Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Peperomin E and its synthetic amino derivatives: potent agents targeting leukaemia stem cells†
Ming Gao‡
a,
Xin-zhi Wang‡ *b,
Ye-ting Shua,
Jie Lianga,
Liang Chena,
Rui Liub,
Jing-yu Liangc and
Hong-mei Wen*a
*b,
Ye-ting Shua,
Jie Lianga,
Liang Chena,
Rui Liub,
Jing-yu Liangc and
Hong-mei Wen*a
aSchool of Pharmacy, Nanjing University of Chinese Medicines, Xianlin Avenue No. 138, Nanjing 210023, The People's Republic of China. E-mail: njwenhm@126.com; Fax: +86-025-858-11839; Tel: +86-025-858-11839
bMarine Drug Research and Development Center of Jiangsu Province, Nanjing University of Chinese Medicines, Xianlin Avenue No. 138, Nanjing 210023, The People's Republic of China. E-mail: wxzatnj@sina.com
cDepartment of Natural Medicinal Chemistry, China Pharmaceutical University, Tongjia Lane No. 24, 210009 Nanjing, The People's Republic of China
First published on 10th November 2017
Abstract
Recent studies have described leukaemia stem cells (LSCs) as being central to the initiation, growth, and relapse of acute myelogenous leukaemia (AML). However, to date, very few small molecules have been shown to directly target this LSC population. The present study, for the first time, demonstrates that peperomin E (PepE), a naturally occurring secolignan, induces apoptosis in cultured AML and LSC-like cell lines, while sparing normal bone marrow stromal cells (hBMSCs). Furthermore, a series of eighteen amino derivatives was synthesised through a diastereoselective conjugate addition of several primary and secondary amines to the α-methylene-γ-butyrolactone moiety of PepE to increase its water-solubility and anti-AML/LSC activity. The synthesised amino analogues were evaluated for in vitro cytotoxicity against AML cell lines (KG-1, HL-60, THP-1) and an LSC-like cell line (KG-1a CD34+CD38−). Most of the secondary open-chain amine derivatives showed enhanced water solubility and high activities against AML/LSC cells. The most potent compound, 6, derived from N-methylethanolamine, exhibited exclusive cytotoxicity against KG-1a CD34+CD38− cells, with an IC50 value of 0.5 ± 0.1 μM, which was 13-fold more potent than PepE. Moreover, compound 6 showed low toxicity against hBMSCs (inhibition of 11.2% at 50 μM). Preliminary mechanistic studies revealed that compound 6 could induce apoptosis of KG-1a CD34+CD38− cells. Therefore, we propose that compound 6 should be tested further for its in vivo activity and safety with the aim to develop it as a new type of drug for LSC-targeted therapy.
Introduction
Acute myelogenous leukaemia (AML) is the most common leukaemia, characterised by the clonal expansion of immature myeloblasts initiating from rare leukemic stem cells (LSCs).1,2 As one of the most common types of lethal leukaemia, AML typically leads to death within weeks to months after clinical presentation. Current chemotherapy for AML commonly uses drugs, such as nucleoside analogues (e.g., cytarabine) and anthracyclines (e.g., idarubicin), which interfere with DNA replication and induce apoptosis primarily in replicating cells.3 Although remission can be achieved in most patients by standard chemotherapy, relapse is common and long-term survival is poor for most cases.2 It has been reported that LSCs are refractory to standard chemotherapy and thereby contribute to relapse. However, there are few compounds that can selectively inhibit LSCs.4 Therefore, novel therapeutic agents with selective activity against LSCs are urgently needed.Peperomin E (PepE) is a naturally occurring secolignan, isolated from Peperomia dindygulensis (Piperaceae), a commonly used folk medicine to treat cancers in southern China.5 Although PepE was first to be extracted from nature plants, it can also be fully synthesized from phosphorylated cyclopropanes, which broadens its further study.6 There has been increasing interest in this compound, due to its remarkable activity against multiple cancer cells in vitro and in vivo with low toxicity against normal cells.7,8 The presence of the α-methylene-γ-butyrolactone moiety in this compound was reported to be essential for its antitumour properties.7 The exo-methylene moiety is considered important because of its reactivity toward biological nucleophiles, especially the thiol groups of cysteines in target proteins, which act as Michael acceptors.9 Recently, our laboratory discovered that PepE could exceptionally target and initiate apoptosis in AML cells (e.g., KG-1 cells) and AML stem cells (e.g., KG-1a CD34+CD38− cells), with high survival rates for normal bone marrow stromal cells (hBMSCs). Studies to elucidate the anti-AML mechanism of this promising agent are underway in our laboratory; however, preliminary experiments have suggested that NF-κB and DNA methyltransferases are the main targets of this compound (unpublished results).
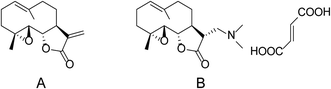
Although PepE has shown potent activity against AML, its naked α-methylene-γ-butyrolactone substructure is unstable in both acidic and basic conditions and imparts poor water solubility and bioavailability;10 these issues have limited its potential therapeutic use in humans. Converting the α-methylene-γ-butyrolactone moiety to its prodrug with a masking group, such as a primary or secondary amine, effectively increases its stability, water solubility, and bioavailability, while maintaining or even enhancing its biological activity.11–15 Amino adducts of α-methylene-γ-butyrolactone have been previously prepared from sesquiterpene lactones, including helenalin, ambrosin, costunolide, alantolactone, and parthenolide (PTL). Among them, the dimethylamino analogue of PTL (DMAPT, Fig. 1) has advanced to a phase I clinical trial for evaluation as a treatment for AML.11,16–20
Therefore, the aim of the present study was to overcome the poor water solubility of PepE, while at least maintaining its activity, by derivatizing it with several primary and secondary amino compounds. These analogues were then converted into water-soluble organic/inorganic salts and assessed for their cytotoxicity against AML cells and LSCs. To the best of our knowledge, there is no published record available concerning the anti-AML or anti-LSC effects of PepE and its amino analogues.
Experimental
Chemistry
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc = 3
EtOAc = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
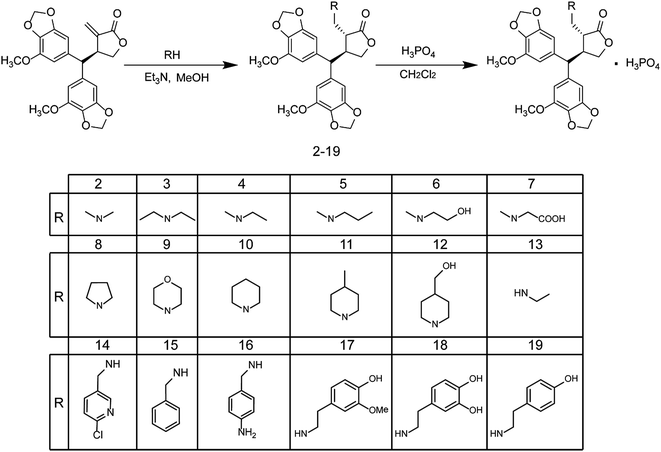
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1] to yield the desired compounds 2–19. Purity of the compounds was established by HPLC analysis, confirming a purity ≥95%. The characterisation data for these compounds are listed below.
1] to yield the desired compounds 2–19. Purity of the compounds was established by HPLC analysis, confirming a purity ≥95%. The characterisation data for these compounds are listed below.
(2S,3S)-6-Dimethylamino-2,6-dihydropeperomin E (2). Pale yellow oil (yield 85%); 98.5% HPLC purity; IR (thin film) νmax 2904.94, 1758.52, 1633.46, 1508.44, 1450.62, 1431.89, 1130.91, 1091.96, 1042.72, 928.07, and 823.67 cm−1; 1H NMR (500 MHz, CDCl3) δH 6.56 (d, J = 2.0 Hz, 1H), 6.51 (d, J = 1.5 Hz, 1H), 6.47 (d, J = 2.0 Hz, 1H), 6.43 (d, J = 1.5 Hz, 1H), 5.93–5.96 (m, 4H), 4.36 (dd, J = 9.0, 12.5 Hz, 1H), 4.01 (dd, J = 4.5, 12.5 Hz, 1H), 3.93 (s, 3H), 3.91 (3H, s), 3.62 (d, J = 15.5 Hz, 1H), 3.36–3.43 (m, 1H), 2.53 (dd, J = 9.5, 15.5 Hz, 1H), 2.42–2.46 (m, 1H), 2.20 (dd, J = 6.5, 15.5 Hz, 1H) and 2.10 (s, 6H) ppm; 13C NMR (125 MHz, CDCl3) δC 178.7, 149.5, 149.2, 143.6, 143.5, 136.6, 136.9, 134.2, 134.1, 108.1, 108.0, 101.9, 101.5 (×2), 101.3, 70.6, 58.1, 57.0, 56.8, 55.4, 45.8 (×2), 44.7 and 41.4 ppm; HRMS-ESI calcd for C24H28NO8 [M + H]+ 458.1815, found 458.1797.
(2S,3S)-6-Diethylamino-2,6-dihydropeperomin E (3). Pale yellow oil (yield 87%); 97.0% HPLC purity; IR (thin film) νmax 2919.07, 1763.85, 1633.47, 1508.71, 1450.46, 1432.51, 1132.31, 1091.86, 1042.90, 926.56, and 818.35 cm−1; 1H NMR (500 MHz, CDCl3) δH 6.54 (d, J = 1.5 Hz, 1H), 6.48 (d, J = 2.0 Hz, 1H), 6.42 (d, J = 1.5 Hz, 2H), 5.93–5.96 (m, 4H), 4.36 (dd, J = 8.5, 12.0 Hz, 1H), 4.05 (dd, J = 3.5, 12.0 Hz, 1H), 3.92 (s, 3H), 3.91 (s, 3H), 3.65 (d, J = 15.5 Hz, 1H), 3.27–3.31 (m, 1H), 2.69 (dd, J = 12.0, 18.0 Hz, 1H), 2.42–2.54 (m, 4H), 2.39 (dd, J = 9.0, 16.5 Hz, 1H), and 0.98 (t, J = 9.5 Hz, 6H) ppm; 13C NMR (125 MHz, CDCl3) δC 178.9, 149.5, 149.2, 143.6, 143.5, 137.1, 136.6, 134.2, 134.1, 108.3, 108.2, 101.8, 101.5, 101.4, 101.3, 70.6, 60.3, 56.9, 56.8, 56.7, 55.1, 44.7, 41.9, 41.8, 20.3 and 11.8 ppm; HRMS-ESI calcd for C26H32NO8 [M + H]+ 486.2128, found 486.2110.
(2S,3S)-6-Ethyl(methyl)amino-2,6-dihydropeperomin E (4). Pale yellow oil (yield 81%); 100.0% HPLC purity; IR (thin film) νmax 2920.97, 1767.51, 1633.26, 1508.64, 1450.71, 1432.08, 1131.95, 1091.38, 1042.65, 927.23, and 827.68 cm−1; 1H NMR (500 MHz, CDCl3) δH 6.56 (d, J = 2.0 Hz, 1H), 6.50 (d, J = 2.0 Hz, 1H), 6.44 (d, J = 1.5 Hz, 2H), 6.42 (d, J = 2.0 Hz, 2H), 5.93–5.97 (m, 4H), 4.37 (dd, J = 9.0, 12.0 Hz, 1H), 4.04 (dd, J = 3.0, 12.0 Hz, 1H), 3.92 (s, 3H), 3.91 (s, 3H), 3.63 (d, J = 15.5 Hz, 1H), 3.30–3.37 (m, 1H), 2.62 (dd, J = 10.5, 16.0 Hz, 1H), 2.44–2.50 (m, 2H), 2.31–2.36 (m, 2H), 2.05 (s, 3H), and 1.03 (t, J = 8.0 Hz, 3H) ppm; 13C NMR (125 MHz, CDCl3) δC 179.0, 149.4, 149.2, 143.6, 143.5, 137.1, 136.6, 134.2, 134.1, 108.4, 108.3, 101.7, 101.5, 101.4, 101.2, 70.6, 57.0, 56.9, 56.8, 54.7, 52.7, 44.9, 42.2, and 11.3 (×2) ppm; HRMS-ESI calcd for C25H30NO8 [M + H]+ 472.1971, found 472.1949.
(2S,3S)-6-Methyl(propyl)amino-2,6-dihydropeperomin E (5). Pale yellow oil (yield 78%); 99.0% HPLC purity; IR (thin film) νmax 2960.34, 1770.08, 1633.32, 1508.85, 1451.98, 1432.89, 1132.41, 1092.48, 1044.01, 929.64, and 828.77 cm−1; 1H NMR (500 MHz, CDCl3) δH 6.55 (d, J = 2.0 Hz, 1H), 6.48 (d, J = 2.0 Hz, 1H), 6.44 (d, J = 2.0 Hz, 1H), 6.42 (d, J = 2.0 Hz, 1H), 5.93–5.96 (m, 4H), 4.36 (dd, J = 9.0, 12.0 Hz, 1H), 4.02 (dd, J = 4.0, 12.0 Hz, 1H), 3.92 (s, 3H), 3.91 (s, 3H), 3.62 (d, J = 15.5 Hz, 1H), 3.36–3.39 (m, 1H), 2.62 (dd, J = 10.0, 15.5 Hz, 1H), 2.46–2.49 (m, 1H), 2.29 (dd, J = 6.5, 16.0 Hz, 1H), 2.24 (t, J = 9.5 Hz, 2H), 2.03 (s, 3H), 1.43–1.50 (m, 2H), 0.92 (t, 3H, J = 9.0 Hz); 13C NMR (125 MHz, CDCl3) δC 178.9, 149.5, 149.2, 143.6, 143.5, 137.1, 136.6, 134.2, 134.1, 108.3, 108.2, 101.8, 101.5, 101.4, 101.3, 70.6, 60.3, 56.9, 56.8, 56.7, 55.1, 44.7, 41.8, 41.7, 20.3, 11.8 ppm; HRMS-ESI calcd for C26H32NO8 [M + H]+ 486.2128, found 486.2102.
(2S,3S)-6-Methyl(hydroxyethyl)amino-2,6-dihydropeperomin E (6). Colourless powder (yield 82%); 100.0% HPLC purity; IR (thin film) νmax 3445.62, 2917.98, 1767.13, 1633.65, 1507.61, 1451.75, 1435.25, 1192.73, 1135.07, 1090.75, 1042.05, 927.53, and 852.33 cm−1; 1H NMR (500 MHz, CDCl3) δH 6.50 (d, J = 2.0 Hz, 1H), 6.48 (d, J = 2.0 Hz, 1H), 6.44 (d, J = 2.0 Hz, 1H), 6.41 (d, J = 2.0 Hz, 1H), 5.95–5.97 (m, 4H), 4.37 (dd, J = 9.0, 12.0 Hz, 1H), 4.00 (dd, J = 5.5, 12.0 Hz, 1H), 3.92 (s, 6H), 3.64 (d, J = 15.5 Hz, 1H), 3.62–3.58 (m, 2H), 3.14–3.20 (m, 1H), 2.54–2.64 (m, 3H), 2.42–2.53 (m, 2H), 2.08 (s, 3H); 13C NMR (125 MHz, CDCl3) δC 178.5, 149.5, 149.3, 143.7, 143.6, 136.6, 136.4, 134.3 (×2), 108.2, 108.1, 101.6, 101.5, 101.2 (×2), 70.5, 59.5, 58.9, 57.4, 56.9 (×2), 55.4, 44.4, 42.4, and 41.7 ppm; HRMS-ESI calcd for C25H30NO9 [M + H]+ 488.1921, found 488.1890.
(2S,3S)-6-Methylglycine-2,6-dihydropeperomin E (7). Pale yellow oil (yield 64%); 98.5% HPLC purity; IR (thin film) νmax 3420.91, 2917.98, 1767.13, 1633.65, 1507.61, 1451.75, 1435.25, 1192.73, 1135.07, 1090.75, 1042.05, and 928.05 cm−1; 1H NMR (500 MHz, CDCl3) δH 6.48 (d, J = 2.0 Hz, 1H), 6.47 (d, J = 2.0 Hz, 1H), 6.40 (d, J = 1.5 Hz, 1H), 6.28 (d, J = 1.5 Hz, 1H), 5.96–5.97 (m, 4H), 4.35 (dd, J = 9.0, 12.0 Hz, 1H), 4.01 (dd, J = 5.5, 12.0 Hz, 1H), 3.92 (s, 3H), 3.91 (s, 3H), 3.68 (d, J = 14.5 Hz, 1H), 3.50–3.59 (m, 1H), 3.09 (q, J = 9.5 Hz, 2H), 2.51 (dd, J = 9.5, 14.5 Hz, 1H), 2.18–2.40 (m, 2H), 2.06 (s, 3H) ppm; 13C NMR (125 MHz, CDCl3) δC 174.0, 170.7, 149.6, 149.3, 143.7, 143.4, 136.1 (×2), 135.9, 108.5, 108.1, 101.6, 101.5 (×2), 101.2, 69.7, 60.7, 60.4, 57.0, 56.9, 55.3, 45.8, 45.6 (×2), and 42.5 ppm; HRMS-ESI calcd for C25H28NO10 [M + H]+ 502.1718, found 502.1720.
(2S,3S)-6-Pyrrolidin-2,6-dihydropeperomin E (8). Pale yellow oil (yield 89%); 99.5% HPLC purity; IR (thin film) νmax 2926.35, 2789.59, 1632.74, 1609.01, 1507.80, 1449.91, 1431.09, 1128.95, 1089.54, 1043.00, 927.88, and 828.35 cm−1; 1H NMR (500 MHz, CDCl3) δH 6.59 (d, J = 2.0 Hz, 1H), 6.57 (d, J = 1.5 Hz, 1H), 6.54 (d, J = 1.5 Hz, 1H), 6.45 (d, J = 1.5 Hz, 1H), 5.89–5.95 (m, 4H), 4.01 (dd, J = 3.5, 12 Hz, 1H), 3.90 (s, 6H), 3.89 (d, J = 15.5 Hz, 1H), 3.52 (dd, J = 4.5, 15.5 Hz, 1H), 3.41–3.48 (m, 1H), 2.47–2.56 (m, 3H), 2.39–2.46 (m, 3H), 2.05–2.13 (m, 1H), 1.94–1.98 (m, 2H), and 1.88–1.92 (m, 2H) ppm; 13C NMR (125 MHz, CDCl3) δC 173.1, 149.2, 148.9, 143.7, 143.4, 138.5, 138.2, 133.7, 133.6, 108.3, 107.7, 101.7, 101.4, 101.2, 101.1, 71.2, 58.2, 57.0, 56.7 (×2), 54.3 (×2), 44.3, 43.7, 23.6 (×2) ppm; HRMS-ESI calcd for C26H30NO8 [M + H]+ 484.1971, found 484.1964.
(2S,3S)-6-Morpholin-2,6-dihydropeperomin E (9). Pale yellow oil (yield 82%); 98.0% HPLC purity; IR (thin film) νmax 2921.57, 1759.13, 1636.71, 1508.44, 1449.62, 1436.61, 1118.32, 1092.17, 1045.22, and 925.09 cm−1; 1H NMR (500 MHz, CDCl3) δH 6.56 (d, J = 2.0 Hz, 1H), 6.50 (d, J = 2.0 Hz, 1H), 6.41 (d, J = 2.0 Hz, 2H), 5.92–5.97 (m, 4H), 4.36 (dd, J = 9.0, 12.0 Hz, 1H), 4.03 (d, J = 4.5, 12.0 Hz, 1H), 3.93 (s, 3H), 3.91 (s, 3H), 3.63–3.69 (m, 4H), 3.61 (d, J = 15.0 Hz, 1H), 3.33–3.40 (m, 1H), 2.59 (dd, J = 10.5, 15.5 Hz, 1H), 2.51–2.62 (m, 1H), 2.32–2.51 (m, 3H), and 2.20–2.26 (m, 2H); 13C NMR (125 MHz, CDCl3) δC 178.5, 149.5, 149.2, 143.6, 143.4, 136.8, 136.6, 134.2, 134.1, 108.7, 108.3, 101.5, 101.4 (×2), 101.1, 70.6, 67.0, 66.8, 57.2, 57.0, 56.9, 55.0, 52.0, 43.9, and 41.8 ppm; HRMS-ESI calcd for C26H30NO9 [M + H]+ 500.1921, found 500.1895.
(2S,3S)-6-Piperidin-2,6-dihydropeperomin E (10). Pale yellow oil (yield 85%); 99.5% HPLC purity; IR (thin film) νmax 2922.04, 2851, 1767.84, 1633.33, 1508.21, 1450.80, 1431.92, 1132.96, 1090.35, 1043.20, 927.59, and 851.45 cm−1; 1H NMR (500 MHz, CDCl3) δH 6.58 (d, J = 2.0 Hz, 1H), 6.51 (d, J = 2.0 Hz, 1H), 6.42 (d, J = 2.5 Hz, 2H), 5.93–5.99 (m, 4H), 4.36 (dd, J = 8.5, 11.5 Hz, 1H), 4.02 (d, J = 6.0, 12.5 Hz, 1H), 3.93 (s, 3H), 3.91 (s, 3H), 3.61 (d, J = 15.5 Hz, 1H), 3.35–3.39 (m, 1H), 3.52 (dd, J = 4.5, 15.5 Hz, 1H), 2.56 (dd, J = 10.5, 15.5 Hz, 1H), 2.48–2.51 (m, 1H), 2.22–2.47 (m, 3H), 2.12–2.16 (m, 1H), 1.52–1.57 (m, 4H), and 1.40–1.45 (m, 2H) ppm; 13C NMR (125 MHz, CDCl3) δC 179.1, 149.4, 149.2, 143.6, 143.4, 137.1, 136.7, 134.2, 134.1, 108.4, 108.2, 101.5, 101.4 (×2), 101.2, 70.7, 57.5, 57.0, 56.8, 55.0, 54.8, 44.2, 41.8, 29.7 (×2), 22.7 ppm; HRMS-ESI calcd for C26H30NO9 [M + H]+ 498.2128, found 498.2119.
(2S,3S)-6-(9-Methylpiperidin)-2,6-dihydropeperomin E (11). Pale yellow oil (yield 78%); 97.0% HPLC purity; IR (thin film) νmax 2922.87, 1763.10, 1633.91, 1509.18, 1450.56, 1433.50, 1131.33, 1090.99, 1043.45, 926.68, and 847.34 cm−1; 1H NMR (500 MHz, CDCl3) δH 6.56 (d, J = 2.0 Hz, 1H), 6.51 (d, J = 1.5 Hz, 1H), 6.42 (d, J = 1.5 Hz, 1H), 6.41 (d, J = 2.0 Hz, 1H), 5.93–5.96 (m, 4H), 4.35 (dd, J = 8.5, 12.0 Hz, 1H), 4.03 (dd, J = 4.5, 13.5 Hz, 1H), 3.93 (s, 3H), 3.91 (s, 3H), 3.61 (d, J = 15.5 Hz, 1H), 3.39–3.41 (m, 1H), 2.57–2.66 (m, 1H), 2.51 (dd, J = 15.5, 10.0 Hz, 1H), 2.46–2.49 (m, 1H), 2.30–2.34 (m, 1H), 2.24 (dd, J = 15.5, 6.0 Hz, 1H), 1.98–2.03 (m, 1H), 1.81–1.84 (m, 1H), 1.61–1.63 (m, 1H), 1.45–1.48 (m, 1H), 1.19–1.25 (m, 2H), and 0.93 (d, J = 8.0 Hz, 3H) ppm; 13C NMR (125 MHz, CDCl3) δC 179.1, 149.4, 149.2, 143.6, 143.4, 137.1, 136.7, 134.1 (×2), 108.4, 108.3, 101.7, 101.6, 101.5, 101.4, 70.7, 60.4, 57.0, 56.9, 55.1, 54.9, 53.6, 44.4, 41.7, 34.1, 30.5, 21.7, 14.2 ppm; HRMS-ESI calcd for C28H34NO8 [M + H]+ 512.2284, found 512.2270.
(2S,3S)-6-(9-Hydroxymethylpiperidin)-2,6-dihydropeperomin E (12). Pale yellow oil (yield 73%); 96.4% HPLC purity; IR (thin film) νmax 3430.79, 2919.26, 1767.42, 1633.44, 1508.59, 1450.63, 1432.37, 1315.59, 1134.16, 1090.66, 1042.21, 925.88, and 828.06 cm−1; 1H NMR (500 MHz, CDCl3) 6.56 (d, J = 2.0 Hz, 1H), 6.50 (d, J = 2.0 Hz, 1H), 6.41 (d, J = 2.0 Hz, 2H), 5.92–5.96 (m, 4H), 4.35 (dd, J = 8.5, 12.0 Hz, 1H), 4.01 (dd, J = 4.5, 12.0 Hz, 1H), 3.93 (s, 3H), 3.91 (s, 3H), 3.61 (d, J = 15.5 Hz, 1H), 3.52 (d, J = 8.0 Hz, 2H), 3.36–3.38 (m, 1H), 2.61–2.72 (m, 1H), 2.57 (dd, J = 15.6, 10.0 Hz, 1H), 2.41–2.45 (m, 1H), 2.35–2.40 (m, 1H), 2.24 (dd, J = 15.6, 5.8 Hz, 1H), 2.00–2.06 (m, 1H), 1.74–1.80 (m, 1H), 1.72–1.74 (m, 2H), 1.59–1.63 (m, 2H), 1.63–1.66 (m, 2H), 1.44–1.47 (m, 1H); 13C NMR (125 MHz, CDCl3) δC 179.0, 149.5, 149.2, 143.6, 143.4, 137.0, 136.7, 134.2, 134.1, 108.5, 108.3, 101.5 (×2), 101.4, 101.2, 70.7, 67.8, 57.1, 57.0, 56.9, 55.1, 54.5, 53.2, 44.4, 41.7, 38.3, 29.0, and 28.5 ppm; HRMS-ESI calcd for C28H34NO9 [M + H]+ 528.2234, found 528.2213.
(2S,3S)-6-Ethylamino-2,6-dihydropeperomin E (13). Pale yellow oil (yield 65%); 98.0% HPLC purity; IR (thin film) 3444.34, 2910.17, 1765.11, 1631.23, 1501.60, 1451.70, 1431.11, 1120.21, 1031.18, 1040.15, 920.20, and 825.12 cm−1; 1H NMR (500 MHz, CDCl3) δH 6.55 (d, J = 2.0 Hz, 1H), 6.49 (d, J = 1.5 Hz, 1H), 6.44 (d, J = 1.5 Hz, 1H), 6.42 (d, J = 2.0 Hz, 1H), 5.92–5.96 (m, 4H), 4.36 (dd, J = 8.5, 12.0 Hz, 1H), 4.01 (dd, J = 4.5, 12.0 Hz, 1H), 3.91 (s, 3H), 3.89 (s, 3H), 3.62 (d, J = 15.0 Hz, 1H), 3.33–3.37 (m, 1H), 2.61 (dd, J = 10.0, 15.5 Hz, 1H), 2.43–2.53 (m, 2H), 1.02 (t, J = 8.5 Hz, 3H); 13C NMR (125 MHz, CDCl3) δC 179.0, 149.4, 149.2, 143.6, 143.4, 137.0, 136.6, 134.2, 134.1, 108.1 (×2), 101.8, 101.6, 101.5, 101.4, 70.6, 56.9, 56.4, 56.8, 55.0, 44.7, 42.1, 41.5, and 12.1 ppm; HRMS-ESI calcd for C24H28NO8 [M + H]+ 458.1815, found 458.1820.
(2S,3S)-6-(4′′′-Chloropyridinemethylamino)-2,6-dihydropeperomin E (14). Pale yellow oil (yield 79%); 100.0% HPLC purity; IR (thin film) νmax 3441.55, 2902.33, 1765.23, 1633.21, 1507.99, 1452.10, 1432.43, 1315.90, 1042.06, 927.21, and 826.15 cm−1; 1H NMR (500 MHz, CDCl3) 8.26 (d, J = 2.7 Hz, 1H), 7.59 (dd, J = 2.7, 10.2 Hz, 1H), 7.31 (d, J = 10.2 Hz, 1H), 6.48 (d, J = 2.0 Hz, 1H), 6.42 (d, J = 2.0 Hz, 1H), 6.40 (d, J = 2.0 Hz, 2H), 5.92–5.95 (m, 4H), 4.35 (dd, J = 9.0, 12.0 Hz, 1H), 3.92 (s, 3H), 3.89 (s, 3H), 3.87 (dd, J = 4.0, 12.0 Hz, 1H), 3.59–3.64 (m, 3H), 3.23–3.28 (m, 1H), 2.67 (dd, J = 5.5, 15.2 Hz, 1H), 2.52–2.55 (m, 1H), and 2.30 (dd, J = 7.5, 15.2 Hz, 1H) ppm; 13C NMR (125 MHz, CDCl3) δC 175.5, 149.1, 149.0, 143.6, 143.5, 139.5, 138.9, 138.3, 133.9, 133.8 (×2), 128.7, 128.6, 128.0, 127.8, 107.8, 107.7, 101.6, 101.4, 101.3, 101.2, 70.9, 56.9, 56.8, 53.7, 51.6, 45.8, and 42.2 ppm; HRMS-ESI calcd for C28H28ClN2O8 [M + H]+ 555.1534, found 555.1508.
(2S,3S)-6-Benzylamino-2,6-dihydropeperomin E (15). Pale yellow oil (yield 87%); 96.4% HPLC purity; IR (thin film) νmax 3404.80, 2922.65, 1762.13, 1633.55, 1508.00, 1451.40, 1431.05, 1313.60, 1194.09, 1130.23, 1089.83, 1043.31, 928.59, and 828.26 cm−1; 1H NMR (500 MHz, CDCl3) 7.31–7.37 (m, 4H), 7.20–7.27 (m, 1H), 6.53 (d, J = 2.0 Hz, 1H), 6.49 (d, J = 2.0 Hz, 1H), 6.44 (d, J = 2.0 Hz, 1H), 6.40 (d, J = 2.0 Hz, 1H), 5.90–5.94 (m, 4H), 4.38 (dd, J = 8.5, 12.0 Hz, 1H), 4.00 (dd, J = 4.5, 12.0 Hz, 1H), 3.91 (s, 3H), 3.90 (s, 3H), 3.80 (d, J = 15.5 Hz, 1H), 3.72–3.75 (m, 1H), 3.59–3.63 (m, 1H), 3.36–3.39 (m, 1H), 3.16–3.18 (m, 1H), 2.95–2.98 (m, 1H), and 2.46–2.48 (m, 1H) ppm; 13C NMR (125 MHz, CDCl3) δC 178.6, 149.5, 149.4, 146.5, 144.0, 143.7, 143.6, 136.8, 136.4, 134.3, 131.8, 121.3, 114.4, 111.3, 107.9, 101.5, 101.3 (×2), 101.1, 70.9, 57.0, 56.9, 56.1, 55.9, 51.4, 48.0, 45.9, 41.6, and 35.9 ppm; HRMS-ESI calcd for C29H30NO8 [M + H]+ 520.1971, found 520.1952.
(2S,3S)-6-(4′′′-Aminobenzylamino)-2,6-dihydropeperomin E (16). Pale yellow oil (yield 72%); 99.0% HPLC purity; IR (thin film) νmax 3435.85, 2924.67, 2793.68, 1770.11, 1622.13, 1508.11, 1451.41, 1431.95, 1315.88, 1042.77, 928.64, and 814.32 cm−1; 1H NMR (500 MHz, CDCl3) 7.35 (d, J = 8.5 Hz, 2H), 7.29 (d, J = 8.5 Hz, 2H), 6.44 (d, J = 2.0 Hz, 1H), 6.41 (d, J = 2.0 Hz, 2H), 6.38 (d, J = 2.0 Hz, 1H), 5.86–5.93 (m, 4H), 4.29 (dd, J = 8.5, 12.0 Hz, 1H), 4.07 (dd, J = 4.5, 12.0 Hz, 1H), 3.89 (s, 3H), 3.83 (s, 3H), 3.59 (d, J = 15.5 Hz, 1H), 3.40–3.51 (m, 2H), 3.12–3.17 (m, 1H), 2.64 (dd, J = 5.7, 15.5 Hz, 1H), 2.42–2.45 (m, 1H), and 2.31–2.34 (m, 1H) ppm; 13C NMR (125 MHz, CDCl3) δC 178.8, 149.4, 149.2, 143.6, 143.4, 138.9, 137.0, 136.7, 134.1 (×2), 129.2, 128.4, 127.4, 108.1 (×2), 101.7, 101.5, 101.4, 101.2, 70.6, 62.9, 56.9, 56.8, 55.6, 54.7, 44.6, 42.6, 42.1 ppm; HRMS-ESI calcd for C29H30N2O8 [M]+ 534.2002, found 534.2092.
(2S,3S)-6-(4′′′-Hydroxy-5′′′-methoxyphenethylamino)-2,6-dihydropeperomin E (17). Pale yellow oil (yield 80%); 98.4% HPLC purity; IR (thin film) νmax 3441.18, 2919.51, 1762.85, 1633.20, 1514.00, 1450.68, 1431.22, 1315.85, 1131.42, 1091.13, 1041.34, 926.12, and 819.81 cm−1; 1H NMR (500 MHz, CDCl3) 6.85 (d, J = 8.5 Hz, 1H), 6.71 (d, J = 2.0 Hz, 1H), 6.69 (dd, J = 2.0, 8.5 Hz, 1H), 6.47 (d, J = 2.0 Hz, 2H), 6.46 (d, J = 2.0 Hz, 1H), 6.43 (d, J = 2.0 Hz, 1H), 5.92–5.96 (m, 4H), 4.30 (dd, J = 10.0, 12.0 Hz, 1H), 3.92 (s, 3H), 3.90 (s, 3H), 3.89 (s, 3H), 3.87 (dd, J = 4.5, 12.0 Hz, 1H), 3.61 (d, J = 14.5 Hz, 1H), 3.24–3.27 (m, 1H), 2.80 (dd, J = 10.0, 15.5 Hz, 1H), 2.72–2.76 (m, 1H), 2.64–2.67 (m, 2H), 2.48–2.51 (m, 2H), and 2.29 (dd, J = 7.0, 15.5 Hz, 1H) ppm; 13C NMR (125 MHz, CDCl3) δC 178.7, 154.2, 149.4 (×2), 143.6 (×2), 136.7, 136.4, 134.3, 131.9, 130.9, 129.8 (×2), 115.5 (×2), 108.1, 107.9, 101.5, 101.3, 101.2 (×2), 70.9, 57.1 (×2), 56.9, 56.1, 51.2, 48.0, 45.9, 41.5, 35.3 ppm; HRMS-ESI calcd for C31H34NO10 [M + H]+ 580.2183, found 580.2151.
(2S,3S)-6-(4′′′,5′′′-Dihydroxyphenethylamino)-2,6-dihydropeperomin E (18). Pale yellow oil (yield 75%); 99.0% HPLC purity; IR (thin film) νmax 3451.21, 2916.40, 1761.75, 1632.11, 1513.21, 1448.38, 1429.12, 1305.87, 1130.12, 1071.03, 1031.54, 925.31, and 816.51 cm−1; 1H NMR (500 MHz, CDCl3) 6.85 (d, J = 2.0 Hz, 1H), 6.70 (d, J = 8.3 Hz, 1H), 6.69 (d, J = 2.0 Hz, 1H), 6.68 (dd, J = 2.0, 8.3 Hz, 1H), 6.47 (d, J = 2.0 Hz, 2H), 6.46 (d, J = 2.0 Hz, 1H), 6.43 (d, J = 2.0 Hz, 1H), 5.92–5.96 (m, 4H), 4.29 (dd, J = 10.0, 12.0 Hz, 1H), 3.90 (s, 3H), 3.89 (s, 3H), 3.85 (dd, J = 4.5, 12.0 Hz, 1H), 3.60 (d, J = 15.0 Hz, 1H), 3.23–3.27 (m, 1H), 2.79 (dd, J = 10.0, 15.5 Hz, 1H), 2.71–2.74 (m, 1H), 2.60–2.65 (m, 3H), 2.49–2.52 (m, 1H), and 2.30 (dd, J = 7.0, 15.5 Hz, 1H) ppm; 13C NMR (125 MHz, CDCl3) δC 178.0, 149.4, 149.3, 146.4, 144.0, 143.6, 143.5, 136.8, 136.4, 134.2 (×2), 131.7, 121.2, 114.3 (×2), 111.2, 107.8, 101.4, 101.2, 101.1 (×2), 70.8, 56.9, 56.8, 55.9, 51.3, 48.0, 45.9, 41.6, 35.8 ppm; HRMS-ESI calcd for C30H32NO10 [M + H]+ 566.2026, found 566.2024.
(2S,3S)-6-(4′′′-Hydroxyphenethylamino)-2,6-dihydropeperomin E (19). Pale yellow oil (yield 75%); 98.5% HPLC purity; IR (thin film) νmax 3420.78, 2926.96, 1762.35, 1633.09, 1513.83, 1450.58, 1432.88, 1316.05, 1132.70, 1091.71, 1042.23, 926.38, and 827.66 cm−1; 1H NMR (500 MHz, CDCl3) 7.03 (s, 1H), 6.79 (s, 1H), 6.77 (s, 1H), 6.74 (s, 1H), 6.45 (d, J = 1.5 Hz, 1H), 6.41 (d, J = 2.0 Hz, 2H), 6.39 (d, J = 2.0 Hz, 1H), 5.92–5.97 (m, 4H), 4.28 (dd, J = 10.0, 12.0 Hz, 1H), 3.92 (s, 3H), 3.89 (s, 3H), 3.83 (dd, J = 8.5, 12.0 Hz, 1H), 3.58 (d, J = 14.5 Hz, 1H), 3.18–3.23 (m, 1H), 2.73–2.80 (m, 2H), 2.52–2.60 (m, 3H), 2.45–2.48 (m, 1H), and 2.26 (dd, J = 7.0, 15.5 Hz, 1H) ppm; 13C NMR (125 MHz, CDCl3) δC 178.4, 150.2, 149.6, 149.4, 149.3, 143.7, 143.5, 138.5, 136.6, 136.1, 136.0, 134.4, 134.3, 134.2, 124.1 (×2), 108.0, 107.8, 101.6, 101.5, 101.1 (×2), 71.0, 57.0 (×2), 56.2, 50.2, 48.0, 45.9, and 42.0 ppm; HRMS-ESI calcd for C30H32NO9 [M + H]+ 550.2077, found 550.2049.
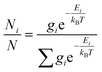 | (1) |
The theoretical calculation was performed using Gaussian 09 (Revision D.01. Gaussian Inc. Wallingford CT, USA). First, the chosen conformer was optimized at 6-31g* using the Hartree–Fock method, and then optimized at B3LYP/6-311G** in methanol using the polarizable continuum model (PCM). The theoretical calculation of ECD was conducted in methanol using time-dependent density functional theory (TD-DFT) at the same theory level. Rotatory strengths for a total of 30 excited states were calculated. The ECD spectrum was simulated in SpecDis22 by overlapping Gaussian functions for each transition, according to (eqn (2)):
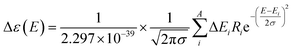 | (2) |
The parameters of σ and UV-shift for each configuration are listed in Table S3.†
Biological assay
Results and discussion
Chemistry
In this manuscript, we report the synthesis of a series of PepE analogues derived from open chain secondary aliphatic (2–7), cyclic secondary aliphatic (8–12), and several open chain or arylalkyl primary (13–19) amines. The choice of amines was based on the results recently reported in studies on PTL derivatives.16,18 The synthesis of PepE derivatives 2–19 was carried out using the procedures outlined in Scheme 1.PepE was obtained from the CH2Cl2 extract of whole P. dindygulensis plants, according to methodology described previously.8 Its structure was unambiguously confirmed by a combination of spectroscopic methods, including infrared spectroscopy, NMR, and high-resolution mass spectrometry.
To obtain a library of water-soluble PepE analogues, 0.2 mM of PepE in methanol was treated with one equivalent of an appropriate primary or secondary amine, using trimethylamine as a catalyst at room temperature (15–20 °C), which led to a significant consumption of PepE within 8 h. The reaction product was purified by flash column chromatography. A good yield (65–92%) of product was obtained, but unreacted PepE persisted even after prolonged reaction time, addition of excess amine, or increased temperature.
The amino PepE analogues thus obtained (2–19) were then converted into water-soluble salts, thereby increasing their water solubility. Multiple inorganic/organic acid were tested for salt formation including fumarate acid, citric acid, sulphate acid, phosphate acid and hydrochloride acid. The phosphate salts have the best crystallinity, and relatively low hygroscopic properties, therefore selected it as the salt-forming acid.
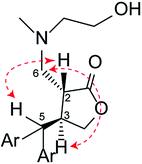
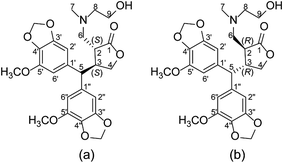
The 1H NMR spectra of the synthesised analogues showed the absence of the two olefinic proton resonances at 6.15 and 4.94 ppm in the PepE molecule.6–8 The appearance of new methine and methylene resonances in the distortionless enhancement by polarization transfer (DEPT) spectrum of 13C NMR was also apparent, indicating that amination at C-6 of the exocyclic double bond had occurred by Michael-addition reactions. The newly formed C-2 chiral carbon was found to be highly stereospecific based on the nuclear Overhauser effect spectroscopy (NOESY) spectrum. The cross-peaks of H-6/H-3 and H-2/H-5, as well as the absence of NOESY correlation between H-6 and H-5 (Fig. 2), indicated a trans configuration at the γ-butyrolactone ring of the synthesised products. Thus, there were only two possible absolute configurations of the γ-butyrolactone ring (Fig. 3), namely (2S,3S) or (2R,3R). The structures of the synthesised products were elucidated by NMR, ultraviolet spectroscopy, and mass spectrometry data; all NMR signals were accordingly assigned based on heteronuclear multiple bond correlation (HMBC) and heteronuclear single-quantum correlation (HSQC) experiments (see Experimental sections).
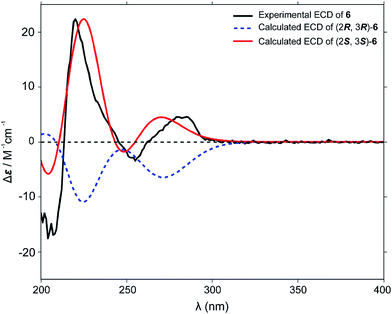
Our repeated attempts to obtain suitable crystals for X-ray diffraction were unsuccessful, and so the absolute configurations of the γ-butyrolactone ring of the analogues were established by comparing the experimental and theoretical electronic circular dichroism (ECD) spectra predicted using the time-dependent density functional theory (TDDFT) at the B3LYP/6-311G** level. Take compound 6 for instance, the energies and populations of all dominative conformers of 6 are provided in Table S1.† The energies of the all conformers of 6 optimized at B3LYP/6-311G** in methanol are provided in Fig. 4 and Table S2.† The ECD spectrum of 6 showed intense positive Cotton effects at 220 and 280 nm, as well as weakly negative Cotton effects at 254 nm, which were similar to those of reported peperomin B.22,23 This indicated that 6 might share the same configuration as peperomin B, which is (2S,3S).24 Moreover, the overall pattern of the simulated ECD spectrum for the (2S,3S) stereoisomer was in good agreement with the measured spectrum of 6 (Fig. 5, the parameters of σ and UV-shift for each configuration are listed in Table S3†). Therefore, the absolute configuration of 6 was unambiguously assigned as 2S,3S. In addition, we have confirmed the (2S,3S) configuration in many of our synthesised products by comparing the experimental and calculated ECD spectra.
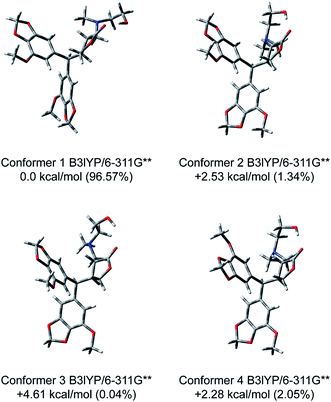 | ||
| Fig. 4 Optimized structures and relative energies of the four lowest-energy conformers of (2S,3S)-6 at the B3LYP/6-311G** level in methanol. | ||
The exclusive formation of the 2S configuration in the γ-butyrolactone ring, according to the literature, is due to the protonation of the enolate formed during Michael addition, which occurs exclusively from the exo face of the γ-butyrolactone ring, resulting in an S configuration at C-2 in the product.16
Biological activity
To evaluate the anti-AML activity of PepE and its amino analogues, all of the synthesised compounds were tested against cultured cell lines, including KG-1 (acute myelogenous leukaemia), HL-60 (acute promyelocytic leukaemia), THP-1 (acute monocytic leukaemia), and KG-1a (acute myelogenous leukaemia), using PTL (a drug that selectively ablates acute leukaemia stem cells and has advanced to a phase I clinical trial for treatment of AML25) as a positive control. hBMSCs were used as a model to test the toxicity of PepE and its derivatives against normal cells. It has been reported that bearing a CD34+CD38− immunophenotype is a characteristic of LSCs.26 From our results of flow cytometry, the percentage of CD34+ cells in the KG-1a cell line was 98.9%, and the percentage of CD34+CD38− was 50.1%. These results corroborated those reported by Liu et al.,27 confirming that KG-1a represents an AML stem cell-like line. Then, CD34+CD38− cells were enriched from the KG-1a cell line using MACS, as reported previously by Zhang et al.28 The percentage of the CD34+CD38− cell population in the KG-1a cells was significantly increased after sorting (92.1 ± 1.3% versus 50.1 ± 5.5% before sorting).Eighteen amino analogues were then studied with a CCK-8 cytotoxicity assay against the KG-1, HL-60, THP-1, and KG-1a CD34+CD38− cell lines. Preliminary cytotoxic screening of the analogues was performed at 10 μM. The analogues that exhibited greater than 50% growth inhibition at 10 μM were further assayed at varying concentrations to generate the IC50 values shown in Table 1. It is evident from Table 1 that the parent molecule, PepE, exhibited similar cytotoxicity against AML and LSC cells as the positive control, PTL, did, while its toxicity at 50 μM against hBMSCs was half that of PTL. It is also evident from Table 1 that several of the amino-PepE products exhibited increased cytotoxicity toward AML and LSC cells compared to that of the parent molecule, while others showed decreased activity. The structure and activity relationship (SAR) of these synthetic derivatives is discussed below.
| Compounds | KG-1 | HL-60 | THP-1 | KG−1a CD34+CD38− | hBMSC | ||||
|---|---|---|---|---|---|---|---|---|---|
| Inhibitiona (%) | IC50b (μM) | Inhibition (%) | IC50 (μM) | Inhibition (%) | IC50 (μM) | Inhibition (%) | IC50 (μM) | Inhibitionc (%) | |
| a % Inhibition refers to growth inhibition measured at 10 μM concentration.b All values are the mean of three independent experiments.c % Inhibition refers to growth inhibition measured at 50 μM concentration.d nd, not determined in the given concentration range.e PTL, parthenolide, used as a positive control. | |||||||||
| PepE | 78.7 | 3.3 ± 0.2 | 77.1 | 4.3 ± 0.3 | 63.7 | 8.3 ± 0.3 | 61.4 | 6.4 ± 0.2 | 19.3 |
| 2 | 54.2 | 6.2 ± 0.9 | 66.5 | 4.0 ± 0.8 | 56.9 | 7.0 ± 2.4 | 65.9 | 7.3 ± 0.2 | 8.3 |
| 3 | 59.9 | 8.0 ± 2.0 | 78.3 | 3.4 ± 0.6 | 64.7 | 8.5 ± 0.2 | 62.1 | 6.9 ± 0.2 | 12.1 |
| 4 | 74.8 | 6.9 ± 0.6 | 73.7 | 4.0 ± 0.4 | 56.3 | 9.1 ± 1.3 | 66.8 | 7.2 ± 0.2 | 16.7 |
| 5 | 81.5 | 3.9 ± 0.5 | 70.4 | 2.4 ± 0.2 | 66.1 | 7.7 ± 1.0 | 59.2 | 10.0 ± 1.0 | 18.7 |
| 6 | 81.2 | 4.9 ± 0.3 | 78.4 | 1.4 ± 0.3 | 73.9 | 2.0 ± 0.6 | 88.2 | 0.5 ± 0.1 | 11.2 |
| 7 | 63.2 | 5.2 ± 0.4 | 65.4 | 3.8 ± 0.3 | 60.8 | 8.6 ± 0.5 | 59.7 | 7.3 ± 0.2 | 16.6 |
| 8 | 38.5 | ndd | 67.8 | 7.1 ± 0.3 | 43.8 | nd | 52.6 | 8.8 ± 0.3 | 9.1 |
| 9 | 36.1 | nd | 64.5 | 8.6 ± 0.5 | 38.1 | nd | 28.5 | nd | 7.2 |
| 10 | 43.3 | nd | 55.8 | 9.6 ± 0.6 | 31.4 | nd | 48.7 | nd | 10.1 |
| 11 | 37.5 | nd | 32.1 | nd | 56.4 | 15.1 ± 0.8 | 60.1 | 9.6 ± 0.2 | 8.3 |
| 12 | 68.9 | 5.8 ± 1.0 | 71.4 | 2.4 ± 0.5 | 68.3 | 6.3 ± 0.3 | 76.3 | 3.3 ± 0.3 | 13.4 |
| 13 | 19.6 | nd | 15.3 | nd | 12.4 | nd | 17.5 | nd | 7.7 |
| 14 | 35.9 | nd | 34.3 | nd | 34.9 | nd | 30.3 | nd | 16.8 |
| 15 | 38.1 | nd | 58 | 9.3 ± 0.3 | 37.8 | nd | 15.9 | nd | 17.2 |
| 16 | 39.5 | nd | 42.9 | nd | 55.2 | 9.3 ± 2.9 | 29.5 | nd | 3.9 |
| 17 | 74.6 | 7.3 ± 0.5 | 71.8 | 3.9 ± 0.4 | 30.1 | nd | 40.4 | nd | 8.4 |
| 18 | 71.1 | 6.2 ± 0.1 | 69.8 | 2.0 ± 0.1 | 38.4 | nd | 37.3 | nd | 11.1 |
| 19 | 56.3 | 9.3 ± 0.7 | 71 | 1.6 ± 0.2 | 42.7 | nd | 55.4 | 9.8 ± 0.3 | 8.6 |
| PTLe | 79.8 | 5.2 ± 0.5 | 73.6 | 4.8 ± 0.9 | 68.9 | 5.7 ± 1.0 | 69.8 | 5.2 ± 0.4 | 40.2 |
The open-chain secondary aliphatic amine series (2–5, 7) was found to be nearly as active as was the parent molecule, PepE. However, compound 6, with a hydroxyl group replacing the 10-methyl group of compound 5, was the exception. The cytotoxicity of compound 6 (IC50 = 0.5 μM) significantly improved up to 13-fold compared to that of the parent molecule against KG-1a CD34+CD38− cells, while there was little effect on the viability of normal hBMSCs (inhibition rate of 11.2% at 50 μM).
The toxicity level against AML cells of the cyclic aliphatic amine series (8–11), in contrast, was significantly decreased when compared with that of the parent molecule. However, replacing the 3-methyl piperidine moiety in compound 11 with a 3-hydroxymethyl piperidine moiety to form compound 12 increased the cytotoxicity against HL-60 and KG-1a CD34+CD38− cells (IC50 = 2.4 and 3.3 μM, respectively) up to 2-fold compared to that of PepE and >3-fold compared to that of compound 11; the safety level of compound 12 was approximately the same as that of PepE (13.4% at 50 μM).
The primary amine series (13–19) was generally less active against AML cells when compared with that of the parent molecule and the secondary amine series; some analogues (13, 14) displayed low growth inhibition at the preliminary screening concentration. However, introduction of tyramine (18) or its hydroxyl derivatives (19) into the PepE structure resulted in a significant increase in cytotoxicity against HL-60 cells (IC50 = 2.0 and 1.6 μM, respectively), increasing cytotoxicity up to 2-fold compared to that of the parent molecule.
In summary, 18 amino-PepE products were synthesised and evaluated for anti-leukaemic activity against AML cell lines in culture. The phosphate salts of all the synthesised compounds were more water soluble than PepE was, especially compound 6, which exhibited 1000-fold greater water-solubility than did PepE. A range of anti-leukaemic activities was observed, with several products exhibiting enhanced activities compared to that of PepE.
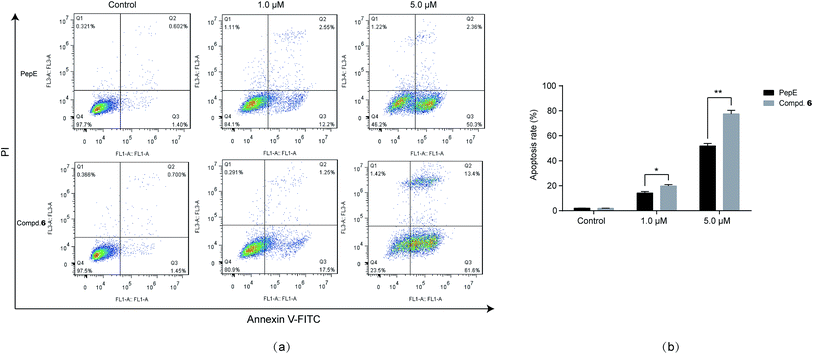
Compound 6 was the most potent compound of all synthesised amino analogues, especially against the immature AML stem cells, KG-1a CD34+CD38−, inducing 80–90% cell death at 10 μM, and affording an IC50 value of 0.5 μM with little effect on hBMSCs. Therefore, the mechanism of action of compound 6 was further explored in preliminary experiments. Apoptosis was analysed by flow cytometric assay. The percentages of apoptotic cells were 18.7% and 75.1% in the KG-1a CD34+CD38− cells after treatment with 1 μM and 5 μM, respectively, and were significantly higher than those of the parent molecule (Fig. 6). This result indicated that 6 can significantly induce apoptosis of leukaemia stem cells.
As reported by many clinicians, the CD34+CD38− AML cell subpopulation is frequently impervious to conventional chemotherapy, making it largely responsible for relapse of AML. Therefore, targeting and eliminating these cells has been proposed as a promising strategy for treatment of AML to decrease the rate of relapse or drug-resistance.2 However, small molecules that can selectively target LSCs are rare and still at early stages of development. As reported in this manuscript, compound 6 may be considered as a new lead compound to eliminate these AML stem cells. Further pharmacological and pharmacokinetic evaluation of this compound is underway.
Conclusions
In conclusion, for the first time, we have reported a newly found naturally occurring agent, PepE, which targets leukaemia stem cells. A series of 18 novel amino-derivatives of PepE was then synthesised and evaluated for anti-AML and anti-LSC activity. All of the compounds showed higher water solubility than the parent molecule. More importantly, this study found that one of the amino derivatives, compound 6, was more potent than was the parent drug, PepE, against the LSC-like cells (KG-1a CD34+CD38−), with no apparent toxicity on hBMSCs. According to the above results, the following SAR of the PepE amino derivatives can be formulated: (1) the secondary open-chain aliphatic amino analogues generally exhibited better activity than the cyclic aliphatic and primary amino analogues did; and (2) introducing a hydroxyl group at the methyl group of the secondary open-chain amine adduct could significantly potentiate the activity against AML stem cells, and result in the most active compound 6.Based on the above results, further optimization and biological evaluation of 6 as a promising agent targeting LSC are ongoing in our laboratory, and the results will be reported shortly.
Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by Grants from the Natural Science Foundation of China (No. 81402812), the Natural Science Foundation of Jiangsu Province (No. BK20130954) and the Priority Academic Program Development of Jiangsu Higher Education Institutions.References
- H. Dohner, D. J. Weisdorf and C. D. Bloomfield, N. Engl. J. Med., 2015, 373, 1136–1152 CrossRef PubMed.
- U. Testa, Ann. Hematol., 2011, 90, 245–271 CrossRef CAS PubMed.
- A. K. Burnett, N. Russell, R. K. Hills, N. Panoskaltsis, A. Khwaja, C. Hemmaway, P. Cahalin, R. E. Clark and D. Milligan, Leukemia, 2015, 29, 1312–1319 CrossRef CAS PubMed.
- D. C. Hassane, M. L. Guzmann, C. Corbert, X. J. Li, R. Abboud, F. Young, J. L. Liesveld, M. Carroll and C. T. Jordan, Blood, 2008, 111, 5654–5662 CrossRef CAS PubMed.
- T. R. Govindachari, G. N. Krishna Kumari and P. D. Partho, Phytochemistry, 1998, 49, 2129–2131 CrossRef CAS.
- M. G. Lloyd, R. J. K. Taylor and W. P. Unsworth, Org. Biomol. Chem., 2016, 14, 8971 CAS.
- X. Z. Wang, Y. Cheng, K. L. Wang, R. Liu, X. L. Yang, H. M. Wen, C. Chai, J. Y. Liang and H. Wu, Cancer Sci., 2016, 10, 1506–1519 CrossRef PubMed.
- X. Z. Wang, Y. Cheng, H. Wu, N. Li, R. Liu, X. L. Yang, Y. Y. Qiu, H. M. Wen and J. Y. Liang, Phytomedicine, 2016, 23, 818–827 CrossRef CAS PubMed.
- A. Ghantous, H. Gali-Muhtasib, H. Vuorela, N. A. Saliba and N. Darwiche, Drug Discovery Today, 2010, 15, 668–678 CrossRef CAS PubMed.
- X. Z. Wang, H. M. Wen, C. Chai, W. Y. Zhang, M. Gao, R. Liu, H. Wu and J. Y. Liang, Biomed. Chromatogr., 2017, 31, e3930 CrossRef PubMed.
- J. R. Woods, H. Mo, A. A. Bieberich, T. Alavanja and D. A. Colby, MedChemComm, 2013, 4, 27–33 RSC.
- A. Kumar, D. Kumar, A. K. Maurya, Y. S. Padwad and V. K. Agnihotri, Phytochem. Lett., 2016, 18, 117–121 CrossRef CAS.
- S. Zimmermann, G. Franche, M. D. Mieri, Y. Yoshimoto, T. Usuki, R. Nthambeleni, C. J. Parkinson, C. van der Westhuyzen, M. Kaiser, M. Hamburger and M. Adams, Molecules, 2014, 19, 3523–3538 CrossRef PubMed.
- X. J. Wang, X. M. Zhang, B. B. Zheng, N. Hu, W. D. Xie and K. Row, Nat. Prod. Res., 2015, 29, 756–763 CrossRef CAS PubMed.
- Y. H. Ding, H. E. Gao, Y. Zhang, Y. Li, N. Vasdev, Y. D. Gao, Y. Chen and Q. Zhang, J. Hematol. Oncol., 2016, 9, 93–99 CrossRef PubMed.
- S. Nasim and P. A. Crooks, Bioorg. Med. Chem. Lett., 2008, 18, 3870–3873 CrossRef CAS PubMed.
- H. Nakshatri, H. N. Appaiah, M. Anjanappa, D. Gilley, H. Tanaka, S. Badve, P. A. Crooks, W. Mathews, C. Sweeney and P. Bhat-Nakshatri, Cell Death Dis., 2015, 6, e1608 CrossRef CAS PubMed.
- M. Moumou, A. El Bouakher, H. Allouchi, A. El Hakmaoui, A. Benharref, V. Mathieu, G. Guillaumet and M. Akssira, Bioorg. Med. Chem. Lett., 2014, 24, 4014–4018 CrossRef CAS PubMed.
- Z. Yang, B. Kuang, N. Kang, Y. Ding, W. Ge, L. Lian, Y. Gao, Y. Wei, Y. Chen and Q. Zhang, Eur. J. Med. Chem., 2017, 127, 296–304 CrossRef CAS PubMed.
- M. L. Guzman, R. M. Rossi, L. Karnischky, X. J. Li, D. R. Peterson, D. S. Howard and C. T. Jordan, Blood, 2005, 105, 4163–4168 CrossRef CAS PubMed.
- N. M. OBoyle, T. Vandermeersch, C. J. Flynn, A. R. Maguire and G. R. Hutchison, J. Cheminf., 2011, 3, 3–8 Search PubMed.
- T. Bruhn, A. Schaumlffl, Y. Hernberger and G. Bringmann, Chirality, 2013, 25, 243–249 CrossRef CAS PubMed.
- C. M. Chen, F. Y. Fan, M. T. Chen and T. J. Lee, Heterocycles, 1989, 29, 411–414 CrossRef.
- T. J. Lee and T. Y. Lee, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 1990, 46, 338–340 CrossRef.
- A. Ghantous, A. Sinjab, Z. Herceg and N. Darwiche, Drug discovery today, 2013, 18, 894–905 CrossRef CAS PubMed.
- M. Terwijn, W. Zeijlemaker, A. Kelder, A. P. Rutten, A. N. Snel, W. J. Scholten and G. J. Ossenkoppele, PLoS One, 2014, 9, e107587 Search PubMed.
- Y. Liu, F. Chen, S. Wang, X. Guo, P. Shi and W. Wang, Cell Death Dis., 2013, 4, e948 CrossRef CAS PubMed.
- Y. Zhang, S. Y. Zhou, H. Z. Yan, D. D. Xu, H. X. Chen, X. Y. Wang, X. Wang, Y. T. Liu, L. Zhang, S. Wang, P. J. Zhou, W. Y. Fu, B. B. Ruan, D. L. Ma, Y. Wang, Q. Y. Liu, Z. Ren, Z. Liu, R. Zhang and Y. F. Wang, Sci. Rep., 2016, 6, 19995 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra09928b |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |