Open Access Article
Open Access ArticleEngineering the metal-binding loop at a type 1 copper center by circular permutation†
Honghui Chenab,
Binbin Suab,
Tongtong Zhangb,
Aiping Huangb,
Haiping Liu*b,
Yang Yu *b and
Jiangyun Wang*c
*b and
Jiangyun Wang*c
aCollege of Biotechnology, Tianjin University of Science and Technology, Tianjin, 300457, China
bTianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin, 300308, China. E-mail: liu_hp@tib.cas.cn; yu_y@tib.cas.cn
cLaboratory of RNA Biology, Institute of Biophysics, Chinese Academy of Sciences, 15 Datun Road, Chaoyang District, Beijing, 100101, China. E-mail: jwang@ibp.ac.cn
First published on 12th December 2017
Abstract
Cupredoxins are electron transfer proteins with a single type 1 copper center. A metal-binding loop in the protein harbors 3 of the copper ligands. Previous research has altered the sequence and length of the loop, changing the spectroscopic properties and reduction potentials of cupredoxins. It remains elusive whether the loop is flexible enough to tolerate long sequences. In this work we extended the loop to infinity by performing circular permutation on azurin, a typical cupredoxin. The mutant protein, Az-loop was characterized by absorption spectroscopy, electron paramagnetic resonance spectroscopy, cyclic voltammetry and X-ray crystallography. The results further confirm the flexibility of the metal-binding loop.
Introduction
Cupredoxins are type 1 copper center-containing electron transfer proteins involved in many biological processes, such as photosynthesis, respiration and cell signaling.1–12 In addition to small electron transfer proteins, the type 1 copper (T1C) center also appears in larger proteins, such as copper-containing nitrite reductase (CuNiR) and laccase. The T1C center has unique spectroscopic characteristics, including intense electronic absorption (∼5000 M−1 cm−1) at around 600 nm and small hyperfine splitting (<100 × 10−4 cm−1) in the electron paramagnetic resonance (EPR) spectrum, which is attributed to its coordination environment.13,14 Cupredoxins and protein domains in larger proteins with a T1C center share great structural similarity: within a common cupredoxin fold, the copper center is located at one end of the protein, and the metal-binding loop connecting the 6th and 7th β strand harbors 3 copper-binding ligands (Fig. 1 and 2A).2 In cupredoxins and related red copper protein and CuA proteins, the metal-binding loop varies in length and sequence (Fig. 1A–G). For cupredoxins and protein domains with a T1C center, the loop sequences are in the general formula of CXmHXnM, where X denotes any amino acid and m and n are the number of residues in between, C, H are conserved copper ligands and M is methionine or another axial residue. Values of m and n are 2–4 for native proteins with T1C center and the value of m could be as large as 8 in domains containing the T1C center, such as in CuNiR.15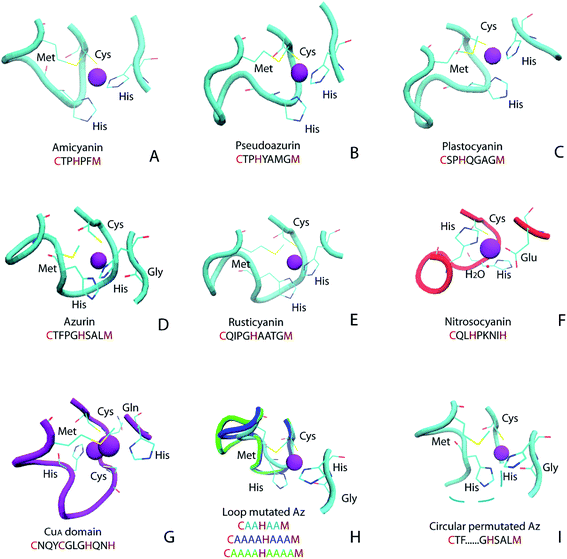 | ||
| Fig. 1 Ligand and metal-binding loop structure in native and engineered cupredoxins: (A) amicyanin (PDB ID 1AAC); (B) pseudoazurin (PDB ID 1PAZ); (C) plastocyanin (PDB ID 1PLC); (D) azurin (PDB ID 4AZU); (E) rusticyanin (PDB ID 1RCY); (F) red copper protein nitrosocyanin (PDB ID 1IBY); (G) CuA domain from Thermus thermophilus heme–copper oxidase (PDB ID 1CUA); (H) loop mutated Az (AZ2A2A, PDB ID 3FS9, AZ4A3A, PDB ID 3FSW, AZ4A4A, PDB ID 3FSZ); (I) circular permutated Az, Az-loop, cyan dotted line connects N- and C-termini (PDB ID 5YT7). Figure (A)–(G) adopted from ref. 11 with permission from American Chemical Society. | ||
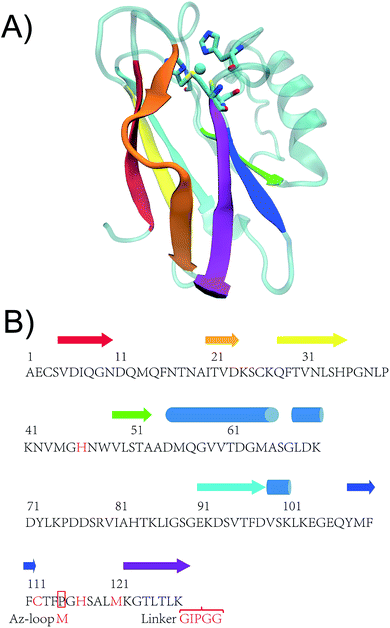 | ||
| Fig. 2 (A) Crystal structure of WT Az (PDB ID: 4AZU). (B) Sequence of WT Az and Az-loop. The amino acid sequence of the mutant starts with a methionine residue, followed by 116th to 128th residue of WT Az, linker sequence (GIPGG) and 1st to 114th residue of WT Az. β strands are shown in arrows while α helices in cylinders. Colors of the secondary structures are matched between crystal structure and the scheme. Adopted from ref. 27 with permission from Wiley. | ||
Due to its importance, the metal-binding loop has been the target for engineering. Two groups have independently replaced the metal-binding loop of cupredoxins with the same region of CuA domain of Cytochrome c Oxidase (CcO), converting the protein with a T1C center into a protein with CuA center, a binuclear copper center (Fig. 2G).16,17 Berry and coworkers have converted the type 1 blue copper center to a red copper center, by replacing the metal-binding loop of azurin (Az) from that of nitrosocyanin.18 The metal-binding loop not only influences the electronic structure of the protein, but also affects the reduction potential. Canters, Dennison and others have replaced the metal-binding loop of one cupredoxin with that of another one.19–25 The hybrid protein maintains T1C characters, and its reduction potential shifts toward that of the loop donor.
Given the diversity of the metal-binding loop of cupredoxins (Fig. 1), researchers have been exploring what sequences can be tolerated in the metal-binding loop, and how they influences reduction potential of the protein, using Az and other cupredoxins as a model. Dennison and coworkers have shortened the loop of Az from C112TFPGH117SALM121 to C112TPH115PFM118 and C112TPH115PM117, while keeping the mutants' T1C characteristics.23 The CTPHPM loop, with m = 2 and n = 1, is the shortest loop presented in a cupredoxin. The same group has connected the copper ligands with different numbers of Ala (Fig. 2H).25 The loop are able to accommodate 2–4 amino acids between copper ligands, while keeping type 1 copper characteristics. As shown in the same work, when only one Ala are present between Cys112, His117 and His117, Met121, the protein does not bind Cu. Changing the 15-residue loop of CuNiR to that of amicyanin (7 residues) resulted in a 200 mV increase of reduction potential of the T1C center, abolishing NiR's enzymatic activity.26 On the other hand, amicyanin can accommodate ligand loop of CuNiR, which has 8 residues between Cys and His and 4 residues between His and Met.15 It is not clear the upper limit of number of residues in the loop. Here, we tested an extreme case, where through circular permutation, Phe114 and Gly116 becomes the first and last residue of the new protein, essentially making the number of residues between Cys112 and His117 approaching infinity (Fig. 1I and 2B). In a previous study, two construct have been built for circular permutation in Az.27 The mutants, cpAz1 and cpAz2, maintain typical T1C spectroscopic properties, suggesting the robustness of Az structure. However, as the metal-binding loop harbors 3 ligands of copper center, the question remains whether circular permutation will perturb the copper center. Here we report the design, spectroscopic and structural characterization of the new protein, termed Az-loop.
Materials and methods
Vector construction
The following primers were used to amplify 346–381 and 1–342 of WT Az gene, with a common linker sequence ggaattccgggtggc.NcoI-116-f:
CAGCCGGCGATGGCCatgggtcactccgcactgatgaaaggtacc
127-linker-r: gccacccggaattcctttcagagtcagggtacctttcatcagtgcg
linker-1-f: gaaaggaattccgggtggcgctgaatgctccgttgatatccag
114-NotI-r:
GTGCTCGAGTGCGGCCGCtcagaaagtgcagaagaacatgtactgttc
The two fragment were used as template to amplify the coding sequence of Az-loop. The linear fragment was digested with NcoI and NotI and cloned into pET22 vector.
Nucleotide sequence of Az-loop:
atgggtcactccgcactgatgaaaggtaccctgactctgaaaggaattccgggtggcgctgaatgctccgttgatatccagggtaatgatcagatgcagttcaacaccaacgccatcaccgtcgacaagagctgcaagcagttcactgttaacctgtctcacccaggtaacctgccgaagaacgttatgggtcacaactgggttctgtccaccgcggctgacatgcaaggcgttgtcactgacggtatggctagcggtctggataaagactacctgaagccggatgactctcgagttatcgcccacaccaagctgatcggatccggtgaaaaagactccgttactttcgacgtttccaagcttaaagaaggtgaacagtacatgttcttctgcactttctga
Protein purification and crystallization
Following the previously reported procedure for Az purification,17,28 the protein was further purified using size-exclusion chromatography (Superdex G-75 10/300 GL, GE Healthcare). The protein was eluted with 0.8 mL min−1 50 mM NH4OAc pH 6.35 monitored at 280 and 625 nm using detectors on a GE Akta purifier. After size exclusion column, fractions corresponding to Az monomer were collected and concentrated to 1 mM and 1.5 mM. Two microliter of 1 mM and 1.5 mM protein was mixed with equal volume of well buffer (0.08 M NaOAc, 0.24 M LiNO3, 0.24 M CaCl2 with 20% polyethylene glycol 8000), and crystalized using the hanging drop method, with 400 μL well buffer in each well of the crystallization tray.Spectroscopic characterization
Absorption spectrum of Az-loop was recorded on a MAPADA UV3100 spectrometer. Amount of copper ion in the protein was analyzed by Inductively Coupled Plasma Mass Spectrometry (ICP-MS) on a PerkinElmer NexION 300X spectrometer, calibrated by standard solutions from Sigma. Extinction coefficient was calculated based on the electronic absorption spectrum and copper concentration of the same sample.X-band EPR spectra were collected on a JEOL JES-FA200 spectrometer at the Analysis Center of Tsinghua University. 1.2 equivalence of Cu(II) ion were added into apo WT Az and Az-loop. Excess Cu(II) ion were removed by a short Sephadex G-25 column. The proteins were further purified by anion exchange (Hitrap Q HP, GE Healthcare) and size exclusion (Superdex G-75 10/300 GL, GE Healthcare) chromatography. The samples containing 3–4 mM Cu(II) and 20% glycerol were run at ∼123 K using liquid nitrogen. EPR spectra were simulated with hyperfine spectrum.29
Data collection and structure determination
Diffraction data were collected at BL17U of the Shanghai Synchrotron Radiation Facility (SSRF).30 HKL2000 was used for data processing.31 The structure was solved by molecular replacement using Phaser in CCP4 (ref. 32 and 33) with WT Az structure (PDB ID: 4AZU) as the searching model. Coot and Refmac5 were used for model building and refinement.34,35Electrochemical characterization
The reduction potential of Az-loop was determined by cyclic voltammetry after verifying the WT Az reduction potential on a CH Instruments 620A potentiostat. Briefly, 10 μL of carbon nanotube (2 mg mL−1) dispersed in ethanol was applied on the surface of glassy carbon electrode and let dry.36 2 μL of Az (1.5 mM) was applied on top of the carbon-nanotube paste. After a short incubation time, the electrode was immersed in either 50 mM NH4OAc, pH 4.0 with 100 mM NaCl, or 50 mM NH4OAc, pH 7.0 with 100 mM NaCl before data collection. The reduction potentials were measured against Ag/AgCl and converted to those against NHE by adding 210 mV to the obtained value calculated based on the value of ferricyanide control.Results and discussion
Design of circularly permutated Az variant
As shown in Fig. 2B, here we applied circular permutation strategy, to connect N- and C-termini with a linker GIPGG and create new termini at Gly116 (N-terminus) and Phe114 (C terminus), similar to what have been reported for cpAz1 and cpAz2.27The Az-loop mutant was constructed by overlap-extension PCR. The protein was purified in a procedure similar to that of the WT protein28 to obtain metal-free apo protein with a yield of ∼40 mg L−1. The identities of the Az-loop protein was verified by electrospray mass spectrometry after purification (Fig. S1†).
Spectroscopic and electrochemical characterization of Az-loop
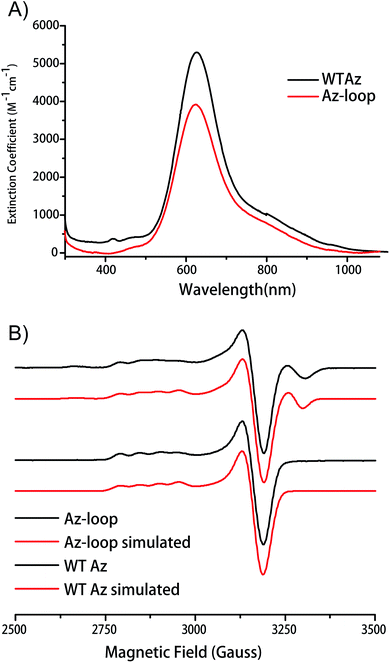
Addition of Cu2+ into the apo protein gave the protein blue color. Absorption spectrum of the protein showed a λmax of 624 nm, only slightly blue-shifted from that of WT Az (627 nm). The absorption at 624 nm is assigned as pπ(Cys)–Cu(II) charge transfer band (Fig. 3A). As the hallmark of cupredoxins, the intense absorption indicates that the mutant protein has an un-perturbed T1C center.EPR spectroscopy provides a more sensitive way to characterize copper coordination environment. As shown in Fig. 3B, EPR spectrum of Az-loop showed more than one species. Simulation of the spectrum showed the majority of the signal (84%) is from a type 1 center with the same parameters as WT Az, while the rest of the signal is from a type 2 center with large hyperfine splitting values (200 × 10−4 cm−1, Fig. S2 and Table S1†). Addition of equal amount of apo WT Az into Cu(II)-loaded Az-loop did not cause significant change in electronic absorption spectrum (Fig. S3†), suggesting that species 2 is not adventitious Cu(II) in the sample. Since species 2 is likely a type 2 center, its absorption at 600 nm region is weak, which accounts for the lower than WT extinction coefficient. Due to its low abundance, no additional feature is observe in electronic absorption spectrum.
The redox potential of Az-loop was measured by cyclic voltammetry using a carbon nanotube modified glassy carbon electrode. Redox potential Az-loop was determined to be 353 ± 2 mV and 239 ± 12 mV at pH 4 and pH 7, respectively, which is 22 and 13 mV higher than that of the WT (Fig. S4†). The potential can be assigned to the species with a type 1 center due to its high abundance and similarity to the WT protein.
In the loop transfer mutants, where loop of a cupredoxin is transferred to another cupredoxin, the spectroscopic properties and reduction potentials of the protein are dictated by the loop donor.24 When the residues between copper ligands in the loop are changed to the same Ala/Gly/Val (Fig. 2H), the copper center in Az still forms.37 As Dennison and coworkers suggested, the reduction potential of the mutants is not affected by the length of the loop, but by the polarity of the copper binding site. In Az-loop, although the length of the loop is infinity due to circular permutation, hydrogen bonding and hydrophobicity of the site does not change since the native residues are unchanged. As expected, the reduction potential of Az-loop is similar to that of WT Az.
The pKa values of C-terminal His in reduced cupredoxins range from 4.3 to 6.7 for most cupredoxins, including amicyanin, plastocyanin, pseudoazurin, stellacyanin, and cucumber basic protein,4 while the value for Az is estimated to be below 2.38 The length of the metal-binding loop has long been proposed to affect the pKa value.39 Dennison and coworkers have found that in Az mutants with engineered loop, the pKa values of C-terminal His increased to 4.4 or higher.23,25,40 A – 60 mV pH−1 change of reduction potential in response to pH between pH 4–7 was observed in the mutants with higher pKa, while in WT Az the reduction potential is not affected by pH below pH 5.5.25 In Az-loop, the difference of reduction potential between pH 4 and pH 7 is 112 mV, similar to that of the WT Az, suggesting the pKa value of His3 (corresponding to the C-terminal His117 in WT Az) is smaller than 4.
Structural characterization
To investigate the effect of circular permutation on the copper coordination, as well as on the overall structure of the protein, we crystallized Az-loop and the crystal was diffracted to 1.66 Å resolution. The structure of Az-loop was refined to an Rfree of 0.217 (Fig. 4 and Table S2†). Coordinates and structure factors were deposited in the Protein Data Bank (accession code: 5YT7).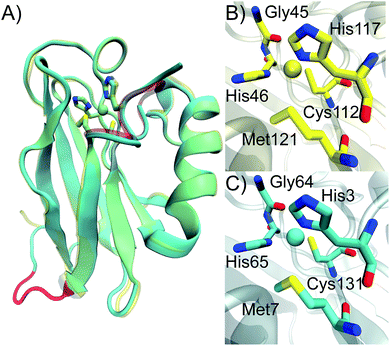 | ||
| Fig. 4 Crystal structure of Az-loop. Overlay of crystal structure of WT Az (yellow, PDB: 4AZU) and Az-loop (cyan, PDB: 5YT7). Active site residues were in licorice style and nearly overlaid. The metal-binding loop in WT Az and the loop connecting the original N- and C-termini in Az-loop were highlighted in red. Comparison of copper center of WT Az (B) and Az-loop (C). | ||
As shown in Fig. 4A, structure of Az-loop overlaid well with that of WT Az with a RMSD of backbone atoms of 0.74 Å. The GIPGG linker forms a loop, connecting the original N- and C-termini. In the metal-binding loop, although the constraint eases because of the creation of the new termini, the loop still follows its original direction.
The distances between copper and its ligands are listed in Table S3.† Cys-Cu distance is 0.06 Å and Met–Cu distance is 0.11 Å longer than these in the WT structure (PDB ID: 4AZU). These distances are within the normal range observed in structures in WT Az.2 The un-perturbed primary ligands leads to the intense 624 nm absorbance of the protein. The structure likely represents species 1 with a type 1 copper center observed in EPR spectrum. Beyond copper ligands, residues interact with copper ligands are also important for reduction potential of Az.6,41 We looked into the distances in the second coordination sphere. Only distance between Nδ of N66 and Sγ of Cys131 changes more than 0.05 Å, compared to that of the WT protein. The change alone is not sufficient to alter the reduction potential significantly. Previously observed spectroscopic and electrochemical properties of Az-loop can be explained by its conserved primary and second coordination sphere structure.
In WT Az, the copper ion is shielded by its ligands and not accessible by solvent or external ligands. Mutating His or Met to smaller residues increases solvent accessibility of the site.1,30 30 equivalence of histidine or N3−, strong ligands known to cause absorption spectrum change, was added into Az-loop (Fig. S5†). The absorption spectrum of Az-loop remain unchanged. The surf rendering of WT Az and Az-loop showed similar structures (Fig. S6†). There is no indication of change in solvent accessibility in Az-loop.
Besides EPR spectroscopy, species 2-the one with type 2 copper center remains silent in all other characterizations. In addition to the fact that it is a minor species, weak electronic absorption and low electron transfer activity associate with type 2 sites may also contribute to its reclusive nature. Interestingly, EPR fitting of species 2 showed a larger Az and smaller gz than typical type 2 sites. As a result, it is hard to determine the exact structure of the species.
Conclusion
In the work, we reported construction of an Az mutant of infinite metal-binding loop length by circular permutation. The mutant, Az-loop, exhibited absorption spectrum typical of T1C proteins. EPR spectrum of Az-loop showed a mixture of two species, and the major species contains a type 1 copper center. The reduction potential of Az-loop is close to that of WT Az, indicating circular permutation has minimal perturbation to the structure. The minimal structure perturbation is further confirmed by structure analysis of the mutant Az-loop, which indicates the overall structure and primary and secondary coordination sphere of Az-loop are similar to that of the WT protein.The finding that circular permutation does not alter the protein is consistent with Dennison and coworker's finding, which suggests that the metal-binding loop is flexible enough tolerate different sequences and length while maintaining protein's T1C characters.24,37 The flexibility is important for metallation of cupredoxins. Lu and coworkers and Blackburn and coworkers have reported intermediates during CuA center formation in an engineered CuAAz and CuA domain of T. thermophilus Cytochrome c Oxidase, suggesting ligand rearrangement and loop movement in this process.42,43 Such arrangement is impossible in a rigid structure. Vila and coworker have determined NMR structure of Az and CuA domain of T. thermophilus CcO, a closely related protein with CuA center, further confirming the flexibility of the loop.44
As a method for protein engineering, circular permutation has been applied in constructing different sensors.45,46 our work opens the possibility to attach a redox active protein or small molecule at the termini of Az while keeping the redox centers close for electron transfer.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This material is based on work supported by National Key Research and Development Program under Award 2016YFA0502400 (to Y. Y.), Youth Innovation Promotion Association of CAS (2015137, to H. L.) and the National Science Foundation of China under award 31500641 and 21503268 (to Y. Y.). Y. Y. is thankful for the China Association for Science and Technology fellowship. The authors would like to thank Lin Yang from University of Science and Technology of China for assistance in crystal diffraction and data collection, Dr Lin Xia from Shenzhen Institute of Advanced Technology, CAS for discussion in protein electrochemistry and Xixi Liang from Tsinghua University for assistance in EPR experiment. X-ray diffraction data were collected at beamline 17U of the Shanghai Synchrotron Radiation Facility (SSRF) in China.References
- G. W. Canters and G. Gilardi, FEBS Lett., 1993, 325, 39–48 CrossRef CAS PubMed.
- H. B. Gray, B. G. Malmström and R. J. P. Williams, J. Biol. Inorg Chem., 2000, 5, 551 CrossRef CAS PubMed.
- L. Banci, R. Pierattelli and A. J. Vila, Adv. Protein Chem., 2002, 60, 397–449 CrossRef CAS PubMed.
- C. Dennison, Coord. Chem. Rev., 2005, 249, 3025–3054 CrossRef CAS.
- Y. Lu, N. Yeung, N. Sieracki and N. M. Marshall, Nature, 2009, 460, 855 CrossRef CAS PubMed.
- N. M. Marshall, D. K. Garner, T. D. Wilson, Y. G. Gao, H. Robinson, M. J. Nilges and Y. Lu, Nature, 2009, 462, 113–116 CrossRef CAS PubMed.
- M. G. Savelieff and Y. Lu, J. Biol. Inorg Chem., 2010, 15, 461–483 CrossRef CAS PubMed.
- K. M. Clark, Y. Yu, N. M. Marshall, N. A. Sieracki, M. J. Nilges, N. J. Blackburn, W. A. van der Donk and Y. Lu, J. Am. Chem. Soc., 2010, 132, 10093 CrossRef CAS PubMed.
- M. Choi and V. L. Davidson, Metallomics, 2011, 3, 140–151 RSC.
- J. J. Warren, K. M. Lancaster, J. H. Richards and H. B. Gray, J. Inorg. Biochem., 2012, 115, 119–126 CrossRef CAS PubMed.
- J. Liu, S. Chakraborty, P. Hosseinzadeh, Y. Yu, S. Tian, I. Petrik, A. Bhagi and Y. Lu, Chem. Rev., 2014, 114, 4366–4469 CrossRef CAS PubMed.
- P. Hosseinzadeh, N. M. Marshall, K. N. Chacón, Y. Yu, M. J. Nilges, S. Y. New, S. A. Tashkov, N. J. Blackburn and Y. Lu, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 262–267 CrossRef CAS PubMed.
- E. I. Solomon, R. K. Szilagyi, S. DeBeer George and L. Basumallick, Chem. Rev., 2004, 104, 419–458 CrossRef CAS PubMed.
- E. I. Solomon, Inorg. Chem., 2006, 45, 8012 CrossRef CAS PubMed.
- C. Buning, G. W. Canters, P. Comba, C. Dennison, L. Jeuken, M. Melter and J. Sanders-Loehr, J. Am. Chem. Soc., 2000, 122, 204–211 CrossRef CAS.
- C. Dennison, E. Vijgenboom, S. de Vries, J. van der Oost and G. W. Canters, FEBS Lett., 1995, 365, 92 CrossRef CAS PubMed.
- M. Hay, J. H. Richards and Y. Lu, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 461–464 CrossRef CAS.
- S. M. Berry, E. L. Bladholm, E. J. Mostad and A. R. Schenewerk, J. Biol. Inorg Chem., 2011, 16, 473 CrossRef CAS PubMed.
- C. Dennison, E. Vijgenboom, W. R. Hagen and G. W. Canters, J. Am. Chem. Soc., 1996, 118, 7406–7407 CrossRef CAS.
- R. Remenyi, L. J. C. Jeuken, P. Comba and G. W. Canters, J. Biol. Inorg Chem., 2001, 6, 23–26 CrossRef CAS PubMed.
- G. Battistuzzi, M. Borsari, G. W. Canters, G. di Rocco, E. de Waal, Y. Arendsen, A. Leonardi, A. Ranieri and M. Sola, Biochemistry, 2005, 44, 9944–9949 CrossRef CAS PubMed.
- C. Dennison, Dalton Trans., 2005, 3436–3442 RSC.
- C. Li, S. Yanagisawa, B. M. Martins, A. Messerschmidt, M. J. Banfield and C. Dennison, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 7258–7263 CrossRef CAS PubMed.
- C. Li, M. J. Banfield and C. Dennison, J. Am. Chem. Soc., 2007, 129, 709–718 CrossRef CAS PubMed.
- C. Li, K. Sato, S. Monari, I. Salard, M. Sola, M. J. Banfield and C. Dennison, Inorg. Chem., 2011, 50, 482–488 CrossRef CAS PubMed.
- K. Sato, S. J. Firbank, C. Li, M. J. Banfield and C. Dennison, Chem.–Eur. J., 2008, 14, 5820–5828 CrossRef CAS PubMed.
- Y. Yu, I. D. Petrik, K. N. Chacón, P. Hosseinzadeh, H. Chen, N. J. Blackburn and Y. Lu, Protein Sci., 2017, 26, 218–226 CrossRef CAS PubMed.
- T. K. Chang, S. A. Iverson, C. G. Rodrigues, C. N. Kiser, A. Y. Lew, J. P. Germanas and J. H. Richards, Proc. Natl. Acad. Sci. U. S. A., 1991, 88, 1325–1329 CrossRef CAS.
- W. R. Hagen, Biomolecular EPR Spectroscopy, CRC Press, Boca Raton, Fl, USA, 2009 Search PubMed.
- G. van Pouderoyen, C. R. Andrew, T. M. Loehr, J. Sanders-Loehr, S. Mazumdar, H. A. O. Hill and G. W. Canters, Biochemistry, 1996, 35, 1397–1407 CrossRef CAS PubMed.
- Z. Otwinowski and W. Minor, in Methods Enzymol., Academic Press, 1997, vol. 276, pp. 307–326 Search PubMed.
- A. J. McCoy, R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni and R. J. Read, J. Appl. Crystallogr., 2007, 40, 658–674 CrossRef CAS PubMed.
- M. D. Winn, C. C. Ballard, K. D. Cowtan, E. J. Dodson, P. Emsley, P. R. Evans, R. M. Keegan, E. B. Krissinel, A. G. W. Leslie, A. McCoy, S. J. McNicholas, G. N. Murshudov, N. S. Pannu, E. A. Potterton, H. R. Powell, R. J. Read, A. Vagin and K. S. Wilson, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2011, 67, 235–242 CrossRef CAS PubMed.
- P. Emsley, B. Lohkamp, W. G. Scott and K. Cowtan, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 486–501 CrossRef CAS PubMed.
- A. A. Vagin, R. A. Steiner, A. A. Lebedev, L. Potterton, S. McNicholas, F. Long and G. N. Murshudov, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2004, 60, 2184–2195 Search PubMed.
- J. J. Davis, R. J. Coles, H. Allen and O. Hill, J. Electroanal. Chem., 1997, 440, 279–282 CAS.
- K. Sato, C. Li, I. Salard, A. J. Thompson, M. J. Banfield and C. Dennison, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 5616–5621 CrossRef CAS PubMed.
- L. J. C. Jeuken, P. van Vliet, M. P. Verbeet, R. Camba, J. P. McEvoy, F. A. Armstrong and G. W. Canters, J. Am. Chem. Soc., 2000, 122, 12186–12194 CrossRef CAS.
- C. Dennison, T. Kohzuma, W. McFarlane, S. Suzuki and A. G. Sykes, J. Chem. Soc., Chem. Commun., 1994, 581–582, 10.1039/C39940000581.
- S. Yanagisawa and C. Dennison, J. Am. Chem. Soc., 2004, 126, 15711–15719 CrossRef CAS PubMed.
- S. Yanagisawa, M. J. Banfield and C. Dennison, Biochemistry, 2006, 45, 8812–8822 CrossRef CAS PubMed.
- T. D. Wilson, M. G. Savelieff, M. J. Nilges, N. M. Marshall and Y. Lu, J. Am. Chem. Soc., 2011, 133, 20778–20792 CrossRef CAS PubMed.
- K. N. Chacón and N. J. Blackburn, J. Am. Chem. Soc., 2012, 134, 16401–16412 CrossRef PubMed.
- M.-E. Zaballa, L. A. Abriata, A. Donaire and A. J. Vila, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 9254–9259 CrossRef CAS PubMed.
- K. Wieligmann, B. Norledge, R. Jaenicke and E. M. Mayr, J. Mol. Biol., 1998, 280, 721–729 CrossRef CAS PubMed.
- G. S. Baird, D. a. Zacharias and R. Y. Tsien, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 11241–11246 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra11512a |
| This journal is © The Royal Society of Chemistry 2017 |