Open Access Article
Open Access ArticleSensing caspase-1 activity using activatable 19F MRI nanoprobes with improved turn-on kinetics†
Kazuki
Akazawa
a,
Fuminori
Sugihara
b,
Masafumi
Minoshima
a,
Shin
Mizukami
c and
Kazuya
Kikuchi
 *ab
*ab
aDivision of Advanced Science and Biotechnology, Graduate School of Engineering, Osaka University, 2-1 Yamadaoka, Suita, Osaka 565-0871, Japan. E-mail: kkikuchi@mls.eng.osaka-u.ac.jp
bWPI-Immunology Frontier Research Center, Osaka University, 2-1 Yamadaoka, Suita, Osaka 565-0871, Japan
cInstitute of Multidisciplinary Research for Advanced Materials, Tohoku University, 2-1-1 Katahira, Aoba-ku, Sendai, Miyagi 980-8577, Japan
First published on 2nd October 2018
Abstract
Activatable 19F MRI nanoprobes for sensing caspase-1 activity were developed. Tandem repetition of substrate peptide sequences improved the turn-on response of nanoprobes, allowing detection of caspase-1 activity by 19F MRI. In vivo immune response was successfully imaged using the new nanoprobe.
Magnetic resonance imaging (MRI) is a promising modality that allows non-invasive in vivo imaging of molecular processes in deep tissues in living animals. Various MR imaging methods such as chemical exchange saturation transfer (CEST),119F MRI,2 hyperpolarized MRI,3 or magnetic particle imaging (MPI)4 have been developed to visualize specific biological phenomena. Among these, 19F MRI is one of the best approaches for specific detection of probe signals due to its high sensitivity (83% of 1H), a broad range of 19F chemical shifts (>350 ppm), and an absence of endogenous background signals.5 To trace the dynamics of biomolecules of interest, especially enzyme activities, activatable 19F MRI probes have been devised in response to enzyme reactions.6 However, few activatable 19F MRI probes have been used to visualize enzyme activities in living animals due to the low intensity of 19F MRI signals.
In previous studies, we established an in vivo imaging platform comprising a highly sensitive perfluorocarbon (PFC)-encapsulated 19F MRI nanoprobe and an OFF/ON-switching system based on the paramagnetic relaxation enhancement (PRE) effect. The nanoprobe, termed FLAME (FLuorine Accumulated silica nanoparticle for MRI contrast Enhancement),7 is a core–shell nanoparticle composed of perfluoro-[15] crown-5 ether (PFCE) core and a robust silica shell, and can be functionalized by modifying small molecules or peptides via a silane coupling reaction. To add signal-activatable function to FLAME, we exploited the PRE effect of Gd3+ complexes to modulate the spin–spin relaxation time (T2), which affects the 19F MRI signal intensity of PFCE molecules in the FLAME core,8 and visualized the endogenous caspase-3/7 activity in a living mouse by 19F MRI.9 In the probe design, the Gd3+ complex-conjugated peptide had a substrate peptide sequence tandemly repeated twice to improve cleavage efficiency by caspase-3. However, the improvement in cleavage efficiency was not quantitatively investigated.
In this study, we developed an activatable 19F MRI nanoprobe with an improved turn-on switch for detecting caspase-1 activity. Caspase-1 is a family of cysteine proteases involved in the maturation and secretion of pro-inflammatory cytokines such as interleukin 1β (IL-1β) and IL-18, which evoke an inflammatory response.10 Caspase-1 is present as an inactive zymogen, and is activated by proteolytic cleavage into a heterodimer upon cellular infection or stress. This activation process is mediated by a multiprotein complex called the inflammasome.11 Recent studies report that caspase-1-mediated release of IL-1β and IL-18 contributes to a variety of inflammatory diseases such as Parkinson's disease,12 gout,13 and obesity.14 Therefore, imaging of caspase-1 activity may facilitate understanding the mechanisms of various inflammatory diseases and evaluating the therapeutic efficacy of anti-inflammatory drugs. Until now, the levels and activation of caspase-1 have mainly been analyzed by ELISA or western blotting. To the best of our knowledge, existing real-time imaging techniques for caspase-1 activity are limited to optical imaging using fluorescent proteins15 or fluorescent nanosensors.16 These fluorescence-based probes do not allow non-invasive detection of caspase-1 activity in deep region of living animals due to limited light penetration. MRI could be thus used for monitoring caspase-1 activity in vivo; however, suitable MRI probes have not been developed. Herein, we describe, for the first time, 19F MR imaging of caspase-1 activity using activatable 19F MRI nanoprobe with an improved turn-on switch.
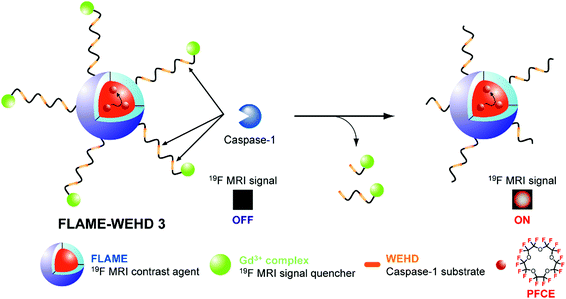
We designed a novel activatable 19F MRI nanoprobe, FLAME-WEHD 3, for sensing caspase-1 activity (Fig. 1). In this sensor, to add signal-activatable function to FLAME in response to caspase-1 activity, Gd3+ complex-conjugated peptides containing the caspase-1 substrate sequence, WEHD,17 were conjugated to the FLAME surface. The T2 of PFCE in FLAME is shortened by PRE effect of the Gd3+ complexes, which quenches the 19F MRI signal of FLAME.
After cleavage of Gd3+ complex-conjugated peptides by caspase-1, the Gd3+ complexes leave the nanoparticle surface, resulting in 19F MRI signal activation. Furthermore, to enhance the activation response of the nanoprobe, we designed the Gd3+ complex-conjugated peptide, WEHD 3, which is a substrate peptide sequence tandemly repeated 3 times. We anticipated that the tandemly repeated substrate peptide sequence could improve enzyme accessibility to substrates on the nanoparticle surface and the cleavage efficiency of peptides by caspase-1, resulting in enhancement of the turn-on response by the nanoprobes.
To test whether the tandemly repeated peptide design strategy could improve the cleavage efficiency of peptides by caspase-1 on the nanoparticle surface, we prepared three types of nanoprobes, FLAME-WEHD X (X = 1–3), modified with the substrate peptide sequence tandemly repeated X times (Fig. S1, ESI†). First, three Gd3+ complex-conjugated peptides, WEHD 1–3, were synthesized by Fmoc chemistry, and purified with reverse-phase high-performance liquid chromatography (HPLC) (Fig. S2 and Scheme S1, ESI†). The nanoparticle surface modification was achieved by the reaction of N-hydroxysuccinimidylated FLAME (FLAME-NHS; Scheme S2, ESI†) with WEHD 1–3. The core–shell structures of FLAME-WEHD 1–3 were confirmed from TEM images (Fig. S3, ESI†). The particle concentrations and hydrodynamic diameters were measured using a tunable resistive pulse sensor (Fig. S4, ESI†).18 The average hydrodynamic diameters of FLAME-WEHD 1–3 were 101 ± 17 nm, 127 ± 31 nm, and 117 ± 18 nm, respectively (Table S1, ESI†). The number of Gd3+ complexes on the FLAME surface was verified by inductively coupled plasma atomic emission spectrometry (ICP-AES). We calculated the number of fluorine atoms (n19F) and Gd3+ ions (nGd) per nanoparticle from the concentrations of PFCE, nanoparticles, and Gd3+ (Table S3 (ESI†); calculations following the previously described method9). The nGd values per nanoparticle were 2.3 × 104, 3.1 × 104, and 5.1 × 104 for FLAME-WEHD 1–3, respectively. Next, the 19F NMR spectra and T2 of FLAME-WEHD 1–3 were measured and compared with FLAME-COOH as a control without the Gd3+ complexes. In the 19F NMR spectra, FLAME-WEHD 1–3 (CPFCE = 10 mM) showed single broad peaks, whereas FLAME-COOH showed a single sharp peak (Fig. S5, ESI†). In accordance with the results of 19F NMR spectra, the T2 values of PFCE in FLAME-WEHD 1–3 were shortened compared to those of FLAME-COOH (432 ms); 61, 63, and 50 ms for X = 1, 2, and 3, respectively. The n19F/nGd values calculated for each nanoparticle indicated that 19F MRI signal quenching efficiency, due to the PRE effect of the Gd3+ complex, was affected by the tandem repeat number (Table S3, ESI†). By tuning the T2 value and the number of Gd3+ complexes, attenuation of 19F MRI signals was achieved even in FLAME-WEHD 3, where the distance between PFCE core and Gd3+ complex is expected to be the longest. In addition, the n19F/nGd value (2.7 × 103) of FLAME-WEHD 3 was larger than that (9.3 × 102) of previously reported FLAME-DEVD 2 (Table S4, ESI†),9 which has two tandem repeats of the amino acid sequence, DEVD. The quenching efficiency of FLAME-WEHD 3 was higher even though the distance between PFCE core and Gd3+ complex was expected to be longer in FLAME-WEHD 3. Furthermore, the decrease in n19F/nGd values corresponding to an increase in the tandem repeat number was less in FLAME-WEHD 1–3 compared to that in FLAME-DEVD 1 and 2. This suggests that the amino acid sequence may also affect the quenching efficiency of PRE effect on the nanoparticle surface.
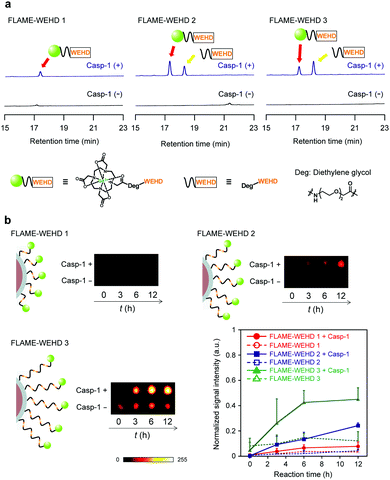
Next, we conducted a reaction of FLAME-WEHD 1–3 with recombinant mouse caspase-1. FLAME-WEHD 1–3 (CPFCE = 6.4 mM) were incubated with caspase-1 in an assay buffer at 37 °C. After 12 h reaction, the supernatant containing cleaved peptide fragments was analyzed using HPLC and electrospray ionization-mass spectrometry (ESI-MS) (Fig. 2). Caspase-1 selectively cleaves the C-terminus of the peptide sequence, i.e., WEHD. The peak around 17 min in all mixtures of FLAME-WEHD 1–3 and caspase-1 was identified by ESI-MS as Gd3+–DOTA–Deg–WEHD, the cleaved peptide derived from the reaction with caspase-1. In addition, a peak around 18.5 min observed in the mixture of FLAME-WEHD 2 or 3 with caspase-1, was identified as Deg–WEHD. Other expected fragments such as Gd3+–DOTA–Deg–WEHD–Deg–WEHD and Gd3+–DOTA–Deg–WEHD–Deg–WEHD–Deg–WEHD were not observed. On the other hand, FLAME-WEHD 1–3 in the absence of caspase-1 gave almost no distinct peptide fragment. These results indicate that the Gd3+ complex-conjugated peptides, WEHD 1–3, on the nanoparticle surface were hydrolyzed at specific sites by caspase-1.
To investigate the cleavage efficiency of the tandemly repeated substrate peptide sequence, we analyzed the time course of caspase-1 reaction with Gd3+ complex-conjugated peptides. WEHD 1–3 (100 μM) were incubated with caspase-1, and the enzyme reaction was traced using HPLC and ESI-MS (Fig. S6, ESI†). ESI-MS analysis of the HPLC peaks showed that caspase-1 selectively cleaved all the substrate peptides at the C-terminus of aspartic acid residues. To compare the cleavage efficiencies of WEHD 1–3 by caspase-1, we calculated the time required to reach 50% consumption of the substrate (t1/2) from the fitting curve of the normalized absorption intensity (280 nm) of WEHD 1–3 (Fig. S7, ESI†). As a result, WEHD 3 showed the shortest half-life (t1/2 = 43 min), compared with that of WEHD 1 (t1/2 = 161 min) and 2 (t1/2 = 63 min) (Table S2, ESI†). This result supports that tandem repetition of the substrate peptide sequence improved the cleavage efficiency of the Gd3+ complex-conjugated peptide based on the repetition number.
Next, we checked whether FLAME-WEHD 1–3 were capable of sensing caspase-1 activity in vitro. After incubation of FLAME-WEHD 1–3 (CPFCE = 6.4 mM) with caspase-1, the 19F MR images were obtained with clear contrasts from FLAME-WEHD 2 and 3 (Fig. 2b). The increase in 19F MRI signal in these images indicate the release of signal-quenching Gd3+ complexes from the FLAME surface, which is consistent with the HPLC analyses of the cleaved peptide fragments. FLAME-WEHD 3 showed a 19F MRI signal intensity of 0.45 ± 0.09 after 12 h incubation with caspase-1. However, FLAME-WEHD 1 and 2 showed 19F MR images with less contrast and low signal intensities of 0.08 ± 0.04 and 0.24 ± 0.02, respectively. This higher 19F MRI signal increase in FLAME-WEHD 3 than in that of FLAME-WEHD 1 and 2 demonstrates that the tandemly repeated peptide design strategy improved enzyme accessibility to the substrates on the nanoparticle surface and enhanced the signal-activation response of the nanoprobe for highly sensitive detection of caspase-1 activity. Moreover, pretreatment with caspase-1 inhibitor (Z-WEHD-FMK) suppressed the 19F MRI signal amplification (98%) of FLAME-WEHD 3 with caspase-1 (Fig. S8, ESI†), indicating that the signal amplification observed in Fig. 2b was induced by caspase-1 activity.
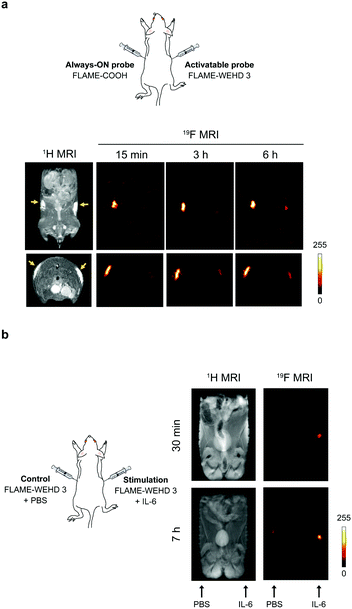
Finally, to demonstrate the feasibility of this sensing system in living animals, we performed in vivo experiments in mice using FLAME-WEHD 3. We tested whether the 19F MRI signals of FLAME-WEHD 3 were quenched in a mouse compared to those of FLAME-COOH, an always ON-type nanoprobe. FLAME-WEHD 3 and FLAME-COOH (CPFCE = 1.67 mM, 120 μL) were subcutaneously injected into a mouse at the right and left flanks, respectively. The coronal and axial 19F MR scans were then acquired after 15 min, 3 h, and 6 h of injection (Fig. 3a). Significant 19F MRI signals from FLAME-COOH were detected at all time points. In contrast, 19F MRI signals of FLAME-WEHD 3 were attenuated at 15 min after injection. Although the 19F MRI signals of FLAME-WEHD 3 were slightly increased after 6 h, the signal intensity was much lower than that of FLAME-COOH.
To test the turn-on ability of our sensor in living animals, we conducted 19F MRI detection of the immune response using FLAME-WEHD 3. To induce the immune response in vivo, we employed interleukin 6 (IL-6), which activates the immune system by promoting population expansion and the differentiation of immune cells.19 FLAME-WEHD 3 (CPFCE = 1.67 mM, 120 μL) was subcutaneously injected with IL-6 (right flank), and 1H/19F MR scanning was then carried out at 30 min and 7 h after injection (Fig. 3b). The 19F MRI signals were clearly detected at the right flank injected with FLAME-WEHD 3 and IL-6 after 30 min. The intensity of these 19F MRI signals increased in a time-dependent manner. In contrast, almost no 19F MRI signal was detected at the left flank injected with only FLAME-WEHD 3. These results demonstrate for the first time that FLAME-WEHD 3 allowed highly sensitive MRI detection of the in vivo immune response induced by IL-6.
In conclusion, we developed an activatable 19F MRI nanoprobe, FLAME-WEHD 3, for sensing caspase-1 activity with an improved turn-on response. We prepared three types of peptides, WEHD 1–3, and three nanoprobes modified using these peptides, FLAME-WEHD 1–3, to investigate the efficacy of tandemly repeated peptide designs in detail. Quantitative analysis using HPLC indicated that cleavage kinetics of peptides by caspase-1 was significantly improved with an increase in the tandem repeat number (Fig. S7, ESI†). Quantification of 19F MRI signal intensity with and without caspase-1 indicated that the tandem repeat number critically contributed to the enhancement of enzyme reaction and signal activation rates (Fig. 2). In addition, we found that the T2 and 19F MRI signals of FLAME-WEHD 3 are sufficiently quenched by the PRE effect of multiple Gd3+ complexes, even though repeated substrate incorporation increased the distance between FLAME and Gd3+ complexes. These results enabled us to design a practical nanoparticle-based probe, FLAME-WEHD 3, for the detection of caspase-1. FLAME-WEHD 3 allowed in vivo imaging of immune response in a mouse by 19F MRI. In future studies, controlled delivery to specific inflammation sites via intravenous injection should be addressed by modifying polyethylene glycol (PEG) derivatives using targeting ligands such as antibodies on the FLAME surface, although PEGylation may affect the efficiency of signal quenching by Gd3+ complexes and enzymatic cleavage. This activatable 19F MRI nanoprobe is thus a practical diagnostic tool for analyzing inflammation in diseased-model animals.
This research was supported by the Grant-in-Aid for Scientific Research (Grant No. 25220207, 16H00768, 18H03935, and 16K01933), and Innovative Areas ‘Frontier Research on Chemical Communications’ (No. 17H06409) of MEXT, Japan; and the Magnetic Health Science Foundation.
Conflicts of interest
There are no conflicts to declare.Notes and references
- (a) B. Yoo and M. D. Pagel, J. Am. Chem. Soc., 2006, 128, 14032 CrossRef CAS PubMed; (b) R. Trokowski, J. M. Ren, F. K. Kalman and A. D. Sherry, Angew. Chem., Int. Ed., 2005, 44, 6920 CrossRef CAS PubMed.
- E. T. Ahrens, R. Flores, H. Y. Xu and P. A. Morel, Nat. Biotechnol., 2005, 23, 983 CrossRef CAS PubMed.
- H. Nonaka, R. Hata, T. Doura, T. Nishihara, K. Kumagai, M. Akakabe, M. Tsuda, K. Ichikawa and S. Sando, Nat. Commun., 2013, 4, 2411 CrossRef PubMed.
- E. Y. Yu, M. I. Bishop, B. Zheng, R. M. Ferguson, A. P. Khandhar, S. J. Kemp, K. M. Krishnan, P. W. Goodwill and S. M. Conolly, Nano Lett., 2017, 17, 1648 CrossRef CAS PubMed.
- I. Tirotta, V. Dichiarante, C. Pigliacelli, G. Cavallo, G. Terraneo, F. B. Bombelli, P. Metrangolo and G. Resnati, Chem. Rev., 2015, 115, 1106 CrossRef CAS PubMed.
- (a) S. Mizukami, R. Takikawa, F. Sugihara, Y. Hori, H. Tochio, M. Walchli, M. Shirakawa and K. Kikuchi, J. Am. Chem. Soc., 2008, 130, 794 CrossRef CAS PubMed; (b) S. Mizukami, R. Takikawa, F. Sugihara, M. Shirakawa and K. Kikuchi, Angew. Chem., Int. Ed., 2009, 48, 3641 CrossRef CAS PubMed; (c) Y. Yuan, H. B. Sun, S. C. Ge, M. J. Wang, H. X. Zhao, L. Wang, L. N. An, J. Zhang, H. F. Zhang, B. Hu, J. F. Wang and G. L. Liang, ACS Nano, 2015, 9, 761 CrossRef CAS PubMed; (d) H. Wang, K. R. Raghupathi, J. M. Zhuang and S. Thayumanavan, ACS Macro Lett., 2015, 4, 422 CrossRef CAS PubMed; (e) X. Y. Yue, Z. Wang, L. Zhu, Y. Wang, C. Q. Qian, Y. Ma, D. O. Kiesewetter, G. Niu and X. Y. Chen, Mol. Pharmaceutics, 2014, 11, 4208 CrossRef CAS PubMed; (f) M. Carril, J. Mater. Chem. B, 2017, 5, 4332 RSC.
- H. Matsushita, S. Mizukami, F. Sugihara, Y. Nakanishi, Y. Yoshioka and K. Kikuchi, Angew. Chem., Int. Ed., 2014, 53, 1008 CrossRef CAS PubMed.
- T. Nakamura, H. Matsushita, F. Sugihara, Y. Yoshioka, S. Mizukami and K. Kikuchi, Angew. Chem., Int. Ed., 2015, 54, 1007 CrossRef CAS PubMed.
- K. Akazawa, F. Sugihara, T. Nakamura, S. Mizukami and K. Kikuchi, Bioconjugate Chem., 2018, 29, 1720 CrossRef CAS PubMed.
- (a) A. Denes, G. Lopez-Castejon and D. Brough, Cell Death Dis., 2012, 3, e338 CrossRef CAS PubMed; (b) L. Franchi, T. Eigenbrod, R. Munoz-Planillo and G. Nunez, Nat. Immunol., 2009, 10, 241 CrossRef CAS PubMed.
- (a) T. Strowig, J. Henao-Mejia, E. Elinav and R. Flavell, Nature, 2012, 481, 278 CrossRef CAS PubMed; (b) H. Guo, J. B. Callaway and J. P. Ting, Nat. Med., 2015, 21, 677 CrossRef CAS PubMed.
- W. Wang, L. T. T. Nguyen, C. Burlak, F. Chegini, F. Guo, T. Chataway, S. L. Ju, O. S. Fisher, D. W. Miller, D. Datta, F. Wu, C. X. Wu, A. Landeru, J. A. Wells, M. R. Cookson, M. B. Boxer, C. J. Thomas, W. P. Gai, D. Ringe, G. A. Petsko and Q. Q. Hoang, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 9587 CrossRef CAS PubMed.
- F. Martinon, V. Petrilli, A. Mayor, A. Tardivel and J. Tschopp, Nature, 2006, 440, 237 CrossRef CAS PubMed.
- M. G. Netea, L. A. B. Joosten, E. Lewis, D. R. Jensen, P. J. Voshol, B. J. Kullberg, C. J. Tack, H. van Krieken, S. H. Kim, A. F. Stalenhoef, F. A. Van de Loo, I. Verschueren, L. Pulawa, S. Akira, R. H. Eckel, C. A. Dinarello, W. Van den Berg and J. W. M. Van der Meer, Nat. Med., 2006, 12, 650 CrossRef CAS PubMed.
- T. Liu, Y. Yamaguchi, Y. Shirasaki, K. Shikada, M. Yamagishi, K. Hoshino, T. Kaisho, K. Takemoto, T. Suzuki, E. Kuranaga, O. Ohara and M. Miura, Cell Rep., 2014, 8, 974 CrossRef CAS PubMed.
- A. Moquin, E. Hutter, A. O. Choi, A. Khatchadourian, A. Castonguay, F. M. Winnik and D. Maysinger, ACS Nano, 2013, 7, 9585 CrossRef CAS PubMed.
- N. A. Thornberry, T. A. Rano, E. P. Peterson, D. M. Rasper, T. Timkey, M. Garcia-Calvo, V. M. Houtzager, P. A. Nordstrom, S. Roy, J. P. Vaillancourt, K. T. Chapman and D. W. Nicholson, J. Biol. Chem., 1997, 272, 17907 CrossRef CAS PubMed.
- R. Vogel, G. Willmott, D. Kozak, G. S. Roberts, W. Anderson, L. Groenewegen, B. Glossop, A. Barnett, A. Turner and M. Trau, Anal. Chem., 2011, 83, 3499 CrossRef CAS PubMed.
- C. A. Hunter and S. A. Jones, Nat. Immunol., 2015, 16, 448 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8cc05381b |
| This journal is © The Royal Society of Chemistry 2018 |