Pw38 | Spry regulation of FGF signaling | 2 |
| Pw39 | Netrin-1 signaling | 12 |
| Pw40 | Toll like receptor 9 (TLR9) cascade | 15 |
| Pw41 | Toll like receptor 3 (TLR3) cascade | 15 |
| Pw42 | Toll like receptor TLR1:TLR2 cascade | 15 |
| Pw43 | Toll like receptor 2 (TLR2) cascade | 15 |
| Pw44 | TRIF(TICAM1)-mediated TLR4 signaling | 15 |
| Pw45 | MyD88-independent TLR4 cascade | 15 |
| Pw46 | Defective HK1 causes hexokinase deficiency (HK deficiency) | 1 |
| Pw47 | Metabolism of angiotensinogen to angiotensins | 4 |
| Pw48 | Regulation of TP53 Activity | 6 |
| Pw49 | Signaling by receptor tyrosine kinases | 81 |
| Pw50 | Cellular senescence | 21 |
| Pw51 | HSP90 chaperone cycle for steroid hormone receptors (SHR) | 3 |
| Pw52 | Interleukin-12 family signaling | 3 |
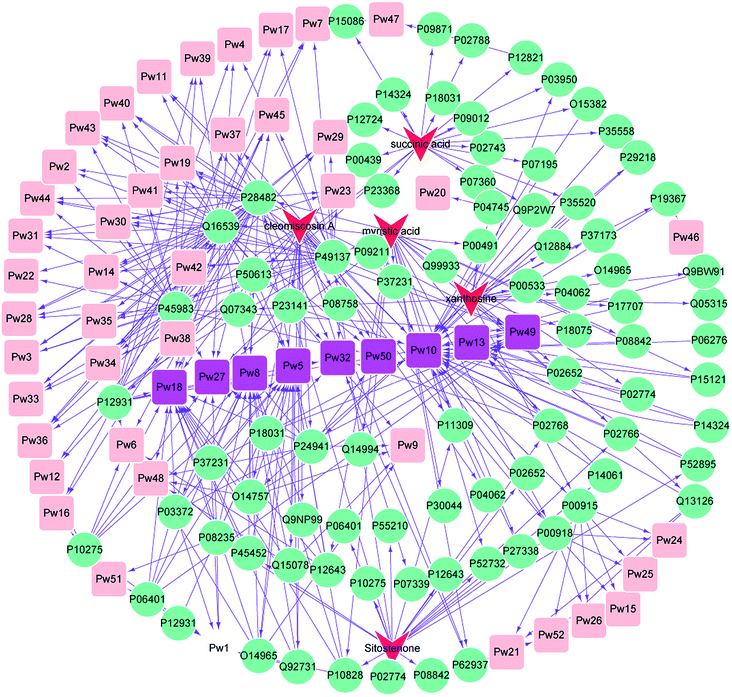
3.3 Main effect component-target protein-pathway network construction for XF
An effect component-target-pathway network model was established using Cytoscape software, and the relationship between the 5 components, 99 targets and 52 pathways is shown in Fig. 3. There were complex network relationships between the effect components of XF and their targets, as well as the targets and pathways.Cleomiscosin A was related to the following pathways: nuclear receptor transcription pathway (Pw1), activation of the AP-1 family of transcription factors (Pw2), MAPK targets/nuclear events mediated by MAP kinases (Pw3), p38MAPK events (Pw4), Generic Transcription Pathway (Pw5), Transcriptional regulation by RUNX2 (Pw6), Signalling to RAS (Pw7), RNA Polymerase II Transcription (Pw8), Regulation of TP53 Activity through Phosphorylation (Pw9), Metabolism (Pw10), Nuclear Events (kinase and transcription factor activation) (Pw11), MAP kinase activation in TLR cascade (Pw14), erythrocytes take up oxygen and release carbon dioxide (Pw15), Signalling to ERKs (Pw17), Gene expression (Transcription) (Pw18), Interleukin-17 signaling (Pw19), Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation (Pw21), DSCAM interactions (Pw22), NGF signalling via TRKA from the plasma membrane (Pw23), Reversible hydration of carbon dioxide (Pw24), O2/CO2 exchange in erythrocytes (Pw25), erythrocytes take up carbon dioxide and release oxygen (Pw26), cellular responses to stress (Pw27), MyD88 cascade initiated on plasma membrane (Pw28), Toll Like Receptor 10 (TLR10) Cascade (Pw29), Toll Like Receptor 5 (TLR5) Cascade (Pw30), TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation (Pw31), platelet activation, signaling and aggregation (Pw32), oxidative stress induced senescence (Pw33), MyD88 dependent cascade initiated on endosome (Pw34), Toll Like Receptor 7/8 (TLR7/8) Cascade (Pw35), MyD88:Mal cascade initiated on plasma membrane (Pw36), Toll Like Receptor TLR6:TLR2 Cascade (Pw37), Netrin-1 signaling (Pw39), Toll Like Receptor 9 (TLR9) Cascade (Pw40), Toll Like Receptor 3 (TLR3) Cascade (Pw41), Toll Like Receptor TLR1:TLR2 Cascade (Pw42), Toll Like Receptor 2 (TLR2) Cascade (Pw43), TRIF(TICAM1)-mediated TLR4 signaling (Pw44), MyD88-independent TLR4 cascade (Pw45), Regulation of TP53 Activity (Pw48), Signaling by Receptor Tyrosine Kinases (Pw49), Cellular Senescence (Pw50) and Interleukin-12 family signaling (Pw52).
Myristic acid was related to the following pathways: nuclear receptor transcription pathway (Pw1), Activation of the AP-1 family of transcription factors (Pw2), MAPK targets/nuclear events mediated by MAP kinases (Pw3), p38MAPK events (Pw4), Generic Transcription Pathway (Pw5), Transcriptional regulation by RUNX2 (Pw6), Signalling to RAS (Pw7), RNA Polymerase II Transcription (Pw8), Metabolism (Pw10), Nuclear Events (kinase and transcription factor activation) (Pw11), RUNX2 regulates osteoblast differentiation (Pw12), Metabolism of steroids (Pw13), MAP kinase activation in TLR cascade (Pw14), RUNX2 regulates bone development (Pw16), Signalling to ERKs (Pw17), Gene expression (Transcription) (Pw18), Interleukin-17 signaling (Pw19), NGF signalling via TRKA from the plasma membrane (Pw23), Cellular responses to stress (Pw27), MyD88 cascade initiated on plasma membrane (Pw28), Toll Like Receptor 10 (TLR10) Cascade (Pw29), Toll Like Receptor 5 (TLR5) Cascade (Pw30), TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation (Pw31), Platelet activation, signaling and aggregation (Pw32), Oxidative Stress Induced Senescence (Pw33), MyD88 dependent cascade initiated on endosome (Pw34), Toll Like Receptor 7/8 (TLR7/8) Cascade (Pw35), MyD88:Mal cascade initiated on plasma membrane (Pw36), Toll Like Receptor TLR6:TLR2 Cascade (Pw37), Spry regulation of FGF signaling (Pw38), Toll Like Receptor 9 (TLR9) Cascade (Pw40), Toll Like Receptor 3 (TLR3) Cascade (Pw41), Toll Like Receptor TLR1:TLR2 Cascade (Pw42), Toll Like Receptor 2 (TLR2) Cascade (Pw43), TRIF(TICAM1)-mediated TLR4 signaling (Pw44), MyD88-independent TLR4 cascade (Pw45), Signaling by Receptor Tyrosine Kinases (Pw49) and Cellular Senescence (Pw50).
Succinic acid was related to the following pathways: p38MAPK events (Pw4), Generic Transcription Pathway (Pw5), Transcriptional regulation by RUNX2 (Pw6), Signalling to RAS (Pw7), RNA Polymerase II Transcription (Pw8), Metabolism (Pw10), RUNX2 regulates osteoblast differentiation (Pw12), Metabolism of steroids (Pw13), RUNX2 regulates bone development (Pw16), Signalling to ERKs (Pw17), Gene expression (Transcription) (Pw18), NGF signalling via TRKA from the plasma membrane (Pw23), Platelet activation, signaling and aggregation (Pw32), Spry regulation of FGF signaling (Pw38), Netrin-1 signaling (Pw39), Metabolism of Angiotensinogen to Angiotensins (Pw47) and Signaling by Receptor Tyrosine Kinases (Pw49).
Xanthosine was related to the following pathways: Metabolism (Pw10), Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation (Pw21), Defective HK1 causes hexokinase deficiency (HK deficiency) (Pw46) and Interleukin-12 family signaling (Pw52).
Sitostenone was related to the following pathways: nuclear receptor transcription pathway (Pw1), Generic Transcription Pathway (Pw5), transcriptional regulation by RUNX2 (Pw6), Signalling to RAS (Pw7), RNA Polymerase II Transcription (Pw8), Metabolism (Pw10), Nuclear Events (kinase and transcription factor activation) (Pw11), RUNX2 regulates osteoblast differentiation (Pw12), Metabolism of steroids (Pw13), RUNX2 regulates bone development (Pw16), Gene expression (Transcription) (Pw18), Interleukin-17 signaling (Pw19), Cellular responses to stress (Pw27), Signaling by Receptor Tyrosine Kinases (Pw49) and HSP90 chaperone cycle for steroid hormone receptors (SHR) (Pw51).
We were surprised to find that the five components have one common pathway – the Metabolism pathway (Pw10). Nine other pathways occurred frequently including Generic Transcription Pathway (Pw5), RNA Polymerase II Transcription (Pw8), Metabolism (Pw10), Metabolism of steroids (Pw13), Gene expression (Transcription) (Pw18), Cellular responses to stress (Pw27), Platelet activation, signaling and aggregation (Pw32), Signaling by Receptor Tyrosine Kinases (Pw49) and Cellular Senescence (Pw50).
4. Discussion
The PharmMapper database can be used to search for potential targets based on small active molecules. This database uses a pharmacophore matching method to obtain drug point information by rapidly searching four major databases. This database is based on 7000 pharmacophore models and can cover most clinical indications.According to the network pharmacological prediction of the five components in XF, all five components can be connected with the same pathway via the same target, and also can be connected with the same pathways with different targets. Different components can produce the same effect through different ways, and also can offer multi-target synergy.
Interestingly, this predicted common pathway is consistent with the result we got from the metabolic pathway analysis experiment (unpublished), which indicates that this result is reliable although it still requires further verification.
5. Conclusion
In this paper, a network pharmacology method has been successfully developed to predict the potential targets of the main components absorbed in serum after oral administration of XF extract. When considered alongside our previous anti-allergic rhinitis metabolomics study, the predicted potential targets and the role of the pathways were considered to have a certain degree of accuracy. This article has established a “multi component-multi target-multi pathway” network model for TCM research, and started to unravel the multidimensional regulatory action of XF, which may provide a reference and basis for studying the molecular mechanism of XF.Conflicts of interest
The authors have declared no conflicts of interest.Acknowledgements
The study is supported by the Natural Science Foundation of China (no. 81573603), “Six Talents Peak Project” of Jiangsu Province (no. SWYY-108), Jiangsu Province Science and Technology Support Plan Project (no. BE2016626) and the Natural Science Foundation of Jiangsu Province (no. BK20161047).References
- J. Wang and X. J. Li, Drug targets discovery based on dynamic signal transduction networks, Acta Pharm. Sin., 2010, 45(1), 1–8 Search PubMed.
- N. Wang, Y. Zheng, J. Gu, Y. Cai, S. Wang, F. Zhang, J. Chen, H. Situ, Y. Lin and Z. Wang, Network-pharmacology-based validation of TAMS/CXCL-1 as key mediator of XIAOPI formula preventing breast cancer development and metastasis, Sci. Rep., 2017, 7(1), 14513–14528 CrossRef PubMed.
- M. Lyu, C. L. Yan, H. X. Liu, T. Y. Wang, X. H. Shi, J. P. Liu, J. Orgah, G. W. Fan, J. H. Han, X. Y. Wang and Y. Zhu, Network pharmacology exploration reveals endothelial inflammation as a common mechanism for stroke and coronary artery disease treatment of Danhong injection, Sci. Rep., 2017, 7(1), 15427–15445 CrossRef PubMed.
- M. Hong, S. Li, H. Y. Tan, F. Cheung, N. Wang, J. Huang and Y. Feng, A Network-Based Pharmacology Study of the Herb-Induced Liver Injury Potential of Traditional Hepatoprotective Chinese Herbal Medicines, Molecules, 2017, 22(4), 632–646 CrossRef PubMed.
- S. J. Yue, L. T. Xin, Y. C. Fan, S. J. Li, Y. P. Tang, J. A. Duan, H. S. Guan and C. Y. Wang, Herb pair Danggui-Honghua: mechanisms underlying blood stasis syndrome by system pharmacology approach, Sci. Rep., 2017, 11(7), 40318–40323 CrossRef PubMed.
- M. Hong, Y. S. Zhang, S. Li, H. Y. Tan, N. Wang, S. Mu, X. Hao and Y. Feng, A Network Pharmacology-Based Study on the Hepatoprotective Effect of Fructus Schisandrae, Molecules, 2017, 22(10), 1617–1628 CrossRef PubMed.
- A. H. Zhang, H. Fang, Y. Y. Wang, G. L. Yan, H. Sun, X. H. Zhou, Y. Y. Wang, L. Liu and X. J. Wang, Discovery and verification of the potential targets from bioactive molecules by network pharmacology-based target prediction combined with high-throughput metabolomics, RSC Adv., 2017, 7(81), 51069–51078 RSC.
- G. L. Kang, S. Li and J. F. Zhang, Entropy-based model for interpreting life systems in traditional Chinese medicine, Evid. Based Complement. Alternat. Med., 2008, 5(3), 273–279 CrossRef PubMed.
- M. Wu, C. H. Ma, Y. Wu and S. Li, Simultaneous LC analysis of five bioactive alkaloids in an anti-angiogenesis herbal formula, Chromatographia, 2008, 68(7–8), 579–585 CAS.
- S. Li, Network systems underlying traditional Chinese medicine syndrome and herb formula, Curr. Bioinf., 2009, 4(3), 188–196 CrossRef CAS.
- S. Li, B. Zhang and N. B. Zhang, Network target for screening synergistic drug combinations with application to traditional Chinese medicine, BMC Syst. Biol., 2011, 5(suppl. 1), S10 CrossRef PubMed.
- Chinese Pharmacopoeia Commission Pharmacopoeia of the People's Republic of China, China Medical Science and Technology Press, Beijing, 2015, p. 162, Part I Search PubMed.
- X. R. Cui, X. B. Ma, Q. Zhang, Q. S. Li, W. Wang, Q. J. Han, H. M. Lei and Q. Li, Research progress on the chemical composition and clinical application of Xanthii Fructus, Drugs Clin., 2012, 27(6), 614–618 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra00186c |
| ‡ Dr Baochang Cai and Dr Hao Cai are responsible for the idea behind this paper. Dr Yanshuang Zhuang is the main implementer of this paper. The remaining authors helped with this paper. |
| This journal is © The Royal Society of Chemistry 2018 |