Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Enhanced sulfamethoxazole degradation in soil by immobilized sulfamethoxazole-degrading microbes on bagasse†
Shengbing Hu,
Huimin Hu,
Wenlong Li,
Yaoyi Ke,
Minghua Li and
Yuechun Zhao *
*
College of Materials and Energy, South China Agricultural University, Guangzhou, Guangdong 510642, PR China. E-mail: yczhao@scau.edu.cn; Fax: +86-20-8528-2366; Tel: +86-20-8528-0323
First published on 5th December 2017
Abstract
The presence of sulfamethoxazole (SMX) in the environment is becoming a serious problem because of its toxicity and high risk to human health and microbial activity. In this study, enhanced degradation of this compound in soil is shown by sulfamethoxazole-degrading microbes when using bagasse to pre-grow these microbial sources. The two sulfamethoxazole-degrading strains, T2 and Z3, isolated from pig manure, were selected for an immobilized microorganism technique. T2 and Z3 were identified as Enterobacter cloacae and a fungal endophyte by detailed morphological, biochemical and molecular characterization. The performance of the immobilized cells on agricultural residues (bagasse) in the biodegradation of sulfamethoxazole in soil was greater than that of cells free in solution. The effects of pH, temperature, indigenous soil microbes, and heavy metals on bioremediation were investigated, as well as the products of SMX degradation by strains, and a series of experiments at an initial concentration of 100 mg kg−1 were performed at various temperatures (23–43 °C) and pHs (2.5–6.5). SMX degradation was affected by the initial temperature and pH because of the effect on the essential groups in the activation center of enzymes and membrane permeability. At 28 °C and pH 3.5, the immobilized strain T2 showed an excellent ability to degrade sulfamethoxazole in soil; 84.14% removal rate of SMX was achieved.
1. Introduction
Sulfamethoxazole (4-amino-N-(5-methylisoxazol-3-yl)-benzenesulfonamide; SMX) is a worldwide antibiotic, which is commonly used in clinical treatment and veterinary medicine.1,2 SMX is also widely used in farming and aquaculture.3–6 Due to the characteristics of SMX, approximately 15–25% is incompletely metabolized and excreted into the surrounding environment from the human or animal body after ingestion.7–10 The wide use of SMX raises particular concerns for human health because its presence in the environment may harm human health and lead to antibiotic resistance,11–13 as well as being a serious threat to soil and water ecosystems. In China, SMX is released from four major sources: industry, hospitals, farms, and households; some SMX is released into water and soil without pre-treatment.6,14 The degradation of SMX in soil, therefore, has become a major environmental concern. To date, different physicochemical techniques have been tested to remove SMX from polluted soil or water including oxidation,15–17 adsorption,18–21 photodegradation22 and hydrolysis.23 Kobayashi et al.24 reported that SMX degradation by zero-valent iron (ZVI) under the anoxic condition was completed within 300 min in the acidic solutions, however remained to less than 70% after 300 min in neutral or moderate alkaline solutions. Yang et al.25 investigated that the degradation rate of SMX by a novel hydrogel catalyst (p(HEA/NMMA)–CuS) with the function of adsorption p(HEA/NMMA)–CuS hydrogel reached 95.91% and the SMX was mineralized with 43.56% in aqueous solution under 500 W visible light achieved balance in 24 h. Al-Hamadani et al.26 examined the feasibility of using two types of fly ash (an industrial waste from power station) as a low-cost catalyst to enhance the ultrasonic (US) degradation of SMX. The results showed that 99% degradation of SMX was achieved by sonication for 60 min at 580 kHz and pH 3.5. Wang et al.27 employed Fenton process and persulfate process to remove sulfamethoxazole from aqueous. The maximal mineralization reached 83% when hydrogen peroxide concentration was 1 mM and Fe(II) was 0.05 mM for Fenton process. Peleyeju et al.28 reported the photoelectrocatalytic degradation of sulfamethoxazole at a TiO2–exfoliated graphite (TiO2–EG) anode. Almost 100% of SMX had been degraded after photoelectrocatalytic process. Wang et al.29 studied the degradation process of SMX in anodic chamber of microbial fuel cell (MFC) reactors. They found that SMX even with high concentration (200 ppm) can be totally degraded rapidly.Compared with physicochemical methods, biodegradative methods are widely preferred, due to lower costs and the possibilities of complete mineralization. Various SMX-degrading microbial strains and their biodegradability are presented in the ESI†. Some studies on biodegradation of sulfonamides in aquatic environment by bacterial showed some remarkable results had been achieved over the last few decades. Especially, Jiang et al.3 studied the biodegradation and metabolic pathway of sulfamethoxazole by a cold-adapted bacterium in waste water. The maximal degradation rate of SMX was 34.3% at 10 °C for 192 h. Larcher et al.30 investigated the biodegradation of sulfamethoxazole by individual and mixed bacteria in waste water. The results showed that the individual bacteria (R. equi) had the greatest ability to remove SMX leading to 29% degradation. Araujo et al.31 declared that approximately 74% of SMX were removed from culture medium by White Rot Fungi (P. ostreatus) after 15 days of cultivation. Xiao et al.32 investigated that SMX were consistently removed at efficiencies of 67.8 ± 13.9% in a lab-scale anaerobic membrane bioreactor (AnMBR) treating with synthetic sewage. When powdered activated carbon (PAC) was added to the AnMBR, the degradation efficiency for SMX was 95.5 ± 4.6%. Jia et al.33 found that the removal of SMX followed the pseudo-zero-order kinetic model with a specific removal rate of 13.2 ± 0.1 μg L−1 d−1 at initial SMX concentration of 100 μg L−1 using sulfate-reducing bacteria (SRB) sludge in a sulfate-reducing up-flow sludge bed reactor. Wang et al.34 isolated and identified a novel strain (Acinetobacter sp.), which can degrade SMX in a range of 5–240 mg L−1 in water with efficiency up to 100%.
The immobilization of microbial cells proved to be a promising method, which offers a suitable environment for the biodegradation process.35,36 Immobilized cultures tend to have several advantages over free cells: a higher level of biodegradation activity; a stronger resilience to environmental perturbations, such as pH, or exposure to toxic chemical concentrations; as well as an increased density of cells in the matrix.37 Furthermore, immobilization of cells is biotechnologically easier because of a facilitated process control.38 In addition, the immobilized microorganism technique has been proven to be highly effective, is widely applied, and results in a production of innocuous end products. However, there are only a few studies on immobilized microorganism removal of SMX in soil.37,39,40
The technique presents a method for preparing and using microbial immobilized material for the remediation of contaminated soil. Bagasse, dry sugar cane pulp, was selected as an optimal bio-carrier for this study. The aims of this study were to isolate, characterize and exploit the SMX biodegradation potential of isolated microbial species. In particular, the cell growth of the two strains was investigated. In addition, the degradation of SMX by isolated strains of T2 and Z3 in a batch culture using free cells was compared to its degradation by immobilized cells. The effects of the concentration of bacterial inoculation, pH, temperature, indigenous soil microbes, and the presence of heavy metals on SMX degradation were determined, as were as the products of SMX degradation.
2. Methods and material
2.1 Chemicals and materials
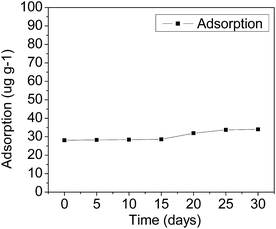
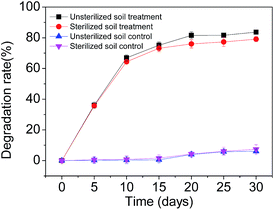
All chemicals used were of analytical grade or HPLC grade and purchased either from Sinopharm Chemical Reagent Co., Ltd (Guangdong, China) or Guangzhou Chemical Reagent Factory (Guangdong, China). SMX of greater than 99% purity was used. Agricultural residues (bagasse, peanut shell, straw) were obtained from the South China Agricultural University farm. Soil samples were collected from a forest garden in the South China Agricultural University. This site had no history of exposure to SMX. The soil sample was air-dried and sieved through Ø 3 mm mesh and stored at room temperature before use.The adsorption capacity of SMX in soil was determined by serial batch-type experiments. Prior to the experiment, the both soil and glass materials employed were autoclaved at 121 °C for 15 min to inhibit microbial degradation processes. 2.0 g of soil was mixed with 10 mL of initial SMX concentrations 20 mg L−1 in centrifuge tube. The obtained SMX suspension with 0.01 mol L−1 CaCl2 (40 mL) was shaken in a thermostatic oscillator at a constant agitation speed (220 rpm) for 24 h. The SMX suspension was centrifuged at 5000 rpm for 15 minutes to separate the liquid from the solid. Finally, the supernatant solution was filtered with 0.22 μm filter membranes.34
During the experiment, temperature was maintained at 28 °C and pH 3.5. 1 mL of aliquots was taken from the liquid phase at various time points (5, 10, 15, 20, 25 and 30 d) in order to check SMX.
The adsorption amounts of per gram experimental soil were shown in Fig. 1.
2.2 Culture conditions
The SMX-degrading strains were isolated from samples of pig manure from a pig farm in Guangdong Province, China. The pigs were administered with SMX for the control of meningitis for more than 5 days before collection. For the selection of microbial strains, standard batch enrichment culture techniques were performed. 2.0 g of samples of pig manure were placed into 250 mL flasks containing 50 mL of SMX at a concentration of 10 mg L−1, and incubated aerobically at 28 °C on a reciprocal shaker at 150 rpm. After 1 h, aliquots were transferred weekly to fresh medium and incubated under the same conditions. This process was repeated at least six times. Pure cultures were obtained by plating the enrichment culture on solid mineral medium. Mineral medium contained (per liter): 0.5 g KH2PO4; 0.5 g K2HPO4, 0.2 g NaCl, 1.0 g NH4NO3, 0.2 g MgSO4, and 10.0 mL of a trace element solution containing (per liter): 0.1 g FeSO4, 0.1 g MnSO4, 0.1 g ZnSO4, 0.01 g Na2MoO4, 0.1 g CaCl2, 3 g MgSO4, 0.1 g CuSO4. Solid media contained 25 g agar per liter.32.3 Identification of SMX-degrading strains
The selected SMX-degrading isolates were identified by biochemical characterization tests and the 16S rDNA gene sequence analytical methods. A total volume of 20 μL of the 16S PCR product was sent to Honortech Corporation in Guangzhou for sequencing. For the identification of bacteria, DNA sequences were analyzed by the Basic Local Alignment Search Tool (BLAST) at the National Center for Biotechnology Information website (NCBI, http://www.ncbi.nlm.nih.gov/).2.4 Choice of optimum residue and cell growth
Several 10 mm disks of the T2 and Z3 actively growing strains on agar were separately combined with a 1 h autoclaved 1.5 g sample of each agricultural residue (bagasse, peanut shell and straw bar straw pole) and 10 mL of mineral medium, which was incubated at 28 °C on a rotary shaker at 150 rpm for 2 days. Mineral media without added agricultural residues were also inoculated with each of the two strains separately, which were used as controls, these will be called ‘free cells’ henceforth. The biomass was analysed by UV-Vis spectrophotometer.Cultures were prepared in 250 mL Erlenmeyer flasks by adding 100 mL of minimal mineral medium amended with peptone (2 g L−1). 5 mL of each of the two SMX-degrading bacterial suspensions and SMX (250 mg L−1) were added to it, and then incubated at 28 °C on a rotary shaker at 150 rpm for two days. Abiotic controls without inoculum were incubated under identical conditions. Experimental treatments and controls were set up in triplicate. The biomass was analysed by UV-Vis spectrophotometer at 0 h, 24 h, 48 h, 72 h, 96 h, 120 h, 144 h, 168 h and 192 h.
2.5 Degradation experiments
To facilitate the bioremediation studies, additional experiments were carried out at 28 °C and pH 3.5. 3.5 mL of each of the two SMX-degrading bacterial suspensions and 7 mL mineral medium were added to 1 g of sterilized agricultural residues, and then incubated at 28 °C on a rotary shaker at 150 rpm. After 2 days, 10.0 g of sterilized soil with an SMX concentration of 100 mg kg−1 was added to it. All experiments were carried out in triplicate. Controls without agricultural residues were prepared for each assay. The SMX concentration was analysed at 0 d, 5 d, 10 d, 15 d, 20 d, 25 d and 30 d.The optimum physiological parameters for SMX degradation of pH,41 temperature, indigenous soil microbes, concentration of bacterial inoculation and heavy metal were investigated.
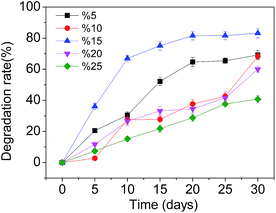
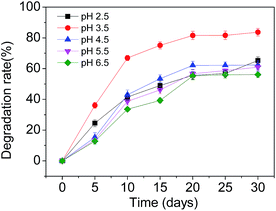
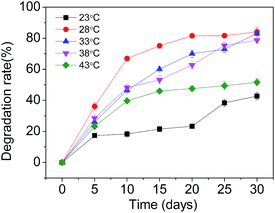
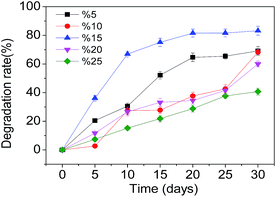
Firstly, experiments were carried out at various pHs (2.5, 3.5, 4.5, 5.5 and 6.5), keeping SMX-degrading bacterial suspensions at 3.5 mL and temperature at 28 °C. At pH 3.5 further experiments were carried out at various temperatures (23 °C, 28 °C, 33 °C, 38 °C and 43 °C) with SMX-degrading bacterial suspensions of 3.5 mL in order to determine the optimal temperature for SMX degradation. Effects of the heavy metal ions Cd2+ and Pb2+ on the degradation of SMX in soil were also investigated. 1 mmol L−1 of Cd2+ and Pb2+ was added separately to the samples, keeping SMX-degrading bacterial suspensions at 3.5 mL, temperature at 28 °C and pH 3.5. To study the effects of indigenous soil microbes on SMX degradation, the additions of unsterilized and sterilized soil were compared, keeping SMX-degrading bacterial suspensions at 3.5 mL, temperature at 28 °C and pH 3.5. In order to ascertain the effect of the concentration of bacterial inoculation on the degradation rate, experiments were carried out at various SMX-degrading bacterial suspensions (1.0 mL, 2.0 mL, 3.5 mL, 4.5 mL, and 6.0 mL) at 28 °C and pH 3.5. These experiments were carried out using pure cultures of T2 immobilized on bagasse.
2.6 Sample pre-treatment
For the measurement of SMX concentrations in soil, each soil sample was immersed in 50 mL buffer solution–methanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and transferred equally to two 50 mL centrifuge tubes. The buffer solution was made by dissolving 21.0 g of citric acid and 20.4 g of MgCl2·H2O in water, and using ammonia to adjust the pH to 5.0 to make 1000 mL. The centrifuge tubes were placed on a rotary shaker at 150 rpm for 5 minutes, ultrasonic extraction for 15 minutes, and then separated by centrifuge at 4500 rpm for 10 minutes. The extracted liquid was transferred to 250 mL round-bottom flasks and the whole process above was repeated two times. The resulting supernatant was evaporated by rotator evaporation and adjusted to 20 mL with methanol–water (3
1) and transferred equally to two 50 mL centrifuge tubes. The buffer solution was made by dissolving 21.0 g of citric acid and 20.4 g of MgCl2·H2O in water, and using ammonia to adjust the pH to 5.0 to make 1000 mL. The centrifuge tubes were placed on a rotary shaker at 150 rpm for 5 minutes, ultrasonic extraction for 15 minutes, and then separated by centrifuge at 4500 rpm for 10 minutes. The extracted liquid was transferred to 250 mL round-bottom flasks and the whole process above was repeated two times. The resulting supernatant was evaporated by rotator evaporation and adjusted to 20 mL with methanol–water (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) for HPLC analysis. Degradation rate of SMX was calculated according to the standard curve of SMX (ESI†).
2) for HPLC analysis. Degradation rate of SMX was calculated according to the standard curve of SMX (ESI†).
2.7 Analytical methods
Cell growth was monitored using UV-Vis spectrophotometer at 600 nm. The measurement was made such that the OD value of the samples was below 0.8 as obtained by sample dilution. The OD value was then converted into the biomass value of the sample.SMX was analysed quantitatively by high-performance liquid chromatography (Agilent 1100 HPLC) equipped with a UV-Vis detector,42 and a reverse-phase column: LiChrospher C-18 (4.6 mm × 250 mm, 5 μm, Athena). The operating conditions for the analysis of SMX were: 20 μL injection volume, λ = 269 nm, eluent acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ultrapure water with 0.3% acetic acid in isocratic conditions of 25%
ultrapure water with 0.3% acetic acid in isocratic conditions of 25%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 75% and at a flow rate of 1 mL min−1.43 The collected samples were filtered immediately through 0.22 μm cellulose acetate filter membranes and stored at 4 °C for future analysis.
75% and at a flow rate of 1 mL min−1.43 The collected samples were filtered immediately through 0.22 μm cellulose acetate filter membranes and stored at 4 °C for future analysis.
All experiments and measurements were performed in duplicate and the arithmetic averages were taken for data analysis.
2.8 Analysis of the degradation products
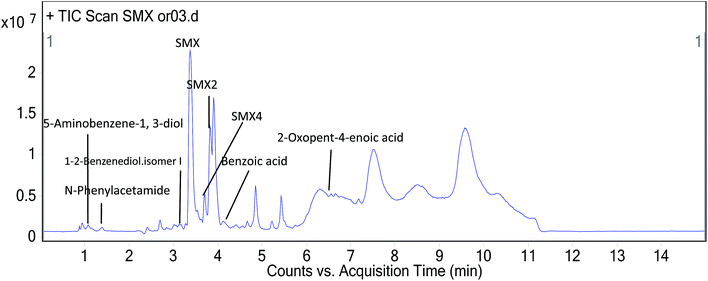
The SMX degradation products from soil samples containing free SMX-degrading bacteria were extracted separately with ethyl acetate and n-butanol three times, then combined the extraction to dryness using nitrogen at 50 °C in a water bath. The dry residues were dissolved in ethyl acetate. The ethyl acetate phase was centrifuged for 10 min at 8000 rpm to collect the upper layer, and concentrated by drying using nitrogen at 50 °C. Finally, the sample was re-dissolved in methanol and diluted to a concentration of 200 ppm. The collected samples were filtered immediately through 0.22 μm filter membranes for UPLC-Q-TOF analysis.33,44 The operating conditions for the analysis of the SMX metabolites were: ECLIPS PLUS C18 (50 × 2.1 mm, 1.8 μm), acetonitrile containing formic acid and ammonium formate (mobile phase), 2.0 μL injection volume. UPLC-Q-TOF was used to attain the primary mass spectrometry data which included accurate mass for various compounds. Find by formula spectrum (FBF) determined the structure of target compounds.3. Results and discussion
3.1 Isolation and characterization of SMX-degrading strains
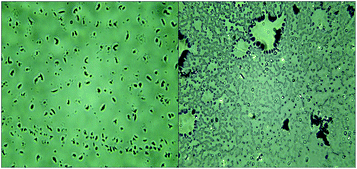
Analysis of the 16S rDNA gene of the two isolates (T2 and Z3) revealed that they belong to the bacterium Enterobacter cloacae and a fungal endophyte, respectively. The morphology of the two isolates under microscope is shown in Fig. 2.45 Biochemical characterization tests of the T2 bacterium are presented in Table 1, which showed that T2 was catalase-positive, motile, indole-negative, facultatively anaerobic or anaerobic, methyl red-negative and starch hydrolysis-positive.| Test | Phenomenon | Result |
|---|---|---|
| Catalase | Oxygen | + |
| Indole | No change in colour | − |
| Methyl red | Faded to yellow | − |
| Kinetic | Bacterial growth is not limited to puncture lines | + |
| Aerobic or anaerobic | Bacteria grows on puncture lines and medium surfaces | + |
| Starch hydrolysis | Hydrolysed | + |
3.2 Cell growth on different agricultural residues
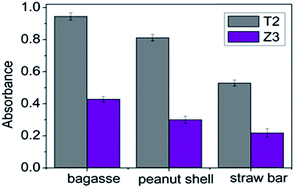
Comparing the growth of the two strains T2 and Z3 immobilized on various biomass materials (straw bar, peanut shell and bagasse), growth was greatest on bagasse in Fig. 3. Growth of T2 and Z3 on bagasse is shown in Fig. 4. Fig. 4 shows that the growth of T2 directly entered into a logarithmic phase and reached a stable phase from 18 h to 36 h. For Z3 there was an initial lag phase followed by a logarithmic phase from 24 h to 56 h. Subsequently, Z3 entered into stable phase from 56 h to 80 h.Studies have shown that the ability of immobilized microbial strains to degrade pollutants in the soil environment is greater than that of free microbes because the material on to which they are immobilized provides a more optimal environment for growth.46
3.3 Degradation of SMX
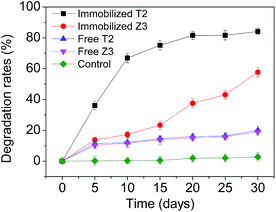 | ||
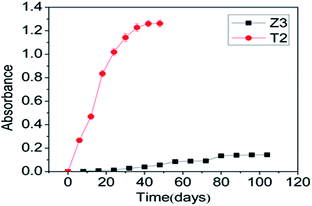
| Fig. 5 SMX degradation comparing strains T2 and Z3 immobilized on bagasse residue with strains grown as free cells. | ||
To overcome the limitations of the use of exogenous bacteria for pollutant bioremediation microbial cells are often immobilized, a modification that can increase the biodegradation rate through protecting the cells from sudden exposure to higher concentrations of toxic substances such as PAHs, POPs and other compounds by avoiding direct contact between cells and pollutants. Strain T2, with the higher degradation rate of the two isolates, was selected for subsequent experiments in this study.
3.4 Analysis of the degradation products
According to the formula obtained by Q-TOF analysis, the structure of SMX, literature sources and general metabolic laws we propose the metabolic process and intermediate products (Table 2) of the degradation of SMX by the T2 bacterium Enterobacter cloacae. Fig. 11 shows the peaks of the partial primary products detected by UPLC. Scheme 1 shows a proposed pathway of SMX-biodegradation based on the analysis of these experimental results. The spectrum of UPLC-Q-TOF can be seen in the ESI.† The initial metabolites of SMX biodegradation follow four pathways. These are: methylation (SMX1); substitution by hydroxyl and double bond oxidation (SMX2); desulphurization (SMX3); and acetylation (SMX4). Among them, the acetylation pathway (SMX4) is significant for further research. Differing from previous thoughts which believed that the initial SMX biodegradation pathways were either hydrolysis or hydroxylation then hydrolysis, the T2 bacterium resists the toxicity of SMX by acetylation. N-(4-Hydroxyphenyl)acetamide might be attributed to hydroxylation of N-phenylacetamide, N-phenylacetamide was also converted to benzoic and phenylacetic acid through other pathways. The final process was the degradation of the benzene ring. Compounds containing a benzene ring are hydroxylated to form catechol and its structural isomers under catalysis of dioxygenase. There was also the combination of two hydroquinone metabolites, under pyrolysis which resulted in peak time diversity. The generation of oxopent-4-enoic acid due to the open-loop reaction of pyrocatechol.16,22,58–61| Chemical structure | RT | Mass | Formula | Tgt mass | Diff (ppm) | Significant ion m/z |
|---|---|---|---|---|---|---|
| 1-2-Benzenediol, isomer IV | 4.367 | 110.03668 | C6H6O2 | 110.03678 | −0.86 | 111.0439 |
| 1-2-Benzenediol, isomer II | 3.506 | 110.03672 | C6H6O2 | 110.03678 | −0.55 | 111.0440 |
| 1-2-Benzenediol, isomer I | 3.126 | 110.03676 | C6H6O2 | 110.03678 | −0.16 | 111.0441 |
| 1-2-Benzenediol, isomer V | 4.797 | 110.03684 | C6H6O2 | 110.03678 | 0.58 | 111.0441 |
| 1-2-Benzenediol, isomer III | 4.201 | 110.03689 | C6H6O2 | 110.03678 | 1.00 | 111.0442 |
| 2-Oxopent-4-enoic acid | 6.485 | 114.03160 | C5H6O3 | 114.03169 | −0.78 | 115.0389 |
| Benzoic acid | 4.119 | 122.03684 | C7H6O2 | 122.03678 | 0.51 | 123.0441 |
| 5-Aminobenzene-1,3-diol | 1.074 | 125.04764 | C6H7NO2 | 125.04768 | −0.30 | 126.0550 |
| N-Phenylacetamide | 1.355 | 135.06826 | C8H9NO | 135.06841 | −1.10 | 136.0755 |
| Phenylacetic acid | 2.960 | 136.05211 | C8H8O2 | 136.05243 | −2.35 | 137.0595 |
| 4-Acetamidophenol | 1.339 | 151.06321 | C8H9NO2 | 151.06333 | −0.82 | 152.0704 |
| SMX3 | 3.374 | 189.09031 | C10H11N3O | 189.09021 | 0.53 | 190.0975 |
| SMX | 3.490 | 253.05315 | C10H11N3O3S | 253.05211 | 4.10 | 254.0606 |
| SMX1 | 3.374 | 256.05198 | C10H12N2O4S | 256.05178 | 0.79 | 257.0594 |
| SMX2, isomer I | 2.944 | 267.06768 | C11H13N3O3S | 267.06776 | −0.32 | 268.0749 |
| SMX2, isomer III | 3.887 | 267.06781 | C11H13N3O3S | 267.06776 | 0.18 | 268.0751 |
| SMX2, isomer II | 3.754 | 267.06794 | C11H13N3O3S | 267.06776 | 0.67 | 268.0753 |
| SMX4 | 3.639 | 295.06275 | C12H13N3O4S | 295.06268 | 0.25 | 296.0700 |
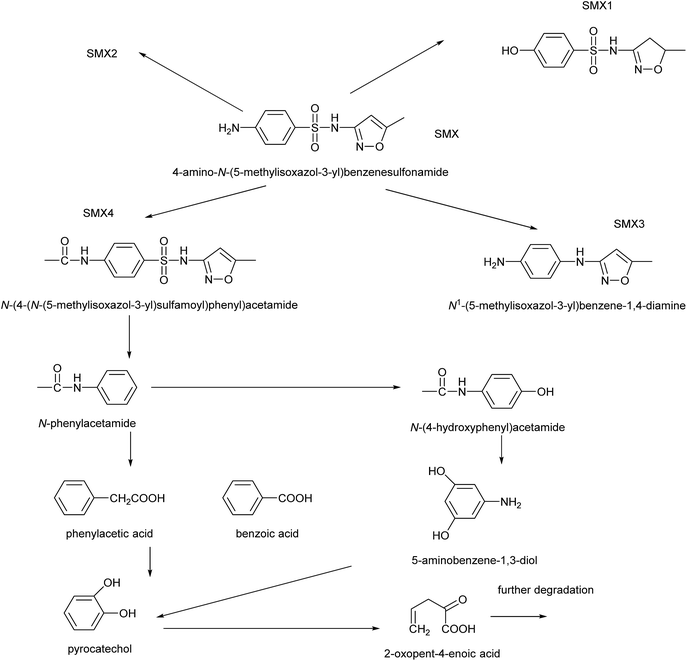 | ||
| Scheme 1 Proposed pathways of SMX degradation by strain the bacterium strain T2, Enterobacter cloacae. | ||
4. Conclusions
Within 30 d of the incubation period, the concentrations of SMX in soil declined rapidly in the first 15 d under all experimental conditions, this decline slowed after this period. This could be due to the decrease in nutrients and microelements and the accumulation of toxic intermediates, which would cause a reduction in the activity of the cells and enzymes. The optimal temperature and pH for the degradation of SMX by the bacterial strain T2 immobilized on bagasse were determined to be 28 °C and 3.5, respectively. We can also conclude that the degradation rate of SMX in the presence of Pb2+ and Cd2+ in soil was significantly lower than that in the control group.62 The participation of indigenous microorganisms improved bioremediation of SMX by the T2 bacterium. Bagasse is an agricultural residue, in this study, we used it as a support for increasing the enzymatic activity by enabling a greater microbial biomass to accumulate and therefore increase the biodegradation of SMX.46 Indeed, the immobilized strains on bagasse exhibited remarkably improved catalytic capacity and stability properties for degrading SMX in soil, the degradation rate reached up to 84.14%. Therefore, these results present a notable potential method for the biodegradation of SMX in soil. Several compounds were identified using HR-MS to analyse the pathways of SMX biodegradation. In summary, selecting bagasse as support for SMX-degrading microbial strains can be used as a guideline for improving the degradation of SMX, which are important economic and environmental aims for remediation of the environment.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This project was supported by the Science and Technology Planning Project of Guangdong Province of China (No. 2015A020209125). We especially thank Dr Sally Gadsdon for the language revision of this manuscript.References
- M. Radke, C. Lauwigi, G. Heinkele, T. E. Muerdter and M. Letzel, Environ. Sci. Technol., 2009, 43, 3135–3141 CrossRef CAS PubMed.
- B. Xu, D. Mao, Y. Luo and L. Xu, Bioresour. Technol., 2011, 102, 7069–7076 CrossRef CAS PubMed.
- B. Jiang, A. Li, D. Cui, R. Cai, F. Ma and Y. Wang, Appl. Microbiol. Biotechnol., 2014, 98, 4671–4681 CrossRef CAS PubMed.
- T.-H. Yu, A. Y.-C. Lin, S. C. Panchangam, P.-K. A. Hong, P.-Y. Yang and C.-F. Lin, Chemosphere, 2011, 84, 1216–1222 CrossRef CAS PubMed.
- F. Bonvin, J. Omlin, R. Rutler, W. B. Schweizer, P. J. Alaimo, T. J. Strathmann, K. McNeill and T. Kohn, Environ. Sci. Technol., 2013, 47, 6746–6755 CrossRef CAS PubMed.
- W. M. M. Mahmoud, N. D. H. Khaleel, G. M. Hadad, R. A. Abdel-Salam, A. Haiss and K. Kuemmerer, Clean: Soil, Air, Water, 2013, 41, 907–916 CrossRef CAS.
- M. Clara, N. Kreuzinger, B. Strenn, O. Gans and H. Kroiss, Water Res., 2005, 39, 97–106 CrossRef CAS PubMed.
- J. Akhtar, N. S. Amin and A. Aris, Chem. Eng. J., 2011, 170, 136–144 CrossRef CAS.
- N. Paxeus, Water Sci. Technol., 2004, 50, 253–260 CAS.
- A. Rodayan, R. Roy and V. Yargeau, J. Hazard. Mater., 2010, 177, 237–243 CrossRef CAS PubMed.
- T. Lin, S. Yu and W. Chen, Chemosphere, 2016, 152, 1–9 CrossRef CAS PubMed.
- B. Ricken, P. F. Corvini, D. Cichocka, M. Parisi, M. Lenz, D. Wyss, P. M. Martínez-Lavanchy, J. A. Müller, P. Shahgaldian and L. G. Tulli, Appl. Environ. Microbiol., 2013, 79, 5550–5558 CrossRef CAS PubMed.
- O. Koba, O. Golovko, R. Kodesova, M. Fer and R. Grabic, Environ. Pollut., 2017, 220, 1251–1263 CrossRef CAS PubMed.
- B. Li and T. Zhang, Environ. Sci. Technol., 2010, 44, 3468–3473 CrossRef CAS PubMed.
- W. D. Oh, Z. Dong, G. Ronn and T. T. Lim, J. Hazard. Mater., 2016, 325, 71–81 CrossRef PubMed.
- R. Yin, W. Guo, X. Zhou, H. Zheng, J. Du, Q. Wu, J. Chang and N. Ren, RSC Adv., 2016, 6, 19265–19270 RSC.
- F. W. Sifuna, F. Orata, V. Okello and S. Jemutai-Kimosop, J. Environ. Sci. Health, Part A: Toxic/Hazard. Subst. Environ. Eng., 2016, 51, 954–961 CrossRef CAS PubMed.
- C. Sheng, A. G. A. Nnanna, Y. Liu and J. D. Vargo, Sci. Total Environ., 2016, 550, 1075–1083 CrossRef CAS PubMed.
- Q. Song, H. Wang, B. Yang, F. Wang and X. Sun, RSC Adv., 2016, 6, 75855–75861 RSC.
- K. K. Shimabuku, J. P. Kearns, J. E. Martinez, R. B. Mahoney, L. Moreno-Vasquez and R. S. Summers, Water Res., 2016, 96, 236–245 CrossRef CAS PubMed.
- F. Bonvin, L. Jost, L. Randin, E. Bonvin and T. Kohn, Water Res., 2016, 90, 90–99 CrossRef CAS PubMed.
- C. Guo, J. Xu, Y. Zhang and Y. He, RSC Adv., 2012, 2, 4720 RSC.
- M. Majewsky, D. Wagner, M. Delay, S. Braese, V. Yargeau and H. Horn, Chem. Res. Toxicol., 2014, 27, 1821–1828 CrossRef CAS PubMed.
- M. Kobayashi, S. Kurosu, R. Yamaguchi and Y. Kawase, J. Environ. Manage., 2017, 200, 88 CrossRef CAS PubMed.
- J. Yang, Z. Li and H. Zhu, Appl. Catal., B, 2017, 217, 603–614 CrossRef CAS.
- Y. A. J. Al-Hamadani, C. M. Park, L. N. Assi, K. H. Chu, S. Hoque, M. Jang, Y. Yoon and P. Ziehl, Ultrason. Sonochem., 2017, 39, 354–362 CrossRef CAS PubMed.
- S. Wang and J. Wang, RSC Adv., 2017, 7, 48670–48677 RSC.
- M. G. Peleyeju, E. H. Umukoro, L. Tshwenya, R. Moutloali, J. O. Babalolac and O. A. Arotiba, RSC Adv., 2017, 7, 40571–40580 RSC.
- L. Wang, Y. C. Wu, Y. Zheng, L. D. Liu and F. Zhao, RSC Adv., 2015, 5, 56430–56437 RSC.
- S. Larcher and V. Yargeau, Appl. Microbiol. Biotechnol., 2011, 91, 211–218 CrossRef CAS PubMed.
- C. A. Vaz de Araujo, G. M. Maciel, E. A. Rodrigues, L. L. Silva, R. F. Oliveira, T. Brugnari, R. M. Peralta and C. G. Marques de Souza, Water, Air, Soil Pollut., 2017, 228, 341 CrossRef.
- Y. Xiao, H. Yaohari, C. D. Araujo, C. C. Sze and D. C. Stuckey, Chem. Eng. J., 2017, 321, 335–345 CrossRef CAS.
- Y. Jia, S. K. Khanal, H. Zhang, G.-H. Chen and H. Lu, Water Res., 2017, 119, 12–20 CrossRef CAS PubMed.
- S. Wang and J. Wang, Appl. Microbiol. Biotechnol., 2017, 1–8 Search PubMed.
- A. Banerjee and A. K. Ghoshal, Int. Biodeterior. Biodegrad., 2011, 65, 1052–1060 CrossRef CAS.
- I. del Castillo, P. Hernandez, A. Lafuente, I. D. Rodriguez-Llorente, M. A. Caviedes and E. Pajuelo, Water Res., 2012, 46, 1723–1734 CrossRef CAS PubMed.
- S. A. Ahmad, N. A. Shamaan, N. M. Arif, G. B. Koon, M. Y. A. Shukor and M. A. Syed, World J. Microbiol. Biotechnol., 2012, 28, 347–352 CrossRef CAS PubMed.
- G. Satchanska, Y. Topalova, R. Dimkov, V. Groudeva, P. Petrov, C. Tsvetanov, S. Selenska-Pobell and E. Golovinsky, Biotechnol. Biotechnol. Equip., 2015, 29, 514–521 CrossRef CAS.
- I. Krallish, S. Gonta, L. Savenkova, P. Bergauer and R. Margesin, Extremophiles, 2006, 10, 441–449 CrossRef CAS PubMed.
- X. Liao, B. Li, R. Zou, S. Xie and B. Yuan, Appl. Microbiol. Biotechnol., 2016, 100, 2439–2447 CrossRef CAS PubMed.
- Y. Liu, X. H. Liu, W. P. Dong, L. L. Zhang, Q. Kong and W. L. Wang, Sci. Rep., 2017, 7, 12 CrossRef PubMed.
- X. Bai and K. Acharya, J. Hazard. Mater., 2016, 315, 70–75 CrossRef CAS PubMed.
- F. I. Hai, X. Li, W. E. Price and L. D. Nghiem, Bioresour. Technol., 2011, 102, 10386–10390 CrossRef CAS PubMed.
- L. Wang, Y. Liu, J. Ma and F. Zhao, Water Res., 2016, 88, 322–328 CrossRef CAS PubMed.
- V. Martinez-Hernandez, R. Meffe, S. Herrera Lopez and I. de Bustamante, Sci. Total Environ., 2016, 559, 232–241 CrossRef CAS PubMed.
- F. Deng, C. Liao, C. Yang, C. Guo and Z. Dang, Int. Biodeterior. Biodegrad., 2016, 110, 46–52 CrossRef CAS.
- D. A. R. Mahmoud and W. A. Helmy, J. Appl. Sci. Res., 2009, 2466–2476 CAS.
- A. Dzionek, D. Wojcieszynska and U. Guzik, Electron. J. Biotechnol., 2016, 23, 28–36 CrossRef CAS.
- M. G. Fernandez-Lopez, C. Popoca-Ursino, E. Sanchez-Salinas, R. Tinoco-Valencia, J. L. Folch-Mallol, E. Dantan-Gonzalez and M. L. Ortiz-Hernandez, MicrobiologyOpen, 2017, 6, 12 CrossRef PubMed.
- K. Zhang, Y. Liu, H. Luo, Q. Chen, Z. Zhu, W. Chen, J. Chen, L. Ji and Y. Mo, Geoderma, 2017, 305, 264–274 CrossRef CAS.
- J. Y. Park and B. Huwe, Environ. Pollut., 2016, 213, 561–570 CrossRef CAS PubMed.
- H. Chen, B. Gao, H. Li and L. Q. Ma, J. Contam. Hydrol., 2011, 126, 29–36 CrossRef CAS PubMed.
- J. Iqbal, C. Metosh-Dickey and R. J. Portier, J. Soils Sediments, 2007, 7, 153–158 CrossRef CAS.
- P. N. Polymenakou and E. G. Stephanou, Biodegradation, 2005, 16, 403–413 CrossRef CAS PubMed.
- G. Bayramoglu and M. Y. Arica, Mater. Sci. Eng., C, 2009, 29, 1990–1997 CrossRef CAS.
- L. Lin, H. Wang and P. Xu, Chem. Eng. J., 2017, 310, 389–398 CrossRef CAS.
- X. Fan, C. Wang, P. Wang, J. Hou and J. Qian, Chem. Eng. J., 2017, 310, 317–327 CrossRef CAS.
- M. Moradi and G. Moussavi, Sep. Purif. Technol., 2018, 190, 90–99 CrossRef CAS.
- E. Muller, W. Schussler, H. Horn and H. Lemmer, Chemosphere, 2013, 92, 969–978 CrossRef CAS PubMed.
- S. Larcher and V. Yargeau, Appl. Microbiol. Biotechnol., 2012, 96, 309–318 CrossRef CAS PubMed.
- S. Yang and D. Che, RSC Adv., 2017, 7, 42233–42241 RSC.
- H. Xu, X. Li, Y. Sun, X. Shi and J. Wu, Water, Air, Soil Pollut., 2016, 227, 120 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra10150c |
| This journal is © The Royal Society of Chemistry 2017 |