A signal amplification strategy and sensing application using single gold nanoelectrodes†
Dongmei
Wang
ab,
Hongmei
Hua
a,
Haoran
Tang
a,
Cheng
Yang
a,
Wei
Chen
a and
Yongxin
Li
 *a
*a
aAnhui Key Laboratory of Chemo/Biosensing, College of Chemistry and Materials Science, Anhui Normal University, Wuhu, 241000, P. R. China. E-mail: yongli@mail.ahnu.edu.cn; Fax: +86-553-386-9303; Tel: +86-553-386-9302
bCollege of Chemistry and Material Engineering, Chaohu University, Chaohu, Anhui 238000, P.R. China
First published on 18th October 2018
Abstract
In this work, a label-free electrochemical apta-nanosensor was fabricated on a single gold nanodisk electrode (AuNDE) for thrombin sensing with high sensitivity via a novel signal amplification strategy. This recognition platform was fabricated via self-assembly of helper DNA (HP-DNA), thrombin-binding aptamer (TBA) and gold nanoparticle (AuNP)-DNA complexes to form a sandwich structure on the AuNDE surface. A novel signal amplification strategy via designed AuNP-DNA complexes was introduced using Ru(NH3)63+ as the signal reporter based on the electrostatic interaction. In the presence of thrombin, the strong interaction between the TBA and target led to the dissociation of sandwich DNA complexes from the AuNDE, which resulted in the reduction current of Ru(NH3)63+. This proposed sensing platform showed a wide detection range of 0.1 pM–5 nM and a low detection limit of 0.02 pM. Considering the small overall dimensions and high sensitivity, this nanosensor can be potentially applied for bioanalysis in living biosystems.
Introduction
Recently, single nanoelectrodes have been extensively utilized in fundamental research and real applications due to their distinct properties, such as low charging current, small RC constants, and high mass-transfer.1–5 Generally, single nanoelectrodes have extraordinary advantages over macro-electrodes in constructing biosensors by minimizing the electrode-induced physical wound in the brain6 or penetrating the membrane of single cells and minimizing cell damage.7 However, a low response signal and the large size of substrates are the main obstacles for the development of nanosensors for application in real sensing in vivo. Some methods have been carried out to solve this problem by using single nanowire electrodes4,8 or electrodepositing dendritic gold structures9 which have a relatively larger specific surface area.To improve the sensitivity of detecting at an ultralow level, various signal amplification methods, such as rolling circle amplification (RCA),7,10 polymerase chain reaction,11 nuclease-assisted target recycling,12 and hybridization chain reaction (HCR),6 have been explored to construct apta-sensors. However, most of the techniques mentioned above involve a time-consuming analysis process, which requires highly-skilled personnel and rigorous experimental conditions.13–15 These shortcomings of complicated procedures and high-cost will prevent their applications in routine analysis and encourage people to develop a new strategy for ATP detection with high sensitivity/selectivity. It is well-known that Au nanoparticle (AuNP)-DNA complexes, which were adapted firstly for bioanalysis by Mirkin's group, have distinctive properties for the application of biosensors due to their large surface area and excellent biocompatibility,16 and they have been applied to the signal amplification strategy.17
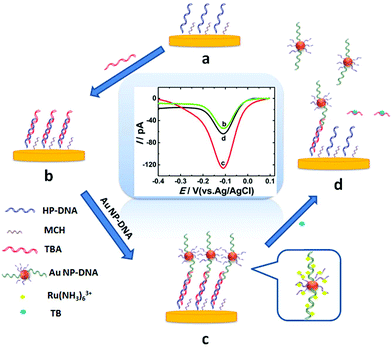
Thrombin, as a coagulation protein in the blood stream plays crucial roles in various life processes and relates to many diseases.18 Therefore, developing a fast, efficient method for ultrasensitive detection of thrombin is essential. In this work, AuNP-DNA complexes were adapted as a signal amplification system to construct a novel label-free sensing platform on gold nano-disk electrodes,19 which showed satisfactory results for sensitive detection of thrombin. Scheme 1 shows the design of the electrochemical assay for thrombin using an AuNP-DNA complex signal amplification system on a gold nanoelectrode. A recognition platform was fabricated via self-assembly of helper DNA (HP-DNA), TBA and AuNP-DNA complexes to form a sandwich structure on the surface of the AuNDE. First, the thiol-modified HP-DNA strands were attached on the gold nanoelectrode via an Au–S bond, and then blocked by 6-mercaptohexanol (MCH, Scheme 1a). Part of the TBA strands was hybridized with HP-DNA (Scheme 1b), and then the AuNPs modified with two different thiol-modified oligonucleotides (noted as S1 and S2) were captured on the nanoelectrode through the hybridization of S1 and the remaining regions of TBA (Scheme 1c). Therefore, the sandwich AuNP-DNA complexes were immobilized on the surface of the Au nanoelectrode. In this signal amplification system, Ru(NH3)63+ was selected as the redox probe which directly avoided the use of any redox labels (such as ferrocene and methylene blue, which are always modified on DNAs and this work is a time-consuming and high-cost process), which could adsorb on all DNAs via electrostatic forces between the anionic phosphate backbone of DNA strands and Ru(NH3)63+. The more DNAs immobilized on the electrode surface, the larger the electrical signal that could be obtained. AuNPs were functionalized with two different thiol-modified oligonucleotides: S1 is responsible for immobilizing on the surface of the nanoelectrode, and S2 is used for loading electroactive probe Ru(NH3)63+ as much as possible. Here, functionalized AuNPs were used to conjugate more DNA strands in order to obtain maximum electrochemical signals. Compared with the absence of AuNP-DNA (Scheme 1b), the value of differential pulse voltammetry (DPV) reduction peak current of Ru3+ increased greatly due to the abundant capture of Ru(NH3)63+ on the sandwich AuNP-DNA complexes (Scheme 1c). Finally, the addition of target thrombin led to the release of AuNP-DNA complexes from the electrode, accompanied by the decrease of the DPV response of Ru(NH3)63+ (Scheme 1d). Thrombin can be detected through monitoring the current signal changes of Ru(NH3)63+ by differential pulse voltammetry (DPV). By combining the advantages of nanoelectrodes and the signal amplification strategy, the label-free nanosensor can be used to detect thrombin in vivo at an extremely low level (0.02 pM).
It is an interesting combination which can both avoid the steric hindrance caused by S1 and capture Ru(NH3)63+ to the utmost to achieve the signal amplification stage. The experimental results show that this designed strategy has a much better amplification effect than AuNPs modified with single DNA (S1), which can be utilized to construct a label-free nanosensor for thrombin sensing with high sensitivity and selectivity, especially for real sample analysis in living systems.
Experimental section
Materials and reagents
Potassium ferricyanide (K3Fe(CN)6, Acros Organics), ferrocene (Fc, Fluka), tris-(2-carboxyethyl)phosphine hydrochloride (TCEP, Sangon, Shanghai), 6-mercaptohexanol (MCH, Sangon, Shanghai), hexaammineruthenium(III)chloride (Ru(NH3)6Cl3, Shanghai Chemical Co., Shanghai, China), sodium chloride, chloroauric acid (HAuCl4·4H2O), and tris(hydroxymethyl)aminomethane (Tris) were purchased from Sigma. IgG, lysozyme, HPR and BSA were provided by Sangon Biotech Co. Ltd (Shanghai, China). The silver-filled epoxy glue, 25 μm diameter Au microwires (99.95%, hard) and glass capillaries (i.d. 80 μm; o.d. 300 μm) were purchased from DuPont Co. Ltd, Alfa-Aesar and Sutter Instrument Co, respectively. Finer grit sandpapers (600 W, 800 W, and 1200 W grits; Sandpaper Inc., Rockland, MA) were used to polish nanoelectrodes. The thrombin-binding aptamer (TBA) and oligonucleotides were synthesized by Sangon Biotech Co. Ltd (Shanghai, China) and the corresponding sequences were listed as follows:The helper probe (HP-DNA): 5′-SH-(CH2)6-AGT CAC CCC AAC CTG CCC TA-3′
TBA: 5′-AGT CCG TGGTAG GGC AGG TTG GGG TGA CT-3′
S1: 5′-CCA CGG ACT TTT TTT-(CH2)6-SH-3′
S2: 5′-ACC CGT TT-(CH2)6-SH-3′
The bold and italics sections represent the binding regions between HP-DNA and TBA as well as TBA and S1. The serum samples were collected from the Anhui Normal University hospital.
Apparatus
The electrochemical tests were done using a CHI 660D Mode Electrochemical Workstation (Chenhua, Shanghai, China). Transmission Electron Microscopy (TEM) images were obtained from a JEM-2100F microscope (Tokyo, Japan). The UV–vis absorption spectra were recorded using a Lambda750 UV–vis spectrophotometer (PerkinElmer). The dynamic light-scattering (DLS) measurements were carried out using a Zetasizer Nano ZSP instrument (Malvern Instruments Ltd, Malvern, UK). Polyacrylamide gel electrophoresis (PAGE) was tested using a DYY-12C electrophoresis apparatus (Junyi-Dongfang Co. Ltd, Beijing, China) and imaged on a Tanon-3500 Gel Image System (Shanghai, China).Fabrication of single Au nanodisk electrodes
The single Au nanodisk electrodes (AuNDEs) were fabricated using an improved laser-assisted pulling technique according to our previous method.19 In short, two needle-shape tips were pulled from a glassy capillary (o.d. = 300 μm; i.d. = 80 μm) sealed with Au wire (25 μm in diameter) using a laser-assisted puller (P-2000, Sutter). The needle-shape tip was inserted into a large glass tube (o.d. = 1.0 mm; i.d. = 0.64 mm) and sealed together using hydrogen–oxygen flame. Then the Au nanowire tip was polished with sandpaper to make an Au nanodisk electrode.Preparation of AuNP-DNA complexes
AuNPs were synthesized following previously reported protocols.20,21 Briefly, 50 mL of 1 mM HAuCl4 solution was heated to boiling, followed by the addition of trisodium citrate solution (38.6 mM, 5 mL) under vigorous stirring. After boiling for 10 min and turning off the heat, the solution was stirred for another 15 min continuously. After the solution was cooled down to room temperature, the obtained Au colloidal solution was stored at 4 °C before use. As seen in TEM images, the diameter of AuNPs is ∼13 nm. AuNP-DNA complexes were prepared according to ref. 22. In short, S1 and S2 were dissolved in 10 mM Tris-HCl buffer (containing 100 mM NaCl, pH 7.4) and incubated in 50 mM tris(2-carboxyethyl)phosphine (TCEP) in the dark for 1 h to reduce disulfide bonds. 1 mL AuNP solution was mixed with 150 μL of 10 μM S1/S2 solution (molar ratios were 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, 1
0, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, 1
4, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7, 1
7, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9, and 1
9, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 11, respectively). After shaking gently for 16 h at room temperature, the two oligonucleotides were self-assembled onto the AuNPs. Next, NaCl solution was added into the above solution gradually until the concentration of NaCl reached 500 mM in 3 days, which was used to conjugate more DNA strands on AuNPs.13 Then the suspension was centrifuged and rinsed five times (pH 7.4, 10 mM PBS, 100 mM NaCl) to remove the unassembled DNA. Finally, the AuNP-DNA was dispersed in 1 mL PBS (pH 7.4, 10 mM, 500 mM NaCl) and stored at 4 °C.
11, respectively). After shaking gently for 16 h at room temperature, the two oligonucleotides were self-assembled onto the AuNPs. Next, NaCl solution was added into the above solution gradually until the concentration of NaCl reached 500 mM in 3 days, which was used to conjugate more DNA strands on AuNPs.13 Then the suspension was centrifuged and rinsed five times (pH 7.4, 10 mM PBS, 100 mM NaCl) to remove the unassembled DNA. Finally, the AuNP-DNA was dispersed in 1 mL PBS (pH 7.4, 10 mM, 500 mM NaCl) and stored at 4 °C.
Fabrication of the nanosensor
The AuNDEs were cleaned electrochemically in a 0.5 M H2SO4 solution as reported previously.19 Prior to immobilizing on the nanoelectrode surface, 10 μL of HP-DNA solution (10 μM) was activated with 10 μL TCEP solution (1 mM) for 1 h to reduce the formation of the disulfide bond, and then diluted to 100 μL with Tris-HCl buffer (10 mM, pH 7.4, 100 mM NaCl, the same below). Herein, TCEP is a disulfide bond reductant.23 When HP-DNA is dissolved in solution, the –SH terminal group on HP-DNA easily formed disulfide bond, which is not beneficial for the modification of HP-DNA on the electrode surface. Therefore, TCEP is added to reduce the formation of the disulfide bond. After that, the AuNDE was immersed in the diluted HP-DNA solution for 12 h, followed by incubating in 1.0 mM MCH for 1 h to eliminate the non-specifically adsorbed ssDNA molecules and to hold a good orientation of probe thiolated ssDNA for its good recognition ability.24 After rinsing with buffer solution, the HP-DNA modified electrode was attached with 2 μM TBA for another 1 h. Finally, AuNP-DNA was immobilized onto the electrode surface by hybridizing with TBA for 2 h, and the nanosensor was thoroughly washed with Tris-HCl buffer for following tests.Electrochemical measurements
A three-electrode system was applied for all tests, including an Ag/AgCl reference electrode, a prepared working electrode, and a Pt wire auxiliary electrode. Electrochemical performance was investigated by DPV in 10 μM Tris-HCl buffer containing 400 μM Ru(NH3)63+. DPV parameters were as follows: −0.5 to 0.2 V, a pulse width of 0.05 s, a pulse period of 0.1 s and an amplitude of 0.05 V. Chronocoulometric (CC) measurements were carried out at a pulse period of 500 ms and a pulse width of 700 mV in Tris-HCl buffer containing 10 μM Ru(NH3)63+. In control experiments, DPV responses were carried out in a 10 μM Tris-HCl buffer containing 400 μM Ru(NH3)63+.Results and discussion
Characterization and electrochemical properties of single AuNDEs
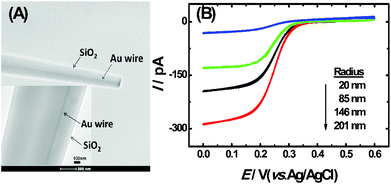
The TEM image of the single AuNDE is shown in Fig. 1A. It is clearly seen that the Au nanowire has been sealed inside a glass layer, the disk is polished smoothly and Au is exposed on a glassy sheath. From the zoom-in image (Fig. 1A, inset), it can be observed that the diameter of the Au nanowire is ∼50 nm and no obvious gap is formed between SiO2 and the Au nanowire. To prove the existence of the gold element, energy-dispersive X-ray spectroscopy (EDS) data and cyclic voltammogram (CV) of the single AuNDE in a 0.5 M H2SO4 solution were collected (shown as Fig. S1 and S2†). From the typical gold energy peaks and characteristic gold reduction peak (∼0.9 V) and oxidation peak (>1.2 V) in H2SO4 solution, it can be confirmed that the AuNDE is fabricated successfully and the surface is electrochemically active.To investigate the electrochemical properties of single AuNDEs, the CVs of different redox species, including K3Fe(CN)6, (Ru(NH)3)6Cl3, and Fc, were recorded, and the results are provided in Fig. 1B, Fig. S3 and S4.† From the above results, it can be seen that all the CVs have well-defined sigmoidal shape with a low charging current, indicating that the AuNDEs were well prepared.4,25 The limiting current of the AuNDE could be calculated according to Bard's book.26
| id = 4nFDCba | (1) |
Characterization of AuNPs and AuNP-DNA complexes
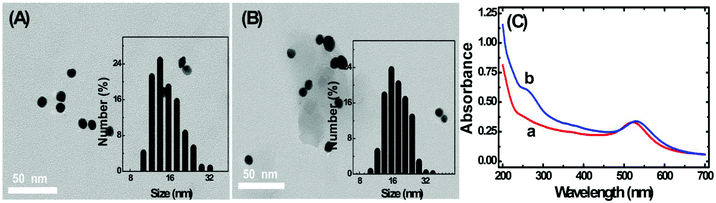
The TEM images of homemade AuNPs and AuNP-DNA complexes are shown in Fig. 2A and B, and we can see that two nanoparticles are monodisperse with a relatively uniform particle size distribution, with no visual differences in dispersity and morphology. The difference in size distribution was obtained by DLS analysis (the insets of Fig. 2A and B). The average hydrodynamic diameters of naked and modified AuNPs were 13 nm and 17 nm, respectively.The modification process of AuNPs with DNA was further assessed by UV-vis absorption spectroscopy. As represented in Fig. 2C, bare AuNPs show a typical absorption peak at 520 nm, which is a characteristic AuNP absorption peak with 13 nm diameter according to previous reports.29 The typical 13 nm AuNP absorption peak matches well with the DLS data (Fig. 2A). When AuNPs are modified with DNA, the 13 nm absorption peak of AuNPs has a slight red shift (from 520 nm to 524 nm) due to the interparticle plasmon coupling.14 In addition, an emerging absorption peak at ∼260 nm was considered as the absorption peak of S1 and S2 on the AuNPs,30,31 indicating that DNA was successfully modified on the AuNPs.
Feasibility of the strategy
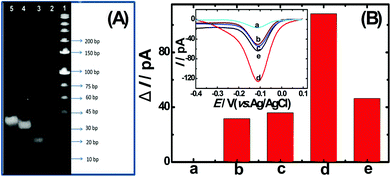
To confirm the fabrication process of the sandwich DNA complex, native polyacrylamide gel electrophoresis (PAGE) and DPV responses of five stages of the modified electrode were recorded. As shown in Fig. 3A, PAGE was used to monitor the hybrid process of DNA strands. HP-DNA1 and S11 used here corresponded to HP-DNA and S1 respectively, which had similar DNA sequences, only without a thiol group. Lanes 2 and 3 represented two single DNA strands (HP-DNA1 and TBA) with different lengths. The hybrid duplexes of HP-DNA1 and TBA (Lane 4) ran slowly and the sandwich DNA hybridization (Lane 5) showed a lower migration rate in PAGE indicating the successful formation of the sandwich DNA. Meanwhile, DPV responses were further recorded to confirm this process. From Fig. 3B, it could be seen that a reduction peak of Ru(NH3)63+ at ∼−0.1 V was observed due to the HP-DNA immobilized on the single AuNDE by Au–S chemistry, followed by MCH blocking (curve b), and then TBA hybridized with HP-DNA increased the electrochemical signal slightly (curve c) because of the formation of duplex DNA. When the AuNP-DNA complex modified S1 and S2 were attached to the remaining region of TBA, a significant current peak appeared owing to the abundant amount of DNA capturing Ru(NH3)63+via electrostatic forces (curve d). Upon the addition of thrombin, the sandwich DNA complexes were dissociated due to the specific affinity of TBA and thrombin, followed by the remarkable reduction of signal (curve e). Moreover, it could be observed that the signal change in Ru(NH3)63+ was directly affected by the concentration of thrombin (shown later). In this way, the proposed nanosensor can be used for the sensitive detection of thrombin. The effect of electrode size on the sensing performance was also investigated and the results are provided in Fig. S6 (ESI†), which showed that the electrochemical signal decreased as the electrode size decreased. When the radius of the electrode was down to 20 nm, almost no signal could be obtained, which might be due to the small surface area of the nanoelectrode that could not be modified with abundant DNA molecules. Furthermore, the packing density of helper-DNA immobilized DNA was measured via CC measurements using Ru(NH3)63+ as a redox marker (Fig. S7, ESI†). ΓDNA can be calculated according to the following eqn (1) and (2):32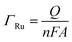 | (1) |
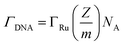 | (2) |
Optimization of experimental conditions
In order to obtain a maximum electrochemical signal of Ru(NH3)63+, the ratio of S1/S2 attached to AuNPs has been optimized. When the concentrations of HP-DNA and TBA were fixed, the influence of five different ratios of S1/S2 was investigated via recording the change in DPV signals (Fig. S8A, ESI†). S1 is the longer DNA sequence, which is responsible for hybridizing with TBA and attaching on the electrode surface. S2 is the shorter DNA sequence, which is responsible for loading more Ru(NH3)63+ species. Decreasing the ratio of S1/S2 can avoid steric hindrance and more S1 is exposed for hybridization with TBA, while too fewer S1 binding with the AuNPs would result in less AuNPs immobilized on the AuNDE through the hybridization reaction between S1 and TBA, and the DPV signal is decreased. As depicted in Fig. S8A (ESI†), the current increased with the decrease in the ratio of S1/S2 until the ratio was down to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9; a further ratio decrease does not make the current increase, but decrease because a too low amount of S1 can cause less AuNPs to be immobilized on the AuNDE. Therefore, the optimal ratio of S1/S2 was 1
9; a further ratio decrease does not make the current increase, but decrease because a too low amount of S1 can cause less AuNPs to be immobilized on the AuNDE. Therefore, the optimal ratio of S1/S2 was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9. Moreover, the dissolving time of the sandwich AuNP-DNA complexes is also a crucial factor for appraising the analytical performance of the designed nanosensor. Fig. S8B (ESI†) shows that the current decreased with the increasing time and reached a minimum value at 2 h. Overtime is by no means favorable for the decreasing current. Therefore, thrombin can be successfully bound to the TBA from the sandwich DNA complex within 2 h, which is comparable to the previous report.17
9. Moreover, the dissolving time of the sandwich AuNP-DNA complexes is also a crucial factor for appraising the analytical performance of the designed nanosensor. Fig. S8B (ESI†) shows that the current decreased with the increasing time and reached a minimum value at 2 h. Overtime is by no means favorable for the decreasing current. Therefore, thrombin can be successfully bound to the TBA from the sandwich DNA complex within 2 h, which is comparable to the previous report.17
Thrombin sensing using a prepared nanosensor
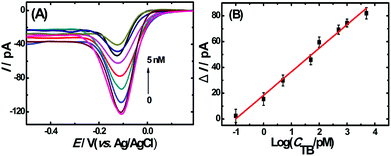
To evaluate the analytical performance of the proposed apta-nanosensor, DPV responses in the presence of TB with different concentrations were investigated under above optimal experimental conditions. As shown in Fig. 4A, the DPV signals decreased accordingly with the increasing thrombin concentrations from 0.1 pM to 5 nM. The results indicated that the sandwich DNA complexes were dissolved with the addition of thrombin, thus no more complex DNA structure was available for the capture of Ru(NH3)63+, resulting in a decrease in DPV response. Fig. 4B represents a linear relationship between the current change and the logarithm of the concentration of the target thrombin with a correlation coefficient of 0.9852. The linear regression equation is ΔI = 18.27log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CTB(pM) + 18.35. Based on IUPAC rules, the limit of detection can be defined as: LOD = 3.29σblank/S, where S is the slope of the linear calibration plot, which indicates the method calibration sensitivity and σblank is the blank standard deviation. The limit of detection was calculated to be 0.02 pM (S/N = 3) which was much lower or comparable to the results reported previously33–38 which are summarized in Table S1 (ESI†).
CTB(pM) + 18.35. Based on IUPAC rules, the limit of detection can be defined as: LOD = 3.29σblank/S, where S is the slope of the linear calibration plot, which indicates the method calibration sensitivity and σblank is the blank standard deviation. The limit of detection was calculated to be 0.02 pM (S/N = 3) which was much lower or comparable to the results reported previously33–38 which are summarized in Table S1 (ESI†).
For comparison, the DPV response of the conventional Au electrode with the radius of 1.5 mm in a 10 μM Tris-HCl buffer containing 400 μM Ru(NH3)63+ was also recorded (shown in Fig. S9†). The current density from our prepared Au nano-sized electrode calculated from Fig. 4A was 0.024 A cm−2, which was greatly larger than that from the Au macro-sized electrode shown in Fig. S9† (1.35 × 10−5 A cm−2), indicating that the prepared nanosensor has its advantage for sensing applications. The successful construction of the label-free, high sensitivity and low LOD sensing platform should be attributed to the signal amplification strategy assisted by AuNP-DNA complexes on the surface of nanoelectrodes. In order to confirm that the decrease in signal is not due to the blocking of the electroactive area by thrombin, control experiments were carried out. S1 was removed from the sensor, and DPV responses in Ru(NH3)63+ solution are shown in Fig. S10 (ESI†). Because S1 and TBA can bind together due to the designed specific affinity, the AuNPs cannot be immobilized on the electrode surface in the absence of S1, and the DPV response of the sensor was greatly decreased compared to the value shown in Fig. 3B and 4.† After incubating in 500 pM thrombin for 2 h, the DPV response was decreased slightly, but it was still greater than the response on helper-DNA, indicating that TBA was partially removed from the electrode surface. These results proved definitively that the DPV signal decrease was not due to the blocking of the electroactive area by TB, but the TBA–TB complex dissolution, which led to the reduction of adsorbed Ru(NH3)63+.
Selectivity, stability and reproducibility
To evaluate the specific responses of the nanosensor, control experiments were done by incubating the nanosensors with several aqueous solutions containing lgG, lysozyme, HPR, BSA (all concentrations are 10 nM) and thrombin (1000 pM) in Tris-HCl buffer. As depicted in Fig. S11 (ESI†), only the signal change caused by thrombin is most obvious as well as those caused by other proteins being very small, which proved the high specificity of the aptamer to the target and the feasibility of the aptasensor in complex samples.The stability and reproducibility of the aptasensor were also investigated. After storing at 4 °C for 4 days, the AuNP-DNA/TBA/HP-DNA/MCH/AuNDE retained 91% of original DPV response in Ru(NH3)63+ aqueous solution, indicating the satisfactory stability of the nanosensors. In addition, three modified electrodes with the same preparation process were used for the detection of thrombin; the relative standard deviation is 1.63%, 3.72% and 2.81%, respectively (Table S2†). All of these results demonstrate that the developed aptasensor possessed satisfactory results.
Application of the nanosensor
To evaluate the applicability and practicability of the nanosensor in a complex system, 1000-fold diluted serum samples were measured with different concentrations of thrombin (1, 10, 100, 1000 pM, shown in Table 1). The recoveries for the added thrombin are 91.1%–113.4% with relative standard deviation (RSD) between 1.1% and 8.1%, implying that the novel aptasensor has great practical application in quantitative assays of thrombin in real samples.| Samples | Added (pM) | Found (pM) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| 1 | 0 | Undetectable | — | — |
| 2 | 1 | 1.07 | 98.0–113.4 | 8.1 |
| 3 | 10 | 9.6 | 91.1–100.7 | 4.8 |
| 4 | 100 | 97.7 | 96.2–100.5 | 2.5 |
| 5 | 1000 | 1006.2 | 99.3–101.4 | 1.1 |
Conclusions
In conclusion, based on the unique properties of nanoelectrodes and the signal amplification strategy, a novel, label-free and sensitive sensing platform for electrochemical detection of thrombin was developed. The signal amplification strategy was realized through AuNP-DNA complexes and Ru(NH3)63+ redox as the indicator. Compared with traditional DNA-based sensors, this nanosensor not only has a wide linear-range, low detection limit and good selectivity, but also has the advantage of small overall dimensions and fast response, which can make it easily applied to detect biomolecules at single molecule per cell level or living biosystem.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Prof. Bo Zhang (Department of Chemistry, University of Washington) for his useful suggestions and discussion. This work is financially supported by the National Natural Science Foundation of China (No. 21775003, 21375002) and the Foundation for Innovation Team of Bioanalytical Chemistry of Anhui Province.Notes and references
- S. M. Oja, M. Wood and B. Zhang, Anal. Chem., 2013, 85, 473–486 CrossRef CAS PubMed.
- J. Clausmeyer and W. Schuhmann, TrAC, Trends Anal. Chem., 2016, 79, 46–59 CrossRef CAS.
- Y. S. Fan, C. Han and B. Zhang, Analyst, 2016, 141, 5474–5487 RSC.
- Y. Zhang, S. Xu, X. Xiao, Y. Liu, Y. Qian and Y. Li, Chem. Commun., 2017, 53, 2850–2853 RSC.
- M. V. Mirkin, T. Sun, Y. Yu and M. Zhou, Acc. Chem. Res., 2016, 49, 2328–2335 CrossRef CAS PubMed.
- S. Lu, T. Hu, S. Wang, J. Sun and X. Yang, ACS Appl. Mater. Interfaces, 2017, 9, 167–175 CrossRef CAS PubMed.
- M. Liu, W. Zhang, Q. Zhang, J. D. Brennan and Y. Li, Angew. Chem., Int. Ed., 2015, 54, 9637–9641 CrossRef CAS PubMed.
- D. Wang, X. Xiao, S. Xu, Y. Liu and Y. Li, Biosens. Bioelectron., 2018, 99, 431–437 CrossRef CAS PubMed.
- J. Liu, S. Wagan, M. Davila Morris, J. Taylor and R. J. White, Anal. Chem., 2014, 86, 11417–11424 CrossRef CAS PubMed.
- C. A. Hutchison 3rd, H. O. Smith, C. Pfannkoch and J. C. Venter, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 17332–17336 CrossRef PubMed.
- Y. Shen, F. Tian, Z. Chen, R. Li, Q. Ge and Z. Lu, Biosens. Bioelectron., 2015, 71, 322–331 CrossRef CAS PubMed.
- X. Y. Lin, L. Cui, Y. S. Huang, Y. Lin, Y. Xie, Z. Zhu, B. C. Yin, X. Chen and C. J. Yang, Chem. Commun., 2014, 50, 7646–7648 RSC.
- E. M. Driggers, H. S. Cho, C. W. Liu, C. P. Katzka, A. C. Braisted, H. D. Ulrich, D. E. Wemmer and P. G. Schultz, J. Am. Chem. Soc., 1998, 120, 1945–1958 CrossRef CAS.
- X. Jing, X. Cao, L. Wang, T. Lan, Y. Li and G. Xie, Biosens. Bioelectron., 2014, 58, 40–47 CrossRef CAS PubMed.
- T. Wang, E. Viennois, D. Merlin and G. Wang, Anal. Chem., 2015, 87, 8173–8180 CrossRef CAS PubMed.
- M. D. Massich, D. A. Giljohann, A. L. Schmucker, P. C. Patel and C. A. Mirkin, ACS Nano, 2010, 4, 5641–5646 CrossRef CAS PubMed.
- L. Wang, R. Ma, L. Jiang, L. Jia, W. Jia and H. Wang, Biosens. Bioelectron., 2017, 92, 390–395 CrossRef CAS PubMed.
- B. Johann, J. A. Heit and G. O. Whyte, Thromb. Res., 1996, 84, 289–294 CrossRef.
- Y. Zhang, S. Xu, Y. Qian, X. Yang and Y. Li, RSC Adv., 2015, 5, 77248–77254 RSC.
- K. C. Grabar, R. G. Freeman, M. B. Hommer and M. J. Natan, Anal. Chem., 1995, 67, 735–733 CrossRef CAS.
- Q. Shao, P. Wu, P. Gu, X. Xu, H. Zhang and C. Cai, J. Phys. Chem. B, 2011, 115, 8627–8637 CrossRef CAS PubMed.
- H. Pei, Y. Zheng, R. Kong, L. Xia and F. Qu, Biosens. Bioelectron., 2016, 85, 76–82 CrossRef CAS PubMed.
- M. Zhou, J. E. Dick and A. J. Bard, J. Am. Chem. Soc., 2017, 139, 17677–17682 CrossRef CAS PubMed.
- A. Asadzadeh-Firouzabadi, H. R. Zare and N. Nasirizadeh, J. Electrochem. Soc., 2016, 163, B43–B48 CrossRef CAS.
- P. Sun and M. V. Mirkin, Anal. Chem., 2006, 78, 6526–6534 CrossRef CAS PubMed.
- A. J. Bard and L. R. Faulkner, Electrochemical Methods: Fundamentals and Applications, John Wiley & Sons, Inc, Hoboken, NJ, 2001 Search PubMed.
- Y. Li, Q. Wu, S. Jiao, C. Xu and L. Wang, Anal. Chem., 2013, 85, 4135–4140 CrossRef CAS PubMed.
- B. Zhang, Y. Zhang and H. S. White, Anal. Chem., 2006, 78, 477–483 CrossRef CAS PubMed.
- J. J. Storhoff, R. Elghanian, R. C. Mucic, C. A. Mirkin and R. L. Letsinger, J. Am. Chem. Soc., 1998, 120, 1959–1964 CrossRef CAS.
- G. Raschke, S. Kowarik, T. Franzl, C. Sonnichsen, T. A. Klar, J. Feldmann, A. Nichtl and K. Kurzinger, Nano Lett., 2003, 3, 935–938 CrossRef CAS.
- H. Zhang, H. Dong, G. Yang, H. Chen and C. Cai, Anal. Chem., 2016, 88, 11108–11114 CrossRef CAS PubMed.
- L.-D. Li, H.-T. Zhao, Z.-B. Chen, X.-J. Mu and L. Guo, Sens. Actuators, B, 2011, 157, 189–194 CrossRef CAS.
- Y. J. Li, Y. Q. Li, N. Xu, J. H. Pan, T. F. Chen, Y. W. Chen and W. H. Gao, Sens. Actuators, B, 2017, 240, 742–748 CrossRef CAS.
- K. Shi, B. Dou, J. Yang, R. Yuan and Y. Xiang, Biosens. Bioelectron., 2017, 87, 495–500 CrossRef CAS PubMed.
- Y. Wang, L. Bao, Z. Liu and D. W. Pang, Anal. Chem., 2011, 83, 8130–8137 CrossRef CAS PubMed.
- J. Yang, B. Dou, R. Yuan and Y. Xiang, Anal. Chem., 2017, 89, 5138–5143 CrossRef CAS PubMed.
- F. Ahour and M. K. Ahsani, Biosens. Bioelectron., 2016, 86, 764–769 CrossRef CAS PubMed.
- Z. J. Xi, R. R. Huang, Y. Deng, E. B. Su and N. Y. He, Sci. Adv. Mater., 2016, 8, 1678–1682 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8an01474d |
| This journal is © The Royal Society of Chemistry 2019 |