Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Construction of core-shell hybrid nanoparticles templated by virus-like particles†
A. Liua,
L. Yang a,
M. Verwegena,
D. Reardonb and
J. J. L. M. Cornelissen
a,
M. Verwegena,
D. Reardonb and
J. J. L. M. Cornelissen *a
*a
aLaboratory for Biomolecular Nanotechnology (BNT), MESA+ Institute of Nanotechnology, University of Twente, 7500 AE Enschede, The Netherlands. E-mail: J.J.L.M.Cornelissen@utwente.nl
bDSM Materials Science Center, B. V., P. O. Box 18, 6160 MD Geleen, The Netherlands
First published on 14th December 2017
Abstract
Plant viruses have been widely used as templates for the synthesis of organic–inorganic hybrids. However, the fine-tuning of hybrid nanoparticle structures, especially the control of inorganic particle size as well as where the silication occurs (i.e. outside and/or inside of the capsid), by simply tuning the pH remains a challenge. By taking advantage of the templating effect of Cowpea Chlorotic Mottle Virus (CCMV) protein cages, we show that the silication at the exterior or interior surface of protein capsids, as well as the resulting structures of silica/virus hybrid nanoparticles can be fine-tuned by pH. At pH 4.0, only small silica particles (diameter of 2.5 nm) were formed inside the protein cages; at pH 6.0, silication mainly takes place inside of the protein cages, leading to monodisperse silica nanoparticles with diameters of 14 nm; and at pH 7.5, silica deposition takes place both at the interior and exterior surfaces of protein cages in aqueous conditions. Under these reaction conditions, multiple component hybrid virus/nanoparticulate systems, such as CCMVAu/silica and Au/silica nanoparticles were prepared step-by-step. Upon removal of the CCMV template by thermal degradation a single gold nanoparticle can be encapsulated in a hollow silica shell emulating the structure of a baby's rattle with an unattached solid particle within a hollow particle. The Au/silica core-hollow shell nanoparticles can then be further used as a stable catalyst. It is anticipated that these synthetic methods provide a versatile methodology to prepare core–shell nanomaterials with well-designed structure and functionality.
Introduction
Organic–inorganic hybrid nanoparticles (NPs) with combined functionalities have attracted great interest in recent years.1–8 In particular, environmentally friendly and low-cost plant virus based hybrid materials have been widely studied9–27 due to their promising applications in biomedicine, electronic devices and vaccine storage.28–41 The advantages of using viruses as templates are: (i) these are monodisperse NPs with a high level of symmetry, which is suitable for the construction of uniform nanoparticles; and (ii) many of the virus NPs are stable over wide pH and temperature ranges, which makes them applicable for the synthesis of hybrid NPs. In particular hybrid protein cage-silica NPs are of interest as these combine the robust properties of silica, e.g. structural integrity, with the high level of definition of virus-based protein cages.Spherical and tube-like viruses have been used as templates for the synthesis of silica-virus hybrid NPs.42–50 Silication has been achieved on the genetically modified protein shell of the cowpea mosaic virus (CPMV)43 however, the size of the particles was hard to control due to particle cross-linking of silanol groups. By mimicking natural silica growth, positively charged APTES ((3-aminopropyl)triethoxysilane) was pre-adsorbed onto the negatively charged viral NPs surface and hydrolyzed/condensed to form silica nuclei, then TEOS (tetraethoxysilane) was added to promote the growth of the silica shell.49,51 Silication on tube-like virus particles, such as the tobacco mosaic virus (TMV) was also reported.44,47 In this case, the modification of the outer-surface of the virus particles with a polymer is necessary, in order to obtain silica shells with sufficient thickness to tolerate the deposition of metal nanoparticles.44,45
Although the construction of silica-virus hybrid NPs was successful, the fine-tuning of hybrid NP structures, especially the control of silication outside or inside the virus capsid, remains challenging. When the exterior surface of the capsid is negatively charged it can interact with positively charged silica precursors, for example APTES, and thus facilitate the deposition of silica on the exterior surface of capsid. Meanwhile, the porous structure of the protein shell of virus NPs allows foreign molecules, such as the silica precursors/or silica nuclei, to get access inside the virus cage. In view of these considerations, virus silication can not only take place at the exterior surface but also inside the capsid, leading to structurally diverse hybrid NPs.52 As a representative model system, the Cowpea Chlorotic Mottle Virus (CCMV) consists of 180 protein units, which assemble into a porous capsid shell with single-stranded RNA encapsulated.40–43,53,54 The interaction between negatively charged CCMV capsid (pI = 3.8) and APTES can be controlled by pH, due to the pH dependence of the negative charge density of the exterior capsid surface.52,55,56 Meanwhile, the size of CCMV capsid pores (1–2 nm) is close to silica nuclei (1–2 nm)57,58 and is also pH dependent, which allows the control of accessibility of silica nuclei through the capsid by pH. CCMV NPs are stable at a pH range of 3.0 to 6.0, where the pore size of the capsid is about 1 nm in diameter. The capsid starts to swell at pH ≥ 6.0, at which the pore size increases to 2 nm, resulting in higher accessibility for foreign molecules.59,60 Synthesis of monodisperse inorganic nanoparticles at this pH has been widely reported.61–63 The virus NPs are highly swollen when the pH > 7.5 while the virus disassembles at this pH with high salt concentrations.64 For the accessibility of the CCMV protein cage three stages were recognized: pH < 5.5, low accessibility; pH 5.5–7, medium accessibility; pH ≥ 7.5, high accessibility.65 Therefore, it should be possible to control the deposition of silica in the cavity to control the growth of particle size as well as control the deposition on both sides of the capsid surface by tuning the pH of the solution, resulting in hybrid protein-silica nanoparticles with controlled structure.
To confirm our hypothesis, CCMV NPs were used as a template to construct hybrid nanoparticles under three representative pH conditions, i.e.: pH 4.0, 6.0 and 7.5. The silication at different pH conditions as well as the structures of the formed hybrid nanoparticles were studied. The control of silication was further employed to construct multi-component core–shell nanoparticles, with as a proof of concept, hybrid nanoparticles with core–shell structures including a Au NPs encapsulating hybrid NP (CCMVAu), which was subsequently silicated to form multi-shell hybrid NPs (CCMVAu/silica). Finally after removal of the organic CCMV template by a thermal calcination process, a solid Au nanoparticle-hollow silica NPs (Au/silica) rattle structure was obtained. The catalytic activity of Au NPs combined with the stability of silica shell, enabled the formation of highly stable catalytic materials.66
Results and discussion
To prepare virus/silica hybrid NPs, silication was carried out by treating CCMV with APTES, followed by TEOS, at different pHs. The reaction solutions were incubated while rotating for 3 days at room temperature, washed with buffer solution for 3 days, followed by washing with MilliQ 18 Mohm water another 3 days. Silication and purification of gold containing protein cages was carried out in via similar method. The silication of virus particles was first confirmed by energy dispersive X-ray spectroscopy (EDX), which shows a silicon signal for all samples (Fig. 1). | ||
| Fig. 1 EDX analysis of CCMV templated silication at (a) pH 4.0 (CCMV-silica4), (b) pH 6.0 (CCMV-silica6), and (c) pH 7.5 (CCMV-silica7.5). | ||
Transmission Electron Microscopy (TEM) characterization further confirmed the successful silication of CCMV. Analysis of stained TEM images with negatively stained CCMV-silica4 displayed NP with D = 22 nm in with a dark core of D = 15 nm (Fig. 2b), while non-stained CCMV-silica4 NPs were hardly visible in TEM (data not shown), indicating a low density silica on the virus particles which was also confirmed by EDX elemental mapping (Fig. S1†). Stained CCMV-silica6 showed D = 24 nm in NPs (Fig. 2c), while non-stained CCMV-silica6 NPs showed a much smaller particle size with D = 14 nm (Fig. 2d). Protein shells are hardly visible under TEM without proper staining, but silica, which has a higher contrast than the organic protein shell, can be observed. Therefore, these 14 nm sized NPs from non-stained CCMV-silica6 are identified as silica particles, indicating that the silication mainly occurred inside of the virus cage at pH 6.0. Further evidence to support this conclusion is that nanoparticles with a size of D = 14 nm were retained after calcination (Fig. S2†). For CCMV-silica7.5, the stained sample showed particles with D = 27 nm (Fig. 2e), while the non-stained sample showed particles with average D = 30 nm (Fig. 2f), suggesting silication might have taken place not only inside the protein cage, but also at the exterior surface of capsid or at the interface of the coat proteins. Although some smaller particles with D = 18 nm were also observed which may mainly have silication inside protein cages. Particle sizes analysed by dynamic light scattering (DLS) are consistent with TEM analysis. Both CCMV-silica4 and CCMV-silica6 show average particle sizes around D = 25 nm with narrow distributions. For CCMV-silica7.5 particles, a relative broader peak with an average diameter of D = 30 nm was found in the DLS analysis (Fig. S3†).
The structures of silicated NPs were further studied by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis, which shows CCMV protein bands at 20 kDa for CCMV-silica4 and CCMV-silica6 (Fig. S4†).23 The presence of these CCMV protein bands indicates that proteins were denatured and separated without the protection of silica shell. For CCMV-silica7.5, however, no significant protein band at 20 kDa was observed, instead aggregates remained on top of the stacking gel (Fig. S4,† line 4), suggesting a robust silica shell that prevents proteins from being released. This increased silica precipitation on the exterior surface could be ascribed to a higher electrostatic interaction between the capsid and APTES at a higher pH.
Removal of the protein part by calcination resulted in silica nanoparticles with height of 2.5 nm, 14 nm and 20 nm from CCMV-silica4, CCMV-silica6 and CCMV-silica7.5 respectively as observed by Atomic Force Microscopy (AFM), as shown in Fig. 3. For CCMV-silica4 and CCMV-silica6, the obtained silica nanoparticles were almost homogeneous in size from AFM images, suggesting a well-controlled silication in the confined viral cavity.
The formation of diverse structures of silicated nanoparticles formed at different pH values in the presence of CCMV is attributed to the pH dependence of pore size and charge state of the exterior virus surface. Three stage were recognized for the accessibility of the CCMV protein cage: pH < 5.5, low accessibility; pH 5.5–7, medium; pH ≥ 7.5, high accessibility.65 Therefore, the access of silica nuclei into the protein cages can presumably be controlled by varying the pH of the system. Moreover, the electrostatic interaction between the negatively charged exterior of the protein shell and the positively charged APTES induces the silication on the exterior surface of the capsids. The exterior surface of capsids becomes more negatively charged when the pH is increased, since the pKa of CCMV is 3.8.67 Combination of these two factors leads to a competition of silica layer growth between the interior and exterior of the protein cages, resulting in silicated virus particles with a fine-tuned core–shell structures.
To construct multifunctional hybrid NPs, we employed the above procedure at neutral or slightly alkaline pH to gold nanoparticles in CCMV capsids (CCMVAu) yielding CCMVAu/silica, and Au/silica materials after removal of protein component.
Commercial Au NPs with diameter of 7 nm stabilized by tannic acid were encapsulated in the protein capsid as previously reported.68 The assembly resulted in hybrid CCMVAu NPs with a core–shell structure with average size of 18 nm, in combination with capsids containing only tannic acid (Fig. 4a). Silication of CCMVAu resulted in CCMVAu/silica multiple core–shell materials (Fig. 4b). According to TEM images, the average diameter of CCMVAu/silica was found around D = 28 nm with a thickness of the silica shell of 5 nm (Fig. S5†). In this step, additional tannic acid was added to the CCMVAu solution, the mixed solution was incubated on a rotor for 0.5 h, silica precursor was then added to produce CCMVAu/silica hybrid NPs. Tannic acid contains a large number of phenol groups which can easily combine with carboxyl groups on the exterior surface of capsid through hydrogen bonding.69,70 Moreover, tannic acid has been widely used as a template for silica growth.71,72 Therefore, the extra tannic acid in this case can absorb on the exterior surface of capsids and act as a guiding reagent, resulted in uniform silica coatings on protein capsids, as shown in Fig. 4b. Elemental mapping showed a thin layer of silica formed around the capsid layer, a minimal amount of silica was found at the interface of the capsid, and no silica was found in the core of the particles (Fig. S6†). Silication in the absence of tannic acid only resulted in irregular NPs (Fig. S7†), indicating that tannic acid plays an essential role in directing silica growth on the exterior surface of CCMV capsids in aqueous solution.
 | ||
| Fig. 4 TEM images of (a) gold NPs encapsulated inside the CCMV protein cage (CCMVAu) with staining; (b) silicified CCMVAu shows Au core/capsid/silica double shell hybrid structures (CCMVAu/silica). | ||
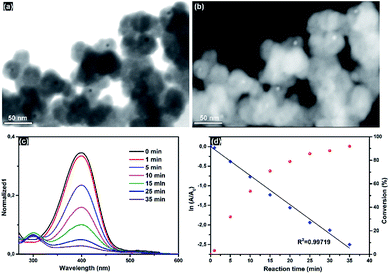
Calcination of CCMVAu/silica resulted in Au/silica core–shell NPs, as the organic components were removed by heating at 550 °C for 3 h. The obtained Au/silica materials were characterized by STEM (Fig. 5a and b), showing the core–shell structure. The core particles with a higher contrast correspond to AuNPs, while the shell consists of silica. No individual AuNPs nor aggregates were observed by STEM analysis, suggesting that the protein cage induced silica shell acts as a protecting layer to prevent aggregation during the calcination at high temperature.
The removal of the vaporized organic materials from the silica network, suggests that these are sufficiently porous to allow small molecules to reach the gold core. This prompted us to study the catalytic activity of the encapsulated Au NPs to explore their potential application as nanoreactors.73–75 As a typical example, the reduction of 4-nitrophenol (4NP) was catalyzed by Au/silica and the progress of the reactions was monitored by UV-vis spectroscopy (Fig. 5c). The characteristic absorption band of deprotonated 4NP at λ = 400 nm decreased, while a new absorption band at λ = 300 nm – indicating the formation of aminophenol – appears during the reaction. The correlation of ln(A/A0) versus reaction time (Fig. 5d) points to pseudo-first-order kinetics with a rate constant of 1.23 × 10−3 s−1 that is comparable to previous studies.76,77 Under the protection of porous silica shell, Au NPs are well stabilized without aggregation, while Au NPs that are stabilized only by tannic acid tend to aggregate after reaction.26 Furthermore, it's facile to coat silica shell protected Au NPs on substrates to prepare easy to handle catalytic thin films, which is a challenge for naked gold nanoparticles.
Conclusions
Core-shell structures of silica and virus based materials can be constructed by controlling the pH during the silication process. We have confirmed that at pH = 6 silica preferable forms on the inside of the CCMV virus, while at pH = 7.5, a silica shell surrounding the protein cage is observed. By further reducing the pH the penetration of silica precursors is virtually blocked, so only small particles (D < 3 nm) are observed. This empirical control over the silica formation with the virus as a template allowed us the construct hybrid silica/gold materials, based on gold nanoparticles encased in the CCMV protein cage devoid of the genetic material. The resultant Au/protein shell/silica materials could be transformed to gold inside silica core–shell networks that are catalytically active in the reduction of 4-nitrophenol as an example. Considering the diversity of virus (-like) particles nowadays studied in materials science and their potential cargos,78–80 this methodology is not only limited to the specific Au/silica system, but also applicable for the preparation of a series of core–shell nanomaterials. It is anticipated that in that way it paves the way for preparing core–shell hybrid NPs with well-designed structures, towards advanced applications in catalysis, drug delivery, bio-sensor and bio-imaging.Experimental
Chemicals
All chemicals were of analytical quality, and used as purchased without further purification. Ethanol, (3-aminopropyl)triethoxysilane (APTES), and tetraorthosilicate (TEOS), were all purchased from Sigma-Aldrich. Buffers were prepared with ultrapure water (Milli Q 18 MOhm water). CCMV NPs were isolated from infected cowpea plant and stored in virus buffer solution under 4 °C.13Synthetic procedures
Preparation of silica/virus hybrid NPs: native CCMV (4 mg mL−1) in 10 mM sodium phosphate (PB) buffer of pH 6 was treated with TEOS and APTES (90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 mol%) ethanol solution with final silica concentration of 0.08 wt%, and left on the rotator for 3 days at room temperature. Considering the limited stability of virus nanoparticles in organic solvent, the final ethanol concentration in water was controlled at 9%. The reaction solution was then dialyzed against buffer for 3 days (refresh buffer 3 times per day), additionally dialyzed against Milli-Q (MQ 18 Mohm) for another 3 days to completely remove the non-reacted silane and salt. The silicified CCMV nanoparticles were separated from non-templated silica by centrifugation at 10k rpm for 10 min, and the supernatant was collected and named as CCMV-silica6. Samples under pH 4 of 10 mM sodium acetate buffer and pH 7.5, 10 mM Tris-HCl buffer were prepared in the same way, and were named as CCMV-silica4 and CCMV-silica7.5, respectively.
10 mol%) ethanol solution with final silica concentration of 0.08 wt%, and left on the rotator for 3 days at room temperature. Considering the limited stability of virus nanoparticles in organic solvent, the final ethanol concentration in water was controlled at 9%. The reaction solution was then dialyzed against buffer for 3 days (refresh buffer 3 times per day), additionally dialyzed against Milli-Q (MQ 18 Mohm) for another 3 days to completely remove the non-reacted silane and salt. The silicified CCMV nanoparticles were separated from non-templated silica by centrifugation at 10k rpm for 10 min, and the supernatant was collected and named as CCMV-silica6. Samples under pH 4 of 10 mM sodium acetate buffer and pH 7.5, 10 mM Tris-HCl buffer were prepared in the same way, and were named as CCMV-silica4 and CCMV-silica7.5, respectively.
Preparation of gold encapsulated CCMV NPs and silication: encapsulation of gold NPs into CCMV protein cages was carried out according to references,26 hybrid NPs were named as CCMVAu. Then gold core–protein–silica–double shell hybrid nanoparticles (CCMVAu/silica) were prepared and templated by CCMVAu NPs. In detail, CCMVAu NPs were washed against MQ water with centrifuge filter for three times to completely remove salt, then a certain amount of tannic acid was added into the solution, the mixture was left on the rotor for 0.5 h (silication without tannic acid was also conducted). Further silication method is same as the preparation of silica/virus hybrid NPs. CCMVAu/silica without tannic acid as directing agent was prepared in a similar method.
Catalytic activity of Au/silica: drop 100 μL CCMVAu/silica solution (contain 0.0127 mg mL−1 of Au NPs) on silicon wafer and dried under vacuum, calcination was then performed to form Au/silica NPs. This substrate was then used for catalytic reaction by mixing with 510 μL of Milli-Q water, 90 μL of 2 mM of 4NP and 450 μL of sodium borohydride. UV-vis monitor was started immediately after mixing.
Characterization
Poly Acrylamide Gel Electrophoresis (PAGE) was performed in the presence of sodium dodecyl sulfate (SDS). Samples were heated at 99 °C for 10 min in the presence of 2-mercaptoethanol and 1% SDS.Transmission electron microscopy (TEM) analysis was performed using Philips CM300 microscope. Samples for imaging were deposited onto a 200 mesh copper grid and left for 2 min, after which the excess buffer or water was blotted away with filter paper. Samples were negatively stained by applying 5 μL of stain (1% w/v uranyl acetate in Milli Q water) onto the grid for 30 s and the excess water was blotted away with filter paper as well. Samples were dried overnight before imaging. Samples without staining were prepared in a similar way.
Dynamic light scattering (DLS) analysis was performed with Nanotrac by Anaspec operating with a Microtrac FLEX operating software at 25 °C, using a laser wavelength of 780 nm at a scattering angle of 180°. The observed sizes and standard derivations of the nanoparticles were calculated by taking an average of 5 measurements.
Scanning transmission electron microscopy (STEM) analysis was taken with a Carl-Zeiss 1550 Gemini field emission scanning electron microscope. Samples were prepared in the same way as for TEM analysis.
Atomic Force Microscopy (AFM) measurements were performed using a Multimode AFM (Bruker Nano Surfaces, Santa Barbara, CA, USA) with a NanoScope V controller with tapping mode.
Muffle furnace calcination of particles were carried out with increasing temperature of 2 °C min−1, hold at 550 °C for 3 h. Samples were drop-casted on the silicon wafers which were pre-cleaned with oxygen plasma and dried overnight in the vacuum oven before calcination.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research forms part of the research program of the Dutch Polymer Institute (DPI), Project #777t (DPI, P. O. Box 902, 5600 AX Eindhoven, the Netherlands) and the European Research Council is acknowledged for financial support by the grant ProtCage (616907).Notes and references
- J. N. Culver, A. D. Brown, F. Zang, M. Gnerlich, K. Gerasopoulos and R. Ghodssi, Virology, 2015, 479, 200–212 CrossRef PubMed.
- Y. Liu, Y. Xu, Y. Zhu, J. N. Culver, C. A. Lundgren, K. Xu and C. Wang, ACS Nano, 2013, 7, 3627–3634 CrossRef CAS PubMed.
- X. Chen, K. Gerasopoulos, J. Guo, A. Brown, C. Wang, R. Ghodssi and J. N. Culver, ACS Nano, 2010, 4, 5366–5372 CrossRef CAS PubMed.
- K. Zhou, F. Li, G. Dai, C. Meng and Q. Wang, Biomacromolecules, 2013, 14, 2593–2600 CrossRef CAS PubMed.
- Y. Liu, W. Zhang, Y. Zhu, Y. Luo, Y. Xu, A. Brown, J. N. Culver, C. A. Lundgren, K. Xu, Y. Wang and C. Wang, Nano Lett., 2013, 13, 293–300 CrossRef CAS PubMed.
- C.-Y. Chiang, J. Epstein, A. Brown, J. N. Munday, J. N. Culver and S. Ehrman, Nano Lett., 2012, 12, 6005–6011 CrossRef CAS PubMed.
- D. Fernandes, M. J. Krysmann and A. Kelarakis, Chem. Commun., 2016, 52, 8294–8296 RSC.
- E. Ertem, N. Murillo-Cremaes, R. P. Carney, A. Laromaine, E.-R. Janecek, A. Roig and F. Stellacci, Chem. Commun., 2016, 52, 5573–5576 RSC.
- D. P. Cormode, P. A. Jarzyna, W. J. M. Mulder and Z. A. Fayad, Adv. Drug Delivery Rev., 2010, 62, 329–338 CrossRef CAS PubMed.
- S. W. Lee, S. K. Lee and A. M. Belcher, Adv. Mater., 2003, 15, 689–692 CrossRef CAS.
- P.-Y. Chen, X. Dang, M. T. Klug, J. Qi, N.-M. Dorval Courchesne, F. J. Burpo, N. Fang, P. T. Hammond and A. M. Belcher, ACS Nano, 2013, 7, 6563–6574 CrossRef CAS PubMed.
- A. Cumbo, B. Lorber, P. F. X. Corvini, W. Meier and P. Shahgaldian, Nat. Commun., 2013, 4, 1503 CrossRef PubMed.
- A. R. Parker and H. E. Townley, Nat. Nanotechnol., 2007, 2, 347–353 CrossRef CAS PubMed.
- G. G. Daaboul, C. A. Lopez, J. Chinnala, B. B. Goldberg, J. H. Connor and M. S. Ünlü, ACS Nano, 2014, 8, 6047–6055 CrossRef CAS PubMed.
- R. J. Tseng, C. Tsai, L. Ma, J. Ouyang, C. S. Ozkan and Y. Yang, Nat. Nanotechnol., 2006, 1, 72–77 CrossRef CAS PubMed.
- S. Kanbak-Aksu, M. Nahid Hasan, W. R. Hagen, F. Hollmann, D. Sordi, R. A. Sheldon and I. W. C. E. Arends, Chem. Commun., 2012, 48, 5745–5747 RSC.
- Y. Shin, A. Dohnalkova and Y. Lin, J. Phys. Chem. C, 2010, 114, 5985–5989 CAS.
- T. Ueno, M. Suzuki, T. Goto, T. Matsumoto, K. Nagayama and Y. Watanabe, Angew. Chem., Int. Ed., 2004, 43, 2527–2530 CrossRef CAS PubMed.
- M. Suzuki, M. Abe, T. Ueno, S. Abe, T. Goto, Y. Toda, T. Akita, Y. Yamada and Y. Watanabe, Chem. Commun., 2009, 4871–4873, 10.1039/b908742g.
- J. M. Slocik, R. R. Naik, M. O. Stone and D. W. Wright, J. Mater. Chem., 2005, 15, 749–753 RSC.
- S. Hong, M. Y. Lee, A. O. Jackson and L. P. Lee, Light: Sci. Appl., 2015, 4, 267 CrossRef.
- L. M. Bronstein, Small, 2011, 7, 1609–1618 CrossRef CAS PubMed.
- M. Brasch, R. M. Putri, M. V. de Ruiter, D. Luque, M. S. T. Koay, J. R. Castón and J. J. L. M. Cornelissen, J. Am. Chem. Soc., 2017, 139, 1512–1519 CrossRef CAS PubMed.
- A. Liu, L. Yang, C. H. H. Traulsen and J. J. L. M. Cornelissen, Chem. Commun., 2017, 53, 7632–7634 RSC.
- L. Yang, A. Liu, S. Cao, R. M. Putri, P. Jonkheijm and J. J. L. M. Cornelissen, Chem.–Eur. J., 2016, 22, 15570–15582 CrossRef CAS PubMed.
- A. Liu, C. H. H. Traulsen and J. J. L. M. Cornelissen, ACS Catal., 2016, 6, 3084–3091 CrossRef CAS.
- F. Setaro, M. Brasch, U. Hahn, M. S. T. Koay, J. J. L. M. Cornelissen, A. de la Escosura and T. Torres, Nano Lett., 2015, 15, 1245–1251 CrossRef CAS PubMed.
- E. Jackson, M. Ferrari, C. Cuestas-Ayllon, R. Fernández-Pacheco, J. Perez-Carvajal, J. M. de la Fuente, V. Grazú and L. Betancor, Langmuir, 2015, 31, 3687–3695 CrossRef CAS PubMed.
- E. Ravera, T. Martelli, Y. Geiger, M. Fragai, G. Goobes and C. Luchinat, Coord. Chem. Rev., 2016, 15, 110–122 CrossRef.
- D. Ma, M. Li, A. J. Patil and S. Mann, Adv. Mater., 2004, 16, 1838–1841 CrossRef CAS.
- S. Kerkhofs, F. Leroux, L. Allouche, R. Mellaerts, J. Jammaer, A. Aerts, C. E. A. Kirschhock, P. C. M. M. Magusin, F. Taulelle, S. Bals, G. Van Tendeloo and J. A. Martens, RSC Adv., 2014, 4, 25650–25657 RSC.
- Z. Liu, X. Xu and R. Tang, Adv. Funct. Mater., 2016, 26, 1862–1880 CrossRef CAS.
- S. Heinemann, T. Coradin and M. F. Desimone, Biomater. Sci., 2013, 1, 688–702 RSC.
- M. L. Foglia, D. E. Camporotondi, G. S. Alvarez, S. Heinemann, T. Hanke, C. J. Perez, L. E. Diaz and M. F. Desimone, J. Mater. Chem. B, 2013, 1, 6283–6290 RSC.
- G. Navarro-Tovar, G. Palestino and S. Rosales-Mendoza, Expert Rev. Vaccines, 2016, 5, 1449–1462 CrossRef PubMed.
- W. K. Cho and S. H. Yang, Eur. J. Inorg. Chem., 2015, 2015, 4481–4494 CrossRef CAS.
- J. Lee, J. Choi, J. H. Park, M.-H. Kim, D. Hong, H. Cho, S. H. Yang and I. S. Choi, Angew. Chem., Int. Ed., 2014, 53, 8056–8059 CrossRef CAS PubMed.
- D. Arcos and M. Vallet-Regí, Acta Biomater., 2010, 6, 2874–2888 CrossRef CAS PubMed.
- S. Wang, X. Wang, F. G. Draenert, O. Albert, H. C. Schröder, V. Mailänder, G. Mitov and W. E. G. Müller, Bone, 2014, 67, 292–304 CrossRef CAS PubMed.
- M. T. Klem, M. Young and T. Douglas, J. Mater. Chem., 2008, 18, 3821–3823 RSC.
- C. Jolley, M. Klem, R. Harrington, J. Parise and T. Douglas, Nanoscale, 2011, 3, 1004–1007 RSC.
- K. Kumar, S. Kumar Doddi, M. Kalle Arunasree and P. Paik, Dalton Trans., 2015, 44, 4308–4317 RSC.
- N. F. Steinmetz, S. N. Shah, J. E. Barclay, G. Rallapalli, G. P. Lomonossoff and D. J. Evans, Small, 2009, 5, 813–816 CrossRef CAS PubMed.
- E. S. Royston, A. D. Brown, M. T. Harris and J. N. Culver, J. Colloid Interface Sci., 2009, 332, 402–407 CrossRef CAS PubMed.
- E. Royston, S.-Y. Lee, J. N. Culver and M. T. Harris, J. Colloid Interface Sci., 2006, 298, 706–712 CrossRef CAS PubMed.
- B. Liu, Y. Cao, Z. Huang, Y. Duan and S. Che, Adv. Mater., 2015, 27, 479–497 CrossRef CAS PubMed.
- C. E. Fowler, W. Shenton, G. Stubbs and S. Mann, Adv. Mater., 2001, 13, 1266–1269 CrossRef CAS.
- K. Altintoprak, A. Seidenstücker, A. Welle, S. Eiben, P. Atanasova, N. Stitz, A. Plettl, J. Bill, H. Gliemann, H. Jeske, D. Rothenstein, F. Geiger and C. Wege, Beilstein J. Nanotechnol., 2015, 6, 1399–1412 CrossRef CAS PubMed.
- G. Wang, H.-J. Wang, H. Zhou, Q.-G. Nian, Z. Song, Y.-Q. Deng, X. Wang, S.-Y. Zhu, X.-F. Li, C.-F. Qin and R. Tang, ACS Nano, 2015, 9, 799–808 CrossRef CAS PubMed.
- Z. Niu, S. Kabisatpathy, J. He, L. A. Lee, J. Rong, L. Yang, G. Sikha, B. N. Popov, T. S. Emrick, T. P. Russell and Q. Wang, Nano Res., 2009, 2, 474–483 CrossRef CAS.
- G. Wang, H. Zhou, Q.-G. Nian, Y. Yang, C.-F. Qin and R. Tang, Chem. Sci., 2016, 7, 1753–1759 RSC.
- R. Konecny, J. Trylska, F. Tama, D. Zhang, N. A. Baker, C. L. Brooks and J. A. McCammon, Biopolymers, 2006, 82, 106–120 CrossRef CAS PubMed.
- C. A. Hommersom, B. Matt, A. van der Ham, J. J. L. M. Cornelissen and N. Katsonis, Org. Biomol. Chem., 2014, 12, 4065–4069 CAS.
- G. J. Bedwell, Z. Zhou, M. Uchida, T. Douglas, A. Gupta and P. E. Prevelige, Biomacromolecules, 2015, 16, 214–218 CrossRef CAS PubMed.
- P. A. Suci, M. T. Klem, T. Douglas and M. Young, Langmuir, 2005, 21, 8686–8693 CrossRef CAS PubMed.
- B. Michen and T. Graule, J. Appl. Microbiol., 2010, 109, 388–397 CAS.
- C. C. M. C. Carcouët, M. W. P. van de Put, B. Mezari, P. C. M. M. Magusin, J. Laven, P. H. H. Bomans, H. Friedrich, A. C. C. Esteves, N. A. J. M. Sommerdijk, R. A. T. M. van Benthem and G. de With, Nano Lett., 2014, 14, 1433–1438 CrossRef PubMed.
- P. W. J. G. Wijnen, T. P. M. Beelen, C. P. J. Rummens and R. A. van Santen, J. Non-Cryst. Solids, 1991, 136, 119–125 CrossRef CAS.
- K. W. Adolph, J. Gen. Virol., 1975, 28, 137–145 CrossRef.
- J. A. Speir, S. Munshi, G. Wang, T. S. Baker and J. E. Johnson, Structure, 1995, 3, 63–78 CrossRef CAS PubMed.
- T. Douglas, E. Strable, D. Willits, A. Aitouchen, M. Libera and M. Young, Adv. Mater., 2002, 14, 415–418 CrossRef CAS.
- T. Douglas and M. Young, Nature, 1998, 393, 152–155 CrossRef CAS.
- T. Douglas and M. Young, Science, 2006, 312, 873–875 CrossRef CAS PubMed.
- L. Lavelle, J.-P. Michel and M. Gingery, J. Virol. Methods, 2007, 146, 311–316 CrossRef CAS PubMed.
- K. W. Adolph, Eur. J. Biochem., 1975, 53, 449–455 CrossRef CAS PubMed.
- J. Chen, R. Zhang, L. Han, B. Tu and D. Zhao, Nano Res., 2013, 6, 871–879 CrossRef CAS.
- R. F. Bruinsma, M. Comas-Garcia, R. F. Garmann and A. Y. Grosberg, Phys. Rev. E, 2016, 93, 032405 CrossRef CAS PubMed.
- A. Liu, M. Verwegen, M. V. de Ruiter, S. J. Maassen, C. H. H. Traulsen and J. J. L. M. Cornelissen, J. Phys. Chem. B, 2016, 120, 6352–6357 CrossRef CAS PubMed.
- H. I. Oh, J. E. Hoff, G. S. Armstrong and L. A. Haff, J. Agric. Food Chem., 1980, 28, 394–398 CrossRef CAS.
- J. P. Van Buren and W. B. Robinson, J. Agric. Food Chem., 1969, 17, 772–777 CrossRef CAS.
- J. Gao, H. Lei, Z. Han, Q. Shi, Y. Chen and Y. Jiang, Sci. Rep., 2017, 7, 45215 CrossRef CAS PubMed.
- Z. Gao and I. Zharov, Chem. Mater., 2014, 26, 2030–2037 CrossRef CAS.
- Y. Chen, H. Chen, Y. Sun, Y. Zheng, D. Zeng, F. Li, S. Zhang, X. Wang, K. Zhang, M. Ma, Q. He, L. Zhang and J. Shi, Angew. Chem., 2011, 123, 12713–12717 CrossRef.
- M. Colombo, S. Carregal-Romero, M. F. Casula, L. Gutierrez, M. P. Morales, I. B. Bohm, J. T. Heverhagen, D. Prosperi and W. J. Parak, Chem. Soc. Rev., 2012, 41, 4306–4334 RSC.
- J. G. Croissant, D. Zhang, S. Alsaiari, J. Lu, L. Deng, F. Tamanoi, A. M. AlMalik, J. I. Zink and N. M. Khashab, J. Controlled Release, 2016, 229, 183–191 CrossRef CAS PubMed.
- R. Ciganda, N. Li, C. Deraedt, S. Gatard, P. Zhao, L. Salmon, R. Hernandez, J. Ruiz and D. Astruc, Chem. Commun., 2014, 50, 10126–10129 RSC.
- K. Kuroda, T. Ishida and M. Haruta, J. Mol. Catal. A: Chem., 2009, 298, 7–11 CrossRef CAS.
- R. M. Putri, J. J. L. M. Cornelissen and M. S. T. Koay, ChemPhysChem, 2015, 16, 911–918 CrossRef CAS PubMed.
- M. Brasch, A. de la Escosura, Y. Ma, C. Uetrecht, A. J. R. Heck, T. Torres and J. J. L. M. Cornelissen, J. Am. Chem. Soc., 2011, 133, 6878–6881 CrossRef CAS PubMed.
- A. de la Escosura, M. Verwegen, F. D. Sikkema, M. Comellas-Aragones, A. Kirilyuk, T. Rasing, R. J. M. Nolte and J. J. L. M. Cornelissen, Chem. Commun., 2008, 1542–1544 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra11310b |
| This journal is © The Royal Society of Chemistry 2017 |