Open Access Article
Open Access ArticleA DNA-based parity generator/checker for error detection through data transmission with visual readout and an output-correction function†
Daoqing
Fan
ab,
Erkang
Wang
ab and
Shaojun
Dong
*ab
aState Key Laboratory of Electroanalytical Chemistry, Changchun Institute of Applied Chemistry, Chinese Academy of Sciences, Changchun, Jilin, 130022 China. E-mail: dongsj@ciac.ac.cn
bUniversity of Chinese Academy of Sciences, Beijing, 100039 China
First published on 26th October 2016
Abstract
During any type of binary data transmission, the occurrence of bit errors is an inevitable and frequent problem suffered. These errors, which have fatal effects on the correct logic computation, especially in sophisticated logic circuits, can be checked through insertion of a parity generator (pG) at the transmitting end and a parity checker (pC) at the receiving end. Herein, taking even pG/pC as a model device, we constructed the first DNA-based molecular parity generator/checker (pG/pC) for error detection through data transmission, on a universal single-strand platform according to solely DNA hybridization. Compared with previous pG/pC systems, the distinct advantage of this one is that it can present not only fluorescence signals but also visual outputs, which can be directly recognized by the naked eye, using DNA inputs modulated split-G-quadruplex and its DNAzyme as signal reporters, thus greatly extending its potential practical applications. More importantly, an “Output-Correction” function was introduced into the pC for the first time, in which all of the erroneous outputs can be adequately corrected to their normal states, guaranteeing the regular operation of subsequent logic devices. Furthermore, through negative logic conversion towards the constructed even pG/pC, the odd pG/pC with equal functions was obtained. Furthermore, this system can also execute multi-input triggered concatenated logic computations with dual output-modes, which largely fulfilled the requirements of complicated computing.
Introduction
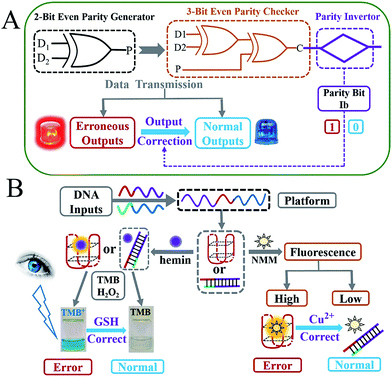
As molecular-level computers carry out binary Boolean operations for data processing, untraditional molecular computing has attracted increasing interest across various research fields in recent years.1–12 During any type of binary data transmission, the occurrence of bit errors13 is an inevitable and frequent problem suffered. These errors, which have fatal effects on the correct logic computation, especially in complicated logic circuits,9,14,15 can be checked through insertion of a parity generator (pG) at the transmitting end and a parity checker (pC) at the receiving end.16 According to the parity generating/checking principle, pG/pCs can be divided into two types: even and odd. As illustrated in the definition,16,17 even pGs can produce an extra parity bit, P, and add it to the original binary bit, Dn, making the total number of 1’s (Σ) in the DnP string even. Taking the transmission of two bits of data (D1 and D2) as an example, the 2-bit even pG will assign the binary value to P (output of pG) according to the truth table of an XOR logic gate18 (see Scheme 1A and Fig. 1A). Consequently, the D1D2P string produced by the 2-bit even pG is transmitted to a 3-bit even pC and subsequently checked by it (Scheme 1A). In the case of an erroneous transmission, the transmitted bits in the D1D2P string will be changed by the disturbances during data transmission (occurrence of bit errors), the number of 1’s in the received wrong string will be altered to an odd value (see Table 1), then the pC will give an “alarm” signal (red light in Scheme 1A), producing the output C = 1. In contrast, if the transmission is normal, the three bits will not be changed and the number of 1’s in the D1D2P string will still be even. As a result, the pC shows a “normal” signal (blue light in Scheme 1A), producing the output C = 0. Therefore, normal/erroneous transmissions can be easily distinguished according to the outputs of the pC. All the above illustrates the “error detection” principle of the even pG/pC system. Analogously, the odd pG/pC system possesses identical functions for error detection through data transmission.Though pG/pC systems have shown great advancements with scientists' efforts, there are several non-negligible limitations that remain further endeavours to break. Firstly, most of the previously developed pG/pC systems have been based on semiconductor substrates, in which the optical inputs were processed by a Mach–Zehnder interferometer or an optical amplifier and so on.17,19–23 The indirect optical outputs of most semiconductor pG/pC systems were often recorded by cumbersome instruments and were difficult to visualize by the naked eye, which restricted the potential practical applications. Secondly, in 2013, Pischel and coworkers pioneered the first molecular implementation of pG/pC using the fluorescence of artificially synthesized organic molecules as the output.16 However, this just showed a general signal-to-noise (S/N) ratio, and organic molecules also usually require a sophisticated synthesis/purification procedure. Molecular pG/pC systems for error detection through data transmission were barely reported after that. Finally, and most importantly, the “Output-Correction” function was never integrated into pCs previously. As commonly used devices in data transmission, pG/pC systems were frequently integrated into complicated logic circuits.14,15 For the erroneous outputs produced by the pC, in addition to giving an “alarm” signal for the occurrence of errors through data transmission, the application of them in subsequent logic devices is fatal. Only the normal outputs of preceding pCs could guarantee the regular operation of later devices (see Scheme S1† and corresponding explanations), which makes the correction of erroneous outputs necessary. Considering the potential applications of pG/pC systems in molecular computation, an easily operated, cost-effective and efficient molecular pG/pC system, with an “Output-Correction” function and a satisfactory S/N ratio, that can present not only fluorescent signals, but also visual outputs recognized by the naked eye, is highly desirable and remains the subject of further study.
Due to its sequence-specificity, well-ordered, flexible-design, predictable-structure and low-cost, DNA is regarded as an outstanding candidate for molecular logic computation,4,5,24–29 in which the G-quadruplex (G4) and its peroxidase-like G4 DNAzyme (G4zyme) are broadly used as signal reporters.8,9,14,15 On the one hand, the fluorescence of N-methyl mesoporphyrin IX (NMM) can be dramatically enhanced after binding to G4;9 on the other hand, upon the stacking of hemin, the formed G4zyme can catalyze the oxidation of 3,3′,5,5′-tetramethylbenzidine (TMB) by H2O2 to produce a colorimetric signal (oxidized TMB, TMB+),8,9 which can be readily distinguished by the naked eye as shown in Scheme 1B. It has been reported that single-stranded DNA, duplex DNA and even incomplete G4 exhibit obvious weaker fluorescence enhancement towards NMM and a much lower peroxidation activity after binding to hemin, in comparison with the intact G4.30,31 Inspired by the above merits and taking the even pG/pC system as a model device, we herein construct the first DNA-based molecular pG/pC system for error detection through data transmission, with an “Output-Correction” function based on a universal single-strand platform, using DNA inputs pre-hybridization modulated split-G4/G4zyme as label-free signal reporters (which can simplify the operation and reduce the costs). It can exhibit not only fluorescent signals that enable easy remote reading,32 but also visual outputs that can be directly viewed by the naked eye. Through negative logic conversion (a broadly used conversion in logic device operation)33,34 towards the constructed even pG/pC system, the odd pG/pC system with equal functions was obtained. Furthermore, this platform can also perform multi-input triggered concatenated logic computations.
Results and discussion
Operation of the 2-bit even pG
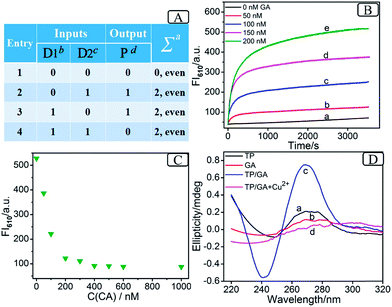
As the prerequisite of a 3-bit even pC, a 2-bit even pG should be implemented at first (truth table, Fig. 1A). A two-input XOR logic gate properly meets these requirements.16Scheme 2 outlines the operation mode of the DNA-based 2-bit even pG, in which pre-hybridization between different DNA inputs and a split G4 (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
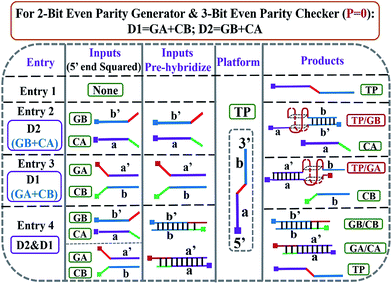
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) strategy are utilized. The pre-hybridization between different inputs can not only prohibit the unnecessary background signal effectively, but also bring flexibility to the operation of the pG/pC system. The single-strand TP, whose middle part (red region) is integrated with GGGTGGG, functions as the universal platform of pG and pC. The mixture of strands GA and CB (GA + CB) acts as the input D1 and that of strands GB and CA (GB + CA) acts as the input D2. As presented in entry 3 of Scheme 2, the segment (TGGG)2 (red region) is integrated into the 5′ end of GA, and its purple region (a′) can hybridize with the purple part of TP (a) to form intact G4 (TP/GA, see products of entry 3). The obviously enhanced fluorescence intensity at 610 nm (FI610) of NMM in the presence of TP and the increasing concentrations of GA (Fig. 1B) and the characteristic peaks9 (Fig. 1D(c)), prove the formation of parallel G4. Similarly, the 3′ end of GB (entry 2 of Scheme 2) is integrated with the segment (GGGT)2 (red region), and the blue region of GB (b′) can hybridize with the blue part of TP (b) to form intact G4 (TP/GB, see products of entry 2), Fig. S2 (ESI, SI†). In contrast, the segment CCCA (green region) is integrated into the 3′ end of CA, and its purple region (a) can hybridize with the a′ part of GA to form a more stable duplex GA/CA (higher Tm value35 than TP/GA, Fig. S3 and Table S2†), see products of entry 4. Thus the pre-hybridization between GA and CA before the addition of TP inhibits the formation of G4 (TP/GA), which is verified in Fig. 1C. Analogously, the 5′ end of CB (entry 3 of Scheme 2) is integrated with the segment ACCC (green region), and the blue region of CB (b) can also hybridize with the b′ part of GB to form a more stable duplex GB/CB than TP/GB, thus inhibiting the formation of G4 (TP/GB) (Table S2†). While, the mix of CB and GA (or the mix of CA and GB) influences neither the interaction between TP and GA (or GB), nor the formation of G4, Fig. S1, S2D.† All the interactions among different DNA strands were verified by native polyacrylamide gel electrophoresis (PAGE) results, Fig. S1.† The sequences of the oligonucleotides used are presented in Table S1.†
2) strategy are utilized. The pre-hybridization between different inputs can not only prohibit the unnecessary background signal effectively, but also bring flexibility to the operation of the pG/pC system. The single-strand TP, whose middle part (red region) is integrated with GGGTGGG, functions as the universal platform of pG and pC. The mixture of strands GA and CB (GA + CB) acts as the input D1 and that of strands GB and CA (GB + CA) acts as the input D2. As presented in entry 3 of Scheme 2, the segment (TGGG)2 (red region) is integrated into the 5′ end of GA, and its purple region (a′) can hybridize with the purple part of TP (a) to form intact G4 (TP/GA, see products of entry 3). The obviously enhanced fluorescence intensity at 610 nm (FI610) of NMM in the presence of TP and the increasing concentrations of GA (Fig. 1B) and the characteristic peaks9 (Fig. 1D(c)), prove the formation of parallel G4. Similarly, the 3′ end of GB (entry 2 of Scheme 2) is integrated with the segment (GGGT)2 (red region), and the blue region of GB (b′) can hybridize with the blue part of TP (b) to form intact G4 (TP/GB, see products of entry 2), Fig. S2 (ESI, SI†). In contrast, the segment CCCA (green region) is integrated into the 3′ end of CA, and its purple region (a) can hybridize with the a′ part of GA to form a more stable duplex GA/CA (higher Tm value35 than TP/GA, Fig. S3 and Table S2†), see products of entry 4. Thus the pre-hybridization between GA and CA before the addition of TP inhibits the formation of G4 (TP/GA), which is verified in Fig. 1C. Analogously, the 5′ end of CB (entry 3 of Scheme 2) is integrated with the segment ACCC (green region), and the blue region of CB (b) can also hybridize with the b′ part of GB to form a more stable duplex GB/CB than TP/GB, thus inhibiting the formation of G4 (TP/GB) (Table S2†). While, the mix of CB and GA (or the mix of CA and GB) influences neither the interaction between TP and GA (or GB), nor the formation of G4, Fig. S1, S2D.† All the interactions among different DNA strands were verified by native polyacrylamide gel electrophoresis (PAGE) results, Fig. S1.† The sequences of the oligonucleotides used are presented in Table S1.†
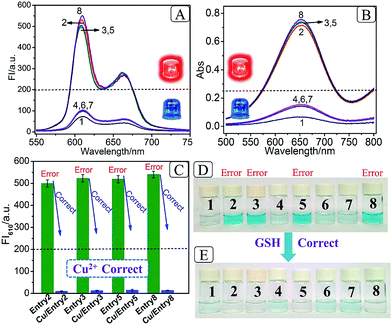
Based on the above mechanism, a 2-bit even pG was operated, as shown in Scheme 2. The FI610 of NMM and the color changes of TMB oxidized by H2O2 under the catalysis of G4zyme, were used as fluorescent and visual outputs P, respectively. In the absence of any input (entry 1), the final product only consists of strand TP with half of G4. After interaction with NMM or the catalytic oxidation of TMB upon binding with hemin, a negligible fluorescent signal and a pale blue color were obtained (P = 0) as shown in Fig. S4A(a), B(a).† In the presence of any input (D1 or D2), the pre-hybridization between GB and CA, or between GA and CB, produced no duplexes. After reacting with TP, the final products were TP/GB and CA (entry 2) or TP/GA and CB (entry 3), accompanied with the formation of G4, yielding a dramatically enhanced fluorescent signal and a dark blue color (P = 1), Fig. S4A(b, c), B(b, c).† However, for the co-existence of both inputs (entry 4), GB in D2 hybridized with CB in D1 and GA in D1 hybridized with CA in D2 during input pre-hybridization. After the addition of TP, the final products were GB/CB, GA/CA and TP. The formation of G4 was inhibited, producing a weak fluorescent signal and a pale blue color (P = 0), Fig. S4A(d), B(d).† A FI610 of 200 a.u. (NMM) and an Abs652 of 0.25 (TMB+) were set as the constant threshold values for fluorescent and visual outputs of the pG/pC system to judge the positive and negative signals with high S/N ratios. The input and output states accorded well with the truth table (Fig. 1A). Thus, the operation of a 2-bit even pG, which could present fluorescent and visual outputs with a high S/N ratio, was accomplished.
Fabrication of the 3-bit even pC
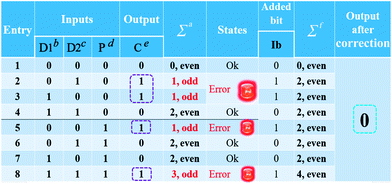
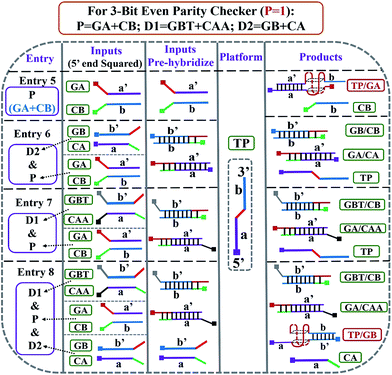
After the fabrication of the DNA-based 2-bit even pG, a 3-bit even pC was conceptually operated, into which the third input P was introduced to properly mimic its function. The corresponding truth table presented in Table 1 can be separated into two parts. The first part is the P = 0 states (entry 1 to 4 in Table 1), whose introduction will not change the final output of the 3-bit even pC. The logic circuit still equates to an XOR logic gate, and the DNA reactions have been discussed for the 2-bit even pG (now the fluorescent and visual outputs are referred to as output C). The later part refers to the P = 1 states (entry 5 to 8 in Table 1), in which the third input P plays the XNOR function which is complementary to inputs D1 and D2.16 The DNA hybridization mode of different entries is shown in Scheme 3. The platform in this case (P = 1 states) is still the TP strand, but the mixture of two new DNA strands, GBT and CAA, was introduced as another input to fulfil the requirements. Strand GBT is 5-thymine (grey region) longer than strand GB at the 5′ end, and strand CAA is 5-adenine (black region) longer than strand CA at the 5′ end, which indicates that GBT and CAA possess similar properties to GB and CA, respectively (see Fig. S5 and S6†). Just like the previous report,16 to implement the 3-bit even pC (under P = 1 states) properly, the three inputs were defined as follows: the mixture of GBT and CAA acts as input D1 and that of GB and CA acts as D2, while the mixture of GA and CB is referred to as input P. For entry 5 (input P solely), the reaction was the same as that of entry 3 in Scheme 2, producing the output C = 1. The operation of entry 6 (input D2 and P) was also identical to that of entry 4, yielding the output C = 0. For the co-existence of inputs D1 and P (entry 7), GBT in D1 hybridized with CB in P, and GA in P hybridized with CAA in D1 after inputs pre-hybridization. After reacting with TP, the final products were GBT/CB, GA/CAA and TP, without the formation of G4 (see the products of entry 7), producing a weak fluorescent signal and a pale blue color (C = 0). For the coexistence of D1, D2 and P (entry 8), taking the pre-mixture of P and D1 as an example (the pre-mixture of P and D2 has the same effect), the reaction was similar to that of entry 7. The products after input pre-hybridization were GBT/CB, GA/CAA, GB and CA. After reacting with TP, b′ of GB (blue part) hybridized with b of TP to form G4. The final products were GBT/CB, GA/CAA, CA and TP/GB (G4), exhibiting a strong fluorescent signal and a dark blue color (C = 1). Fig. 2A, B and D present the fluorescence and absorbance spectra and visual photos of all eight entries of the 3-bit even pC, which proves its successful fabrication. The erroneous entries (entries 2, 3, 5 and 8) could be easily distinguished by the naked eye.“Output-Correction” function for 3-bit even pC
As discussed before, to guarantee the regular operation of downstream logic devices in sophisticated logic circuits, the “Output-Correction” function needs to be introduced into the pC. While the outputs of the pC were obtained according to the parity of 1’s in the DnP string, in order to correct the erroneous outputs and to make all the outputs of the pC normal, the wrong parity of 1’s in the erroneous DnP string should be inverted to their correct states, and the correct parity of 1’s in the normal string should be kept unchanged. Therefore, a “parity invertor”, which plays the function of parity inverting through adding another “parity bit” (Ib) to the received DnP string was inserted after the pC, as shown in Scheme 1A (“1” for erroneous outputs and “0” for normal ones). Taking the even pC for instance, if the 3-bit D1D2P string after data transmission is normal (011, e.g., see entry 6 in Table 1; Σa = 2, even), the parity invertor will add “0” to the 3-bit string and produce a 4-bit D1D2PIb string (0110). The number of 1’s in the 4-bit string (Σf) is still even (2), therefore not changing the normal output “0”. Conversely, if errors occur during the transmission of the above 3-bit string (011 changed to 111, one bit error, e.g., see entry 8 in Table 1; Σa = 3, odd), the parity invertor will add “1” to the erroneous 3-bit string and this yields the 4-bit D1D2PIb string (1111), in which the Σf is even (4), therefore inverting the erroneous output “1” to “0”. Herein, to invert the wrong parity of 1’s of the above DNA-based pC, the abilities of Cu2+ to dissociate G4 and quench the fluorescence of NMM,9 and the property of glutathione (GSH) to reduce blue colored TMB+ to colorless TMB36 (eqn (S1)†), were used to correct the erroneous fluorescence and visual signals (entry 2, 3, 5 and 8) respectively, (Ib = “1” in the presence of Cu2+ or GSH for erroneous entries; Ib = “0” in the absence of them for normal entries). Corresponding verification experiments, as shown in Fig. 1D(d) and S7, S8†, fully proved the aforementioned abilities of Cu2+ and GSH. After the addition of Cu2+, the fluorescent signals of the erroneous entries were all corrected to their normal states, as shown in Fig. 2C. The visual outputs of all eight entries, after correction of the erroneous entries by GSH (Fig. 2E), also changed to normal, which were readily recognized by the naked eye. The column absorbance bars (Fig. S8†) further verified what the naked eye monitored. All the above phenomena demonstrate the efficient operation of the “Output-Correction” function for both fluorescence and visual outputs.It is worth mentioning that the 2-bit odd pG and 3-bit odd pC with fluorescent/visual outputs were also obtained through negative logic conversion33,34 towards the outputs of the above even ones (output “0” was defined as “1”, output “1” was defined as “0”). The logic circuits and truth tables presented in Fig. S9† illustrate its reasonable operation. The odd pC can also perform the “Output-Correction” function, using strand T30695 (an intact G4)37 or its G4zyme as the corrective element, as shown in Fig. S10.†
Concatenated logic computation
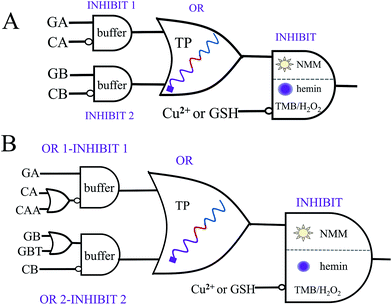
There is no doubt that a molecular logic system, which could not only execute parity generating/checking but also perform concatenated logic computations, would greatly fulfil the requirements of computational complexity.8,9 On the basis of the above system, five-input and seven-input concatenated logic circuits with dual output-modes (fluorescence/visual) were conceptually fabricated, as shown in Fig. 3. For the five-input concatenated logic circuit, strands GA, CA, GB and CB act as four individual inputs, and Cu2+ or GSH functions as the fifth input. The definitions of the fluorescent and visual outputs are identical to the above even pG/pC system. This concatenated circuit is composed of two parallel INHIBIT logic gates (INHIBIT 1 and INHIBIT 2), connected in series by an OR logic gate and another INHIBIT logic gate (Fig. 3A). Herein, strands GA and CA are the two inputs of the INHIBIT 1 gate, in which CA plays the role of an inhibitory element. Analogously, CB is the inhibitory input of the INHIBIT 2 gate (another input is GB). As discussed above, either GA or GB can hybridize with TP to form intact G4, which indicates that the outputs of two parallel INHIBIT gates (different mixtures of GA, CA for INHIBIT 1 and of GB, CB for INHIBIT 2) could act as the two inputs of the subsequent OR gate, using strand TP as the platform. Finally, the output of the OR gate (formation of G4 is defined as output “1”, other combinations of the five strands without formation of G4 were defined as output “0”), together with the inhibitory input of Cu2+ or GSH, comprise the last INHIBIT gate. All four logic gates compose the five-input concatenated logic circuit (INHIBT/INHIBIT)-OR-INHIBIT, in which there are 25 (32) input variations. The corresponding truth table and the fluorescence column values shown in Table S3 and Fig. S11† fully demonstrate its complete operation, which largely enriches the computing capacity. Analogously, if strands CAA and GBT are introduced as another two individual inputs, a seven-input concatenated circuit [(OR-INHIBT)/(OR-INHIBIT)]-OR-INHIBIT with 27 (128) input variations is also obtained, as shown in Fig. 3B (corresponding truth tables and results are not shown due to the numerous input variations).Conclusions
In conclusion, we have successfully fabricated the first DNA-based molecular parity generator/checker for error detection through data transmission, on a universal single-strand platform. It exhibits not only fluorescent signals, but also visual outputs which can be directly distinguished by the naked eye using the DNA inputs modulated split-G4/G4zyme as label-free signal reporters, thus greatly extending its potential applications. Significantly, an “Output-Correction” function was introduced into the molecular pC for the first time, in which all the erroneous outputs can be adequately corrected to normal states. Notably, the pG and pC were operated based on DNA hybridization only, without strand displacement or enzyme cleavage, which could simplify the operation and lower the costs. Furthermore, this system could also execute multi-input triggered concatenated logic computation with dual output-modes, thus largely meeting the requirements of complicated computing. This system enlightens the construction of more powerful DNA-based pG/pC systems, and in combination with nanomaterials or aptamers, it could be applied to intelligent drug loading/delivery, disease diagnosis and bioimaging, by virtue of the excellent biocompatibility of DNA.Experimental section
Chemicals
The oligonucleotides were synthesized by Shanghai Sangon Biotechnology Co. (Shanghai, China) and the sequences are given in Table S1.† All the DNAs were dissolved in distilled water as stock solutions and quantified by UV-vis absorption spectroscopy, using extinction coefficients (ε 260 nm, M−1 cm−1): A = 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400, G = 11
400, G = 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, C = 7400, T = 8700. NMM (N-methylmesoporphyrin IX) was purchased from J&K (Beijing, China) and the stock solution of NMM (50 μM) was prepared with dimethyl sulfoxide (DMSO) and stored at −20 °C in darkness. TMB (3,3′,5,5′-tetramethylbenzidine) was obtained from Sigma Aldrich (USA) and dissolved in DMSO as a stock solution (20.804 mM). Tris(tris(hydroxymethyl)aminomethane) and CuSO4 were purchased from Sinopharm Chemical Regent Co. (Shanghai, China). GSH (glutathione) was provided by Shanghai Sangon Biotechnology Co. (Shanghai, China). The water used in the experiments was purified using a Millipore system. Other chemicals were of reagent grade and used without further purification.
500, C = 7400, T = 8700. NMM (N-methylmesoporphyrin IX) was purchased from J&K (Beijing, China) and the stock solution of NMM (50 μM) was prepared with dimethyl sulfoxide (DMSO) and stored at −20 °C in darkness. TMB (3,3′,5,5′-tetramethylbenzidine) was obtained from Sigma Aldrich (USA) and dissolved in DMSO as a stock solution (20.804 mM). Tris(tris(hydroxymethyl)aminomethane) and CuSO4 were purchased from Sinopharm Chemical Regent Co. (Shanghai, China). GSH (glutathione) was provided by Shanghai Sangon Biotechnology Co. (Shanghai, China). The water used in the experiments was purified using a Millipore system. Other chemicals were of reagent grade and used without further purification.
CD spectra and Tm value measurements
CD spectra were measured on a JASCO J-820 spectropolarimeter (Tokyo, Japan) at room temperature. Spectra were recorded from 220 to 320 nm in 1 mm pathlength cuvettes and averaged from three scans.For the collection of thermal denaturation curves of the duplexes, the absorbances of different duplexes were monitored at 260 nm (the interval temperature was 1 °C, the ramp rate was 1 °C min−1 and the temperature range was 20 to 90 °C).
Native polyacrylamide gel electrophoresis (PAGE)
20 μM DNA stock solution was diluted to 10 μM using 2× TE buffer (100 mM Tris, 10 mM EDTA and 200 mM MgCl2) for future use. All the DNA solutions (10 μM) were heated at 90 °C for 10 min and slowly cooled down to room temperature. After that, different combinations of DNA (2 μM) were mixed and suitable volumes of 1× TE buffer (50 mM Tris, 5 mM EDTA and 100 mM MgCl2, pH 8.0) were added into the mixtures up to a final volume of 50 μL. After incubation at room temperature for more than 30 min, the DNA samples were analysed in a 15% native polyacrylamide gel. The electrophoresis was conducted in 1× TBE buffer (17.80 mM Tris, 17.80 mM boric acid and 2 mM EDTA, pH 8.0) at a constant voltage of 120 V for 1 h. The gels were scanned using a UV transilluminator after staining with Gel-Red.DNA hybridization of pG and pC
Fluorescence measurements
After DNA hybridization, the desired volume of 1× TE buffer (50 mM Tris, 5 mM EDTA and 100 mM MgCl2, pH 8.0) was added up to a final volume of 490 μL. Then 10 μL of 50 μM NMM was added and the mixture was incubated at room temperature for about 1 h. After incubation, the fluorescence emission spectra of the different samples were collected on a Cary Eclipse Fluorescence Spectrophotometer (Agilent Technologies, USA) at room temperature. The emission spectra of NMM were collected from 550 to 750 nm after excitation at 399 nm. The slit widths for the excitation and emission were 10 nm and 20 nm, respectively.Colorimetric reactions
Output-correction for the fluorescence and visual outputs of the even pC
For the “Output-Correction” of the fluorescent outputs of the even pC, 10 mM CuSO4 was added to the fluorescent samples of the erroneous entries (entries 2, 3, 5 and 8), while nothing was added to the normal entries. After reaction at room temperature for 1 min, the fluorescence spectra of the corrected entries were recorded.For the “Output-Correction” of the visual outputs of the even pC, 10 mM GSH was added to the visual samples of the erroneous entries (entries 2, 3, 5, 8), while nothing was added to the normal entries. After reaction at room temperature for 1 min, the photos were taken using the rear camera of an iPhone 5 and the corresponding absorbance spectra of the corrected entries were collected.
Output-correction for the fluorescence and visual outputs of the odd pC
To correct the erroneous fluorescent outputs of the odd pC, 200 nM strand T30695 was added to the fluorescent samples of the erroneous entries (entries 1, 4, 6, 7). After reaction at room temperature for 30 min, the fluorescence spectra of the corrected entries were recorded.To correct the erroneous visual outputs of the odd pC, 200 nM strand T30695 was mixed with 1 μM hemin and the mixture was incubated at room temperature for 1 h to form the intact T30695 G4zyme. After, 20 μL of the obtained G4zyme was added to the visual samples of the erroneous entries (entries 1, 4, 6 and 7). After reaction at room temperature for 15 min, the photos were taken using the rear camera of an iPhone 5 and the corresponding absorbance spectra of the corrected entries were collected.
Operation of the five-input and seven-input concatenated logic circuits
The concentrations of the DNAs, CuSO4 and GSH, used in the five-input concatenated logic circuit, are all the same as those used for the even pG/pC system. The corresponding fluorescent and visual outputs are not presented due to the numerous input variations. Notably, the concentrations of CB used in the seven-input logic circuit was 2 times that of the even pG/pC system.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 21375123, 21427811 & 21675151).Notes and references
- P. A. de Silva, N. H. Q. Gunaratne and C. P. McCoy, Nature, 1993, 364, 42–44 Search PubMed.
- L. Adleman, Science, 1994, 266, 1021–1024 CAS.
- G. Seelig, D. Soloveichik, D. Y. Zhang and E. Winfree, Science, 2006, 314, 1585–1588 Search PubMed.
- R. Orbach, F. Wang, O. Lioubashevski, R. D. Levine, F. Remacle and I. Willner, Chem. Sci., 2014, 5, 3381 Search PubMed.
- R. Orbach, F. Remacle, R. D. Levine and I. Willner, Chem. Sci., 2014, 5, 1074 Search PubMed.
- M. You, G. Zhu, T. Chen, M. J. Donovan and W. Tan, J. Am. Chem. Soc., 2015, 137, 667–674 Search PubMed.
- L. Feng, Z. Lyu, A. Offenhausser and D. Mayer, Angew. Chem., Int. Ed., 2015, 54, 7693–7697 Search PubMed.
- J. Chen, S. Zhou and J. Wen, Angew. Chem., Int. Ed., 2015, 54, 446–450 CAS.
- D. Fan, K. Wang, J. Zhu, Y. Xia, Y. Han, Y. Liu and E. Wang, Chem. Sci., 2015, 6, 1973–1978 Search PubMed.
- X. J. Jiang and D. K. Ng, Angew. Chem., Int. Ed., 2014, 53, 10481–10484 Search PubMed.
- S. Mailloux, Y. V. Gerasimova, N. Guz, D. M. Kolpashchikov and E. Katz, Angew. Chem., Int. Ed., 2015, 54, 6562–6566 Search PubMed.
- E. Katz, Biomolecular information processing: from logic systems to smart sensors and actuators, John Wiley & Sons, 2013 Search PubMed.
- R. Mehra, S. Jaiswal and H. K. Dixit, Optik, 2013, 124, 4744–4745 Search PubMed.
- J. Zhu, L. Zhang, T. Li, S. Dong and E. Wang, Adv. Mater., 2013, 25, 2440–2444 CrossRef CAS PubMed.
- J. Zhu, L. Zhang, S. Dong and E. Wang, ACS Nano, 2013, 7, 10211–10217 Search PubMed.
- M. Balter, S. Li, J. R. Nilsson, J. Andreasson and U. Pischel, J. Am. Chem. Soc., 2013, 135, 10230–10233 Search PubMed.
- F. Wang, Z. Gong, X. Hu, X. Yang, H. Yang and Q. Gong, Sci. Rep., 2016, 6, 24433 Search PubMed.
- Y. Liu, A. Offenhausser and D. Mayer, Angew. Chem., Int. Ed., 2010, 49, 2595–2598 Search PubMed.
- A. Bhattacharyya, D. Kumar Gayen and T. Chattopadhyay, Opt. Commun., 2014, 313, 99–105 Search PubMed.
- E. Dimitriadou, K. E. Zoiros, T. Chattopadhyay and J. N. Roy, J. Comput. Electron., 2013, 12, 481–489 Search PubMed.
- S. Mandal, D. Mandal and S. K. Garai, Opt. Fiber Technol., 2014, 20, 120–129 Search PubMed.
- A. Kumar and S. K. Raghuwanshi, Opt. Quantum Electron., 2014, 47, 2117–2140 Search PubMed.
- S. Kumar, Chanderkanta and A. Amphawan, Opt. Commun., 2016, 364, 195–224 Search PubMed.
- H. Liu, J. Wang, S. Song, C. Fan and K. V. Gothelf, Nat. Commun., 2015, 6, 10089 CrossRef CAS PubMed.
- X. He, Z. Li, M. Chen and N. Ma, Angew. Chem., Int. Ed., 2014, 53, 14447–14450 CrossRef CAS PubMed.
- S. Bi, M. Chen, X. Jia, Y. Dong and Z. Wang, Angew. Chem., Int. Ed., 2015, 54, 8144–8148 Search PubMed.
- A. Prokup and A. Deiters, Angew. Chem., Int. Ed., 2014, 53, 13192–13195 Search PubMed.
- B. M. Janssen, M. van Rosmalen, L. van Beek and M. Merkx, Angew. Chem., Int. Ed., 2015, 54, 2530–2533 Search PubMed.
- T. Li, E. Wang and S. Dong, J. Am. Chem. Soc., 2009, 131, 15082–15083 Search PubMed.
- D. M. Kolpashchikov, J. Am. Chem. Soc., 2008, 130, 2934–2935 CrossRef CAS PubMed.
- D. Fan, J. Zhu, Y. Liu, E. Wang and S. Dong, Nanoscale, 2016, 8, 3834–3840 Search PubMed.
- Q.-C. Wang, D.-H. Qu, J. Ren, K. Chen and H. Tian, Angew. Chem., Int. Ed., 2004, 43, 2661–2665 Search PubMed.
- D. Margulies, G. Melman and A. Shanzer, J. Am. Chem. Soc., 2006, 128, 4865–4871 Search PubMed.
- H. Li, S. Guo, Q. Liu, L. Qin, S. Dong, Y. Liu and E. Wang, Adv. Sci., 2015, 2, 1500054 Search PubMed.
- R. M. Wartell and A. S. Benight, Phys. Rep., 1985, 126, 67–107 CrossRef CAS.
- P. Ni, Y. Sun, H. Dai, J. Hu, S. Jiang, Y. Wang and Z. Li, Biosens. Bioelectron., 2015, 63, 47–52 Search PubMed.
- T. Li, S. Dong and E. Wang, J. Am. Chem. Soc., 2010, 132, 13156–13157 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Scheme S1, Tables S1–S3 and Fig. S1–S11. See DOI: 10.1039/c6sc04056j |
| This journal is © The Royal Society of Chemistry 2017 |