Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Comparative lipidomic profiling of the human commensal bacterium Propionibacterium acnes and its extracellular vesicles†
Jinseong Jeon‡
a,
Seung Cheol Park‡ b,
Jin Herc,
Jae Won Lee
b,
Jin Herc,
Jae Won Lee b,
Jin-Kwan Hana,
Yoon-Keun Kimd,
Kwang Pyo Kim
b,
Jin-Kwan Hana,
Yoon-Keun Kimd,
Kwang Pyo Kim b and
Changill Ban
b and
Changill Ban *c
*c
aDepartment of Life Sciences, Pohang University of Science and Technology (POSTECH), 77, Cheongam-Ro, Nam-Gu, Pohang, Gyeongbuk 37673, Republic of Korea
bDepartment of Applied Chemistry, College of Applied Science, Kyung Hee University, Yongin, 17104, Republic of Korea
cDepartment of Chemistry, Pohang University of Science and Technology (POSTECH), 77, Cheongam-Ro, Nam-Gu, Pohang, Gyeongbuk 37673, Republic of Korea. E-mail: ciban@postech.ac.kr
dMyeongdong Medical, Woori Technology Building, World Cup buk-ro 56-gil 9, Mapo-gu, Seoul, 03923, Republic of Korea
First published on 23rd April 2018
Abstract
Propionibacterium acnes is a lipophilic commensal bacterium mainly found on the skin and in the gastrointestinal tract. Pathophysiological effects of P. acnes have recently been reported not only in acne progression but in various diseases. As an emerging mode of bacterial communication, extracellular vesicles (EVs) have been demonstrated to conduct critical pathophysiological functions. To provide information on P. acnes lipid composition for the first time, we conducted a comparative lipidomic analysis of P. acnes and P. acnes EVs and identified 214 lipids with high confidence using triplicated liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) analyses. P. acnes EVs contained substantially more PCs, DGs, PAs, PEs, LPAs, LPCs, and MGs than P. acnes, and contained fewer PSs, SO1Ps, SA1Ps, LPGs, LPIs, and LPSs. Distinctively, P. acnes EVs possessed a markedly reduced amount of TG. These findings will provide useful clues for understanding the biological and pathophysiological mechanisms of P. acnes and for clinical applications such as vaccine development, diagnostics and therapeutics.
Introduction
Propionibacterium acnes is a lipophilic commensal bacterium found on the skin and in the gastrointestinal tract, and has recently been demonstrated to be an underestimated pathogen in various diseases.1,2 P. acnes is known to be beneficial in normal quantities,3 although it is also well characterized as a direct instigator of inflammation in acne, as its secreted products are a significant factor in the etiology of acne.4,5 Several studies have recently reported that P. acnes can cause not only acne but various other diseases, such as chronic prostatitis leading to prostate cancer; synovitis, acne, pustulosis, hyperostosis, and osteitis (SAPHO) syndrome; sarcoidosis; sciatica; and implant-associated infections.2 To understand the etiological role of P. acnes in these diseases, it is important to analyze its secretions, which lead to host inflammatory responses and lipogenesis.Extracellular vesicles (EVs) are an emerging mode of intercellular communication found in all domains of life.6 EVs are lipid bilayer-enveloped organelles of 20–1000 nm diameter that contains various biomolecules such as DNA, RNA, proteins, and lipids.7,8 EVs are thought to be a major transportation system for extracellular lipids.9 Because of their nanoscale size, it is thought that EVs may transfer their cargo in high concentrations to microenvironments that the bacterium of origin cannot reach.10 Moreover, EVs have been shown to play a role in pathogenesis, so research on pathogenic bacteria and their EVs should ideally be performed concurrently.11 Therefore, the isolation of P. acnes EVs and the identification of their lipid components may provide novel insights into the pathophysiological role of P. acnes and potential targets for the development of diagnostics and vaccines against P. acnes.
Liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) is a powerful tool for analyzing lipid classes in biological samples.12–14 Moreover, the introduction of ultra-performance liquid chromatography (UPLC) allows the rapid and effective separation of individual lipid species.15 Multiple reaction monitoring (MRM) based on triple quadrupole (QqQ)/MS can also be used for sensitive and precise quantitative analysis of target compounds.16,17 Optimized UPLC-QqQ/MS-based lipidomics analysis has been used to provide accurate information on a wide variety of biological samples and broaden our understanding of many biological events.18–20
In this study, UPLC-QqQ/MS was used to profile lipids from P. acnes and its EVs for the first time. The goal of this paper was to suggest that P. acnes EV was an active biomolecule and to report the comparative lipid composition of P. acnes and P. acnes EV for the first time.
Experimental
Bacterial cell culture and EV isolation
P. acnes cultivation and EV isolation were performed as previously described.21 Briefly, the P. acnes 6919 type strain was cultured in brain heart infusion (BHI) broth at 37 °C in an anaerobic chamber (90% N2 and 10% CO2). The culture media were harvested at mid-exponential phase (OD600 = 1.0–1.5), centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 15 min at 4 °C, and then filtered through a 0.45 μm bottle-top filter. After ultrafiltration with a QuixStand benchtop system (GE Healthcare, UK) using a 100 kDa hollow fiber membrane, the concentrate was filtered through a 0.22 μm bottle-top filter. P. acnes EVs were purified from the final filtrate by ultra-centrifugation at 150
000 × g for 15 min at 4 °C, and then filtered through a 0.45 μm bottle-top filter. After ultrafiltration with a QuixStand benchtop system (GE Healthcare, UK) using a 100 kDa hollow fiber membrane, the concentrate was filtered through a 0.22 μm bottle-top filter. P. acnes EVs were purified from the final filtrate by ultra-centrifugation at 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 3 h at 4 °C and density gradient ultracentrifugation with 10%, 40%, and 50% OptiPrep solutions (Axis-Shield PoC AS, Norway) at 200
000 × g for 3 h at 4 °C and density gradient ultracentrifugation with 10%, 40%, and 50% OptiPrep solutions (Axis-Shield PoC AS, Norway) at 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 2 h at 4 °C. The diluted OptiPrep solutions were prepared in HEPES-buffered saline (20 mM HEPES, 150 mM NaCl, pH 7.4). The purified P. acnes EVs were reconstituted in Dulbecco's phosphate-buffered saline (DPBS), quantified by bicinchoninic acid (BCA) assay, and divided into several aliquots, which were stored at −80 °C until use.
000 × g for 2 h at 4 °C. The diluted OptiPrep solutions were prepared in HEPES-buffered saline (20 mM HEPES, 150 mM NaCl, pH 7.4). The purified P. acnes EVs were reconstituted in Dulbecco's phosphate-buffered saline (DPBS), quantified by bicinchoninic acid (BCA) assay, and divided into several aliquots, which were stored at −80 °C until use.
Transmission electron microscopy (TEM) and dynamic light scattering (DLS) analysis
Purified P. acnes EVs were characterized using TEM and DLS analysis. For TEM analysis, a single droplet of 100 μg mL−1 P. acnes EVs was loaded onto a 400-mesh copper grid. The grid was allowed to absorb the P. acnes EVs for 5 min, then washed with deionized water and negatively stained using 2% uranyl acetate. TEM images were obtained using a JEM-1011 transmission electron microscope (JEOL, Japan) at an accelerating voltage of 100 kV. The average size of P. acnes EVs was determined using a Zetasizer Nano series instrument (Malvern Instruments, UK) equipped with a 633 nm laser line at a scatter intensity of 10 × 30 s.Mammalian cell culture
The murine macrophage cell line Raw264.7 was obtained from Korean Cell Line Bank (Seoul, South Korea). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 U mL−1 penicillin, and 100 μg mL−1 streptomycin. Cells were incubated at 37 °C in a 5% CO2 humidified incubator.Analysis of in vitro cytokine production
The expression of pro-inflammatory cytokines from mouse peritoneal macrophages (RAW264.7 cells) was measured by enzyme-linked immunosorbent assay (ELISA; R&D Systems, USA) in accordance with the manufacturer's instructions. First, 2 × 105 cells were seeded in 12-well plates. After 24 h, the cells were incubated with 10–1000 ng mL−1 P. acnes EVs for 6–24 h. After incubation, the culture media supernatants were collected, and residual cells and debris were removed by centrifugation at 5000 × g for 5 min.Lipid extraction
All lipid standards were dissolved in methanol, stored at −27 °C, and diluted to the required concentrations for spiking. A modified two-step extraction method, designed for the high efficiency extraction of non-polar and polar lipids, was performed.22 First, for non-polar lipid extraction, 990 μL of methanol/chloroform (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) and 10 μL of 1 μg mL−1 lipid standards including TG (11
1, v/v) and 10 μL of 1 μg mL−1 lipid standards including TG (11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–11
1–11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–11
1–11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), DG (8
1), DG (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0–8
0–8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), MG (15
0), MG (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), ChE (10
1), ChE (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), PC (10
0), PC (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0–10
0–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), LPC (13
0), LPC (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), PE (10
0), PE (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0–10
0–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), LPE (14
0), LPE (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), PS (10
0), PS (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0–10
0–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), LPS (17
0), LPS (17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), PG (10
1), PG (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0–10
0–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), LPG (14
0), LPG (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), PI (8
0), PI (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0–8
0–8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), LPI (13
0), LPI (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), PA (10
0), PA (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0–10
0–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), LPA (14
0), LPA (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), So (d17
0), So (d17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), So1P (d17
1), So1P (d17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), Sa (d17
1), Sa (d17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), Sa1P (d17
0), Sa1P (d17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), SM (d18
0), SM (d18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–12
1–12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), Cer (d18
0), Cer (d18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–12
1–12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), dCer (d18
0), dCer (d18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–12
1–12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), Cer1P (d18
0), Cer1P (d18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–12
1–12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), and dCer1P (d18
0), and dCer1P (d18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–16
1–16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) were added to the cell pellet. The sample was vortexed intensely for 30 s every 3 min and centrifuged (14
0) were added to the cell pellet. The sample was vortexed intensely for 30 s every 3 min and centrifuged (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, 2 min, 4 °C). The supernatant was moved to a new tube. Second, for polar lipid extraction, 750 μL of methanol/chloroform/37% (1 N) HCl (80
000 × g, 2 min, 4 °C). The supernatant was moved to a new tube. Second, for polar lipid extraction, 750 μL of methanol/chloroform/37% (1 N) HCl (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40
40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v/v) was added to the remaining pellet and incubated for 15 min at room temperature with vortexing every 5 min for 30 s. After 15 min, the tube was moved to ice and an additional 250 μL of cold chloroform and 450 μL of 0.1 M cold HCl were added. After vortexing for 1 min, the tube was centrifuged (6500 × g, 2 min, 4 °C) and the lower organic phase was transferred to a new tube.
1, v/v/v) was added to the remaining pellet and incubated for 15 min at room temperature with vortexing every 5 min for 30 s. After 15 min, the tube was moved to ice and an additional 250 μL of cold chloroform and 450 μL of 0.1 M cold HCl were added. After vortexing for 1 min, the tube was centrifuged (6500 × g, 2 min, 4 °C) and the lower organic phase was transferred to a new tube.
Trimethylsilyldiazomethane (TMSD) methylation
For TMSD methylation, lipid extracts were dried and dissolved in 200 μL methanol. TMSD (2 M, dissolved in hexane) was added to the lipid extracts and the samples were vortexed for 30 s and incubated for 15 min at 37 °C. The reaction was quenched with acetic acid (10 μL) and the yellow solution became colorless. The samples were injected into the LC/MS.16LC-MS analysis
For the quantitative profiling of lipids, sequential UPLC-QqQ MS analysis was constructed using MRM. The UPLC (1290 infinity, Agilent Technologies, USA) system was composed of a binary pump (G4220A, USA), an autosampler (G4226A, USA), and a column compartment (G1316C, USA). Samples were injected into the sample loop and separated by a Hypersil GOLD column (2.1 × 100 mm, 1.9 μm, Thermo Science) with a linear gradient of solvents A (19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 19
19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 acetonitrile
2 acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol
methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water, 20 mM ammonium formate, 0.1% formic acid) and B (2-propanol, 20 mM ammonium formate, 0.1% formic acid). The temperatures of the sampler and column oven were set to 4 and 40 °C, respectively. A 33 min gradient was performed as follows: 0–5 min with 5% B, 5–15 min with 5–30% B, 15–22 min with 30–90% B, 22–27 min with 90% B, 27–28 min with 90–5% B, and 28–33 min with 5% B. The flow rate, 250 μL min−1, was adjusted for all gradient times and the injection volume for each run was 4 μL. For MS analysis, an Agilent 6490 Triple Quadrupole mass spectrometer (Agilent Technologies, USA) was used, with the following parameters: 4000 V positive mode capillary voltage, a sheath gas flow of 11 L min−1 (ultra high purity nitrogen), a drying gas flow of 15 L min−1 at various temperatures, and a nebulizer gas flow at 25 psi. Optimized MRM conditions were used to analyze the various lipid species.
water, 20 mM ammonium formate, 0.1% formic acid) and B (2-propanol, 20 mM ammonium formate, 0.1% formic acid). The temperatures of the sampler and column oven were set to 4 and 40 °C, respectively. A 33 min gradient was performed as follows: 0–5 min with 5% B, 5–15 min with 5–30% B, 15–22 min with 30–90% B, 22–27 min with 90% B, 27–28 min with 90–5% B, and 28–33 min with 5% B. The flow rate, 250 μL min−1, was adjusted for all gradient times and the injection volume for each run was 4 μL. For MS analysis, an Agilent 6490 Triple Quadrupole mass spectrometer (Agilent Technologies, USA) was used, with the following parameters: 4000 V positive mode capillary voltage, a sheath gas flow of 11 L min−1 (ultra high purity nitrogen), a drying gas flow of 15 L min−1 at various temperatures, and a nebulizer gas flow at 25 psi. Optimized MRM conditions were used to analyze the various lipid species.
Data processing and statistical analysis
LC-MS data were obtained and processed using Agilent Mass Hunter Workstation Data Acquisition software. MRM data on the target lipids, including the m/z of precursor and product ions, and the retention time were exported using Qualitative Analysis B.06.00 software (Agilent Technologies, USA). Next, an in-house database constructed using the Skyline software package (MacCoss Laboratory, USA) was applied to determine the peak area of assigned lipids from replicate raw data. The extracted areas of lipid species were normalized to the appropriate internal standard. Principal component analysis (PCA) was performed using the MetaboAnalyst website.23Results
P. acnes EV isolation
We examined the characteristics of P. acnes EVs purified by serial ultracentrifugation. As confirmed by TEM imaging and DLS analysis, purified P. acnes EVs had spherical vesicular structures with an average diameter of 42.5 ± 7.0 nm (Fig. 1). The size of P. acnes EV is mostly in the range of 18 to 38 nm, accounting for 91.8% of the total. Structural characteristics of P. acnes EVs conformed to the general description of EVs.24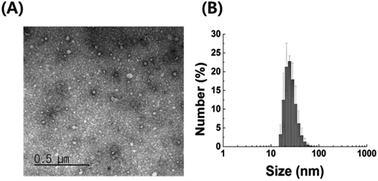 | ||
| Fig. 1 Characterization of EVs isolated from P. acnes culture medium (A) TEM image and (B) DLS analysis of purified P. acnes EVs. | ||
Effect of P. acnes EVs on RAW264.7 cells
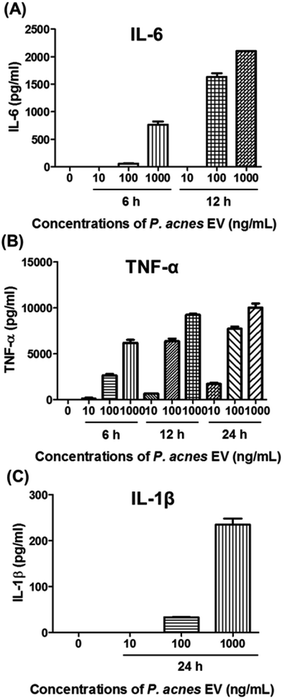
P. acnes can stimulate pro-inflammatory cytokines in host cell lines.25–29 so we investigated whether P. acnes EVs possessed bioactive molecules that could induce inflammatory responses in immune cells. When RAW264.7 cells were incubated with various concentrations of P. acnes EVs, increased expression of the pro-inflammatory cytokines IL-6, TNF-α, and IL-1β was induced in the presence of P. acnes EVs, and increased in a dose- and time-dependent manner (Fig. 2). This suggests that P. acnes EVs possess bioactive molecules that can act as potent immune modulators. Furthermore, the immunogenicity of P. acnes EVs provides an explanation for how the non-motile P. acnes can induce P. acnes-specific immune responses from distant tissues.Lipid identification in P. acnes and P. acnes EVs
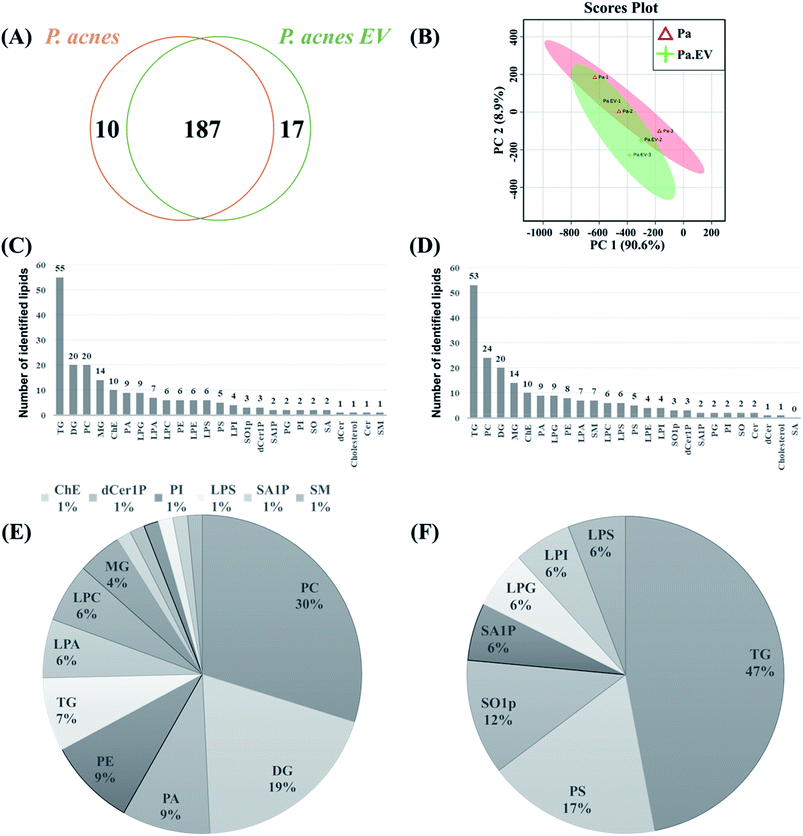
A total of 214 lipids were identified by lipidomic analysis of P. acnes and P. acnes EVs. Among the lipids, 187 (87.38%) were identified in both P. acnes and P. acnes EVs (Fig. 3A), which contained 204 and 197 lipids, respectively As shown in the PCA analysis (Fig. 3B) and ESI Table 1,† although the lipid profiles of P. acnes and P. acnes EVs mostly overlapped, relative lipid quantities of the lipid molecules in two groups showed distinct differences (ESI Tables 1 and 2†). Among the lipid classes, TG was the most common lipid class in both groups, followed by DG, PC, MG, and ChE in P. acnes and PC, DG, MG, and ChE in P. acnes EVs (Fig. 3C and D). Among the lipids belonging to the TG class, in P. acnes, TG(50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), TG(50
1), TG(50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), TG(50
0), TG(50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), TG(54
2), TG(54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), TG(48
3), TG(48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and TG(52
1), and TG(52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) were the most abundant in descending order. In P. acnes EVs, TG(54
2) were the most abundant in descending order. In P. acnes EVs, TG(54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), TG(50
3), TG(50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), TG(50
1), TG(50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), TG(50
0), TG(50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), TG(52
2), TG(52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), and TG(48
2), and TG(48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were the most abundant in descending order. But throughout the lipid classes, PS(28
1) were the most abundant in descending order. But throughout the lipid classes, PS(28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), MG(18
0), MG(18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), MG(16
0), MG(16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), PG(28
0), PG(28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) were the most abundant lipids in both P. acnes and P. acnes EVs (ESI Table 1†). Neutral lipids were predominant in both groups.
0) were the most abundant lipids in both P. acnes and P. acnes EVs (ESI Table 1†). Neutral lipids were predominant in both groups.
Relative quantification of the lipids in the two groups showed 84 differentially regulated lipids (DRLs), including 67 that identified more (fold change > 1.5, p-value < 0.05), and 17 that identified less (fold change < 0.67, p-value < 0.05; ESI Table 2†) in P. acnes EVs compared to P. acnes. P. acnes EVs contained substantially more PCs, DGs, PAs, PEs, LPAs, LPCs, and MGs than P. acnes, and fewer PSs, SO1Ps, SA1Ps, LPGs, LPIs, and LPSs. Lipids in the TG class showed individually different quantities in the two groups, and the amount of total TG was markedly decreased in P. acnes EVs (Fig. 3E and F). Throughout all lipid classes, PC(30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) showed the most dramatic difference, with a level 1210-fold higher in P. acnes EVs than in P. acnes; this was followed by PE(34
1) showed the most dramatic difference, with a level 1210-fold higher in P. acnes EVs than in P. acnes; this was followed by PE(34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), PA(30
2), PA(30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), PC(32
1), PC(32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), PC(34
2), PC(34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), PC(38
2), PC(38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), PA(34
1), PA(34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), PC(28
1), PC(28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), PA(32
0), PA(32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), PC(38
1), PC(38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), among others (ESI Tables 2†) There were also DRLs that showed little difference between P. acnes and P. acnes EVs. The lipids with a difference of less than 5% in the two groups were as follows: TG(46
2), among others (ESI Tables 2†) There were also DRLs that showed little difference between P. acnes and P. acnes EVs. The lipids with a difference of less than 5% in the two groups were as follows: TG(46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), LPE(20
0), LPE(20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), LPS(18
0), LPS(18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), ChE(20
1), ChE(20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), TG(58
1), TG(58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), TG(60
0), TG(60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), TG(56
3), TG(56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), LPS(20
2), LPS(20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), TG(50
1), TG(50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), LPG(20
2), LPG(20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), TG(52
3), TG(52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), TG(50
0), TG(50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), TG(54
0), TG(54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5), and TG(52
5), and TG(52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) (ESI Table 1†).
5) (ESI Table 1†).
Discussion
P. acnes is a lipophilic commensal bacterium that resides on the skin and in the gastrointestinal tract.1,2 P. acnes is usually nonpathogenic and has relatively low virulence, but rarely, it can exhibit serious pathogenicity.5 Although P. acnes has recently received attention as an opportunistic pathogen in various diseases, studies on its pathogenic role have mainly focused on acne.30–32 The pathology of acne vulgaris is a multifactorial process including increased sebum production, altered follicular keratinization, inflammation, and bacterial colonization of the pilosebaceous unit.33,34Up to now, most studies on the mechanism of sebum production have focused only on host lipids in the sebaceous glands, where P. acnes is expected to be present.35,36 Although the potential role of P. acnes-derived lipids in host lipid metabolism and inflammation remains unclear, the most recent information regarding lipid composition, especially fatty acid, of P. acnes was published in 1967.37 We therefore performed P. acnes lipidomic analysis for the first time, to establish a basis for research on lipids derived from P. acnes.
Recent studies have revealed that both Gram-positive and Gram-negative bacteria secrete EVs.24 It is anticipated that even in tissues that bacteria do not directly reach, bacteria-specific host responses can be induced by EVs,11 which contain a large number of biomolecules from their bacterium of origin.6,21 To test this hypothesis, we purified P. acnes EVs from culture medium and incubated them with a mouse macrophage cell line. P. acnes EVs induced the expression of the pro-inflammatory cytokines IL-6, TNF-α, and IL-1β in the macrophages, consistent with the stimulation of pro-inflammatory cytokines by P. acnes in keratinocytes,25,26 sebocytes,27,28 and monocytic cells.29 The results suggest that P. acnes EVs contain bioactive molecules and may affect distant tissues on behalf of the non-motile P. acnes.
To identify potentially bioactive molecules in P. acnes EVs, we analyzed the lipidomes of P. acnes and P. acnes EVs. Interestingly, from a total of 214 identified lipids, the lipid profiles mostly overlapped but the lipid quantities showed distinct differences. In both groups, some lipids such as PS(28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), MG(18
0), MG(18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), MG(16
0), MG(16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), and PG(28
0), and PG(28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) were the most abundant and other lipids such as TG(46
0) were the most abundant and other lipids such as TG(46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), LPE(20
0), LPE(20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), LPS(18
0), LPS(18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), ChE(20
1), ChE(20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), TG(58
1), TG(58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), TG(60
0), TG(60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), TG(56
3), TG(56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), LPS(20
2), LPS(20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), TG(50
1), TG(50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), LPG(20
2), LPG(20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), TG(52
3), TG(52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), TG(50
0), TG(50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), TG(54
0), TG(54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5), and TG(52
5), and TG(52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) showed little difference. The reason why these lipids showed similar configurations in both groups may be because they are essential molecules to support structure, necessary for membrane construction, or they have similar functions. P. acnes EVs contained substantially more PCs, DGs, PAs, PEs, LPAs, LPCs, and MGs than P. acnes and fewer PSs, SO1Ps, SA1Ps, LPGs, LPIs, and LPSs. Distinctively, P. acnes EVs possessed a markedly reduced amount of TG compared to P. acnes and possessed dramatically elevated amounts of PC(30
5) showed little difference. The reason why these lipids showed similar configurations in both groups may be because they are essential molecules to support structure, necessary for membrane construction, or they have similar functions. P. acnes EVs contained substantially more PCs, DGs, PAs, PEs, LPAs, LPCs, and MGs than P. acnes and fewer PSs, SO1Ps, SA1Ps, LPGs, LPIs, and LPSs. Distinctively, P. acnes EVs possessed a markedly reduced amount of TG compared to P. acnes and possessed dramatically elevated amounts of PC(30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), PE(34
1), PE(34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), PA(30
2), PA(30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), PC(32
1), PC(32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), PC(34
2), PC(34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), PC(38
2), PC(38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), among other lipids. (ESI Table 2†). This suggests that P. acnes EVs may have distinctly different roles than the bacteria itself.
1), among other lipids. (ESI Table 2†). This suggests that P. acnes EVs may have distinctly different roles than the bacteria itself.
By identifying these constituents, we hope to ultimately elucidate the mechanism of action of P. acnes and its EVs in the bio-physiology and the pathology of P. acnes. We anticipate that a variety of factors contribute to the mechanisms of action of EVs. First, the biomolecules on the surface of EVs can factor into the mechanism of action. Surface biomolecules may bind to receptors or ligands on the surface of the target cell, or membrane-bound enzymes on EVs may play a role as well. In this process, unlike a single molecule interaction, various membrane biomolecules on the EV surface can interact with their targets simultaneously. Second, biomolecules inside the EVs may also factor into the mechanism of action. After endocytosis, biomolecules such as proteins, carbohydrates, lipids, and nucleic acids in the EVs may function or interact with counterpart molecules in target cells or act as cofactors. Unlike a single molecule interaction, as a cargo ship, EVs can simultaneously transfer numerous biomolecules at high local concentrations. For penetration, we assume that lipids or proteins may mediate the intrusion process. We hope that this data will enable study of previously unreported roles for P. acnes-derived lipids and P. acnes EVs, and provide clues towards unknown pathogenic mechanisms.
Conclusions
In this study, we have performed a comparative lipidomic analysis of P. acnes and P. acnes EVs for the first time, and propose that P. acnes EVs may act as pro-inflammatory modulators in the host, stimulating IL-6, TNF-α, and IL-1β expression. A total of 214 lipids were successfully analyzed by optimized UPLC and MRM. Statistical data analysis indicated that P. acnes EVs had distinct lipid composition compared to P. acnes. We expect these findings to provide useful clues towards understanding the biological and pathophysiological mechanisms of P. acnes and developing clinical applications such as vaccines, diagnostics and therapeutics.Conflicts of interest
There are no conflicts to declare.Abbreviations
| TG | Triacylglycerol |
| DG | Diacylglycerol |
| MG | Monoacylglycerol |
| ChE | Cholesterolester |
| PC | Phosphatidylcholine |
| LPC | Lysophosphatidylcholine |
| PE | Phosphatidylethanolamine |
| LPE | Lysophosphatidylethanolamine |
| PS | Phosphatidylserine |
| LPS | Lysophosphatidylserine |
| PG | Phosphatidylglycerol |
| LPG | Lysophosphatidylglycerol |
| PI | Phosphatidylinositol |
| LPI | Lysophosphatidylinositol |
| PA | Phosphatidic acid |
| LPA | Lysophosphatidic acid |
| So | Sphingosine |
| So1P | Sphingosine-1-phosphate |
| Sa | Sphinganine |
| Sa1P | Sphinganine-1-phosphate |
| SM | Sphingomyelin |
| Cer | Ceramide |
| dCer | Dihydroceramide |
| Cer1P | Ceramide-1-phosphate |
| dCer1P | Dihydroceramide-1-phosphate |
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF-2014K1B1A1073720) and a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI15C2900).Notes and references
- M. E. Portillo, S. Corvec, O. Borens and A. Trampuz, BioMed Res. Int., 2013, 2013, 804391 CrossRef PubMed.
- G. G. Aubin, M. E. Portillo, A. Trampuz and S. Corvec, Med. Mal. Infect., 2014, 44, 241–250 CrossRef CAS PubMed.
- A. L. Cogen, V. Nizet and R. L. Gallo, Br. J. Dermatol., 2008, 158, 442–455 CrossRef CAS PubMed.
- C. G. Burkhart, C. N. Burkhart and P. F. Lehmann, Postgrad. Med. J., 1999, 75, 328–331 CrossRef CAS PubMed.
- B. Dreno, H. Gollnick, S. Kang, D. Thiboutot, V. Bettoli, V. Torres, J. Leyden and G. A. I. Outcomes, J. Eur. Acad. Dermatol. Venereol., 2015, 29, 3–11 CrossRef PubMed.
- L. Brown, J. M. Wolf, R. Prados-Rosales and A. Casadevall, Nat. Rev. Microbiol., 2015, 13, 620–630 CrossRef CAS PubMed.
- B. Gyorgy, T. G. Szabo, M. Pasztoi, Z. Pal, P. Misjak, B. Aradi, V. Laszlo, E. Pallinger, E. Pap, A. Kittel, G. Nagy, A. Falus and E. I. Buzas, Cell. Mol. Life Sci., 2011, 68, 2667–2688 CrossRef CAS PubMed.
- B. Gyorgy, T. G. Szabo, M. Pasztoi, Z. Pal, P. Misjak, B. Aradi, V. Laszlo, E. Pallinger, E. Pap, A. Kittel, G. Nagy, A. Falus and E. I. Buzas, Cell. Mol. Life Sci., 2011, 68, 2667–2688 CrossRef CAS PubMed.
- L. M. Mashburn and M. Whiteley, Nature, 2005, 437, 422–425 CrossRef CAS PubMed.
- L. Gangoda, S. Boukouris, M. Liem, H. Kalra and S. Mathivanan, Proteomics, 2015, 15, 260–271 CrossRef CAS PubMed.
- L. M. Mashburn-Warren and M. Whiteley, Mol. Microbiol., 2006, 61, 839–846 CrossRef CAS PubMed.
- M. Giera, A. Ioan-Facsinay, R. Toes, F. Gao, J. Dalli, A. M. Deelder, C. N. Serhan and O. A. Mayboroda, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2012, 1821, 1415–1424 CrossRef CAS PubMed.
- M. Scherer, K. Leuthauser-Jaschinski, J. Ecker, G. Schmitz and G. Liebisch, J. Lipid Res., 2010, 51, 2001–2011 CrossRef CAS PubMed.
- Y. P. Xiong, Y. Y. Zhao, S. Goruk, K. Oilund, C. J. Field, R. L. Jacobs and J. M. Curtis, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2012, 911, 170–179 CrossRef CAS PubMed.
- X. L. Gao, Q. B. Zhang, D. Meng, G. Isaac, R. Zhao, T. L. Fillmore, R. K. Chu, J. Y. Zhou, K. Q. Tang, Z. P. Hu, R. J. Moore, R. D. Smith, M. G. Katze and T. O. Metz, Anal. Bioanal. Chem., 2012, 402, 2923–2933 CrossRef CAS PubMed.
- J. W. Lee, S. Nishiumi, M. Yoshida, E. Fukusaki and T. Bamba, J. Chromatogr., 2013, 1279, 98–107 CrossRef CAS PubMed.
- N. R. Kitteringham, R. E. Jenkins, C. S. Lane, V. L. Elliott and B. K. Park, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2009, 877, 1229–1239 CrossRef CAS PubMed.
- M. Gaudin, M. Panchal, N. Auzeil, C. Duyckaerts, A. Brunelle, O. Laprevote and D. Touboul, Bioanalysis, 2012, 4, 2153–2159 CrossRef CAS PubMed.
- J. W. Lee, H. Shinohara, J. H. Jung, H. J. Mok, Y. Akao and K. P. Kim, RSC Adv., 2016, 6, 29512–29518 RSC.
- H. J. Mok, H. Shin, J. W. Lee, G. K. Lee, C. S. Suh, K. P. Kim and H. J. Lim, PLoS One, 2016, 11, 0148677 Search PubMed.
- J. Jeon, H. J. Mok, Y. Choi, S. C. Park, H. Jo, J. Her, J. K. Han, Y. K. Kim, K. P. Kim and C. Ban, Proteomics: Clin. Appl., 2017, 11, 1600040 CrossRef PubMed.
- M. Haag, A. Schmidt, T. Sachsenheimer and B. Brugger, Metabolites, 2012, 2, 57–76 CrossRef CAS PubMed.
- J. G. Xia, I. V. Sinelnikov, B. Han and D. S. Wishart, Nucleic Acids Res., 2015, 43, W251–W257 CrossRef CAS PubMed.
- J. H. Kim, J. Lee, J. Park and Y. S. Gho, Semin. Cell Dev. Biol., 2015, 40, 97–104 CrossRef CAS PubMed.
- N. Akaza, H. Akamatsu, M. Kishi, H. Mizutani, I. Ishii, S. Nakata and K. Matsunaga, J. Dermatol., 2009, 36, 213–223 CrossRef CAS PubMed.
- G. M. Graham, M. D. Farrar, J. E. Cruse-Sawyer, K. T. Holland and E. Ingham, Br. J. Dermatol., 2004, 150, 421–428 CrossRef CAS PubMed.
- I. Nagy, A. Pivarcsi, K. Kis, A. Koreck, L. Bodai, A. McDowell, H. Seltmann, S. Patrick, C. C. Zouboulis and L. Kemeny, Microbes Infect., 2006, 8, 2195–2205 CrossRef CAS PubMed.
- Z. J. Li, D. K. Choi, K. C. Sohn, M. S. Seo, H. E. Lee, Y. Lee, Y. J. Seo, Y. H. Lee, G. Shi, C. C. Zouboulis, C. D. Kim, J. H. Lee and M. Im, J. Invest. Dermatol., 2014, 134, 2747–2756 CrossRef CAS PubMed.
- M. Qin, A. Pirouz, M. H. Kim, S. R. Krutzik, H. J. Garban and J. Kim, J. Invest. Dermatol., 2014, 134, 381–388 CrossRef CAS PubMed.
- C. Beylot, N. Auffret, F. Poli, J. P. Claudel, M. T. Leccia, P. Del Giudice and B. Dreno, J. Eur. Acad. Dermatol. Venereol., 2014, 28, 271–278 CrossRef CAS PubMed.
- Y. Achermann, E. J. Goldstein, T. Coenye and M. E. Shirtliff, Clin. Microbiol. Rev., 2014, 27, 419–440 CrossRef CAS PubMed.
- C. Dessinioti and A. D. Katsambas, Clin. Dermatol., 2010, 28, 2–7 CrossRef PubMed.
- E. Contassot and L. E. French, J. Invest. Dermatol., 2014, 134, 310–313 CrossRef CAS PubMed.
- B. R. Vowels, S. J. Yang and J. J. Leyden, Infect. Immun., 1995, 63, 3158–3165 CAS.
- C. C. Zouboulis, J. Invest. Dermatol., 2009, 129, 2093–2096 CrossRef CAS PubMed.
- K. Iinuma, T. Sato, N. Akimoto, N. Noguchi, M. Sasatsu, S. Nishijima, I. Kurokawa and A. Ito, J. Invest. Dermatol., 2009, 129, 2113–2119 CrossRef CAS PubMed.
- C. W. Moss, V. R. Dowell Jr, V. J. Lewis and M. A. Schekter, J. Bacteriol., 1967, 94, 1300–1305 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: See DOI: 10.1039/c7ra13769a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |