 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Selective recognition of c-myc promoter G-quadruplex and down-regulation of oncogene c-myc transcription in human cancer cells by 3,8a-disubstituted indolizinone†
Fengmin
Yang
ab,
Xin
Sun
a,
Lixia
Wang
 *a,
Qian
Li
*a,
Aijiao
Guan
a,
Gang
Shen
ab and
Yalin
Tang
*a,
Qian
Li
*a,
Aijiao
Guan
a,
Gang
Shen
ab and
Yalin
Tang
 *ab
*ab
aNational Laboratory for Molecular Sciences, Center for Molecular Sciences, State Key Laboratory for Structural Chemistry of Unstable and Stable Species, Institute of Chemistry Chinese Academy of Sciences, Beijing, 100190, P. R. China. E-mail: tangyl@iccas.ac.cn; Fax: +86 10 62522090; Tel: +86 10 82617304
bGraduate University of the Chinese Academy of Sciences, Yuquan Road 19 (A), Shijingshan District, Beijing, 100049, P. R. China
First published on 8th November 2017
Abstract
C-myc promoter G-quadruplex is a very important target for developing anti-cancer drugs. However, highly selective recognition of DNA c-myc G-quadruplex with parallel configuration is also a very challenging problem. Here, we showed a new type of N-containing alkaloid, 3,8a-disubstituted indolizinone, which adopted a distorted configuration. 6-Methyl-3-(naphthalen-2-yl)-8a-(4-methylpyridin-2-yl)-indolizinone could selectively recognize DNA c-myc G-quadruplex leading to an obvious fluorescence enhancement by 11-fold, and meanwhile the G-quadruplex can be stabilized up to 90 °C. Further, it could down-regulate the transcription of oncogene c-myc in both human non-small cell line (A549) and human laryngeal epithelial cell line (Hep-2).
Introduction
The G-quadruplex structure formed by tandem guanine (G) nucleotides, which are incorporated in nucleic acid sequences, has illuminated a new role for DNA in biology.1 G-quadruplexes as one of the higher-order DNA and RNA structures have been identified in G-rich human telomeres and gene promoter regions. G-quadruplex as a therapeutic target has attracted a growing interest, due to its profound roles in a wide spectrum of diseases, such as cancer, diabetes and cardiovascular disorders.2 Several organic probes have been reported to recognize DNA G-quadruplex via diverse interaction modes.3 Stabilization of G-quadruplex structures in the promoter region of certain oncogenes is an emerging field in anticancer drug design.4 Human c-myc gene is one of these oncogenes, and G-quadruplexes have been proven to be the transcriptional controller of this gene.Human c-myc gene or its product is a central regulator of cellular proliferation and cell growth, and over-expression of c-myc gene is related to various cancers.5 C-myc is the first and most extensively studied system of promoter G-quadruplex formation. The major G-quadruplex formed in the c-myc promoter is a parallel stranded structure with two G3NG3 single-nt strand-reversal loops, and parallel-stranded structures are found to be more common in the promoter G-quadruplexes.1 Some known G-quadruplex-interactive compounds have become prospective anticancer agents that display relatively low cytotoxicity. In particular, Quarfloxin, a first-in-class G-quadruplex-interactive drug reached phase 2 clinical trials, which was considered to be a c-myc G-quadruplex ligand at first.6 Quarfloxin is a fluoroquinolone derivative with antineoplastic activity, and another known c-myc G-quadruplex ligand quindoline is a derivative of the natural product cryptolepine,4 and both of them are N-containing alkaloids. Here we reported a new type of N-containing alkaloid, indolizinone (Scheme 1), which can selectively recognize DNA c-myc G-quadruplex (DNA MycG4) and down-regulate oncogene c-myc.
 | ||
| Scheme 1 Structure of 6-methyl-3-(naphthalen-2-yl)-8a-(4-methylpyridin-2-yl)-indolizinone (indolizinone). | ||
Results
Indolizinones are N-heterocycles that have only recently appeared in the chemical literature,7 and to the best of our knowledge, any application of which wasn't found to be reported. Recently, we firstly reported a copper-catalyzed domino reaction to construct functionalized 3,8a-disubstituted indolizinones, and this protocol was simple and highly efficient.8 In the solid state, 3,8a-disubstituted indolizinones adopted distorted configuration, and thus they provided no obviously strong fluorescence spectra in solution due to the lack of a larger conjugated π system. Most of DNA MycG4 ligands that have been previously reported are N-heterocycles with planar π systems, which can't distinguish DNA MycG4 from other configurations of G-quadruplexes. In this report, we found 6-methyl-3-(naphthalen-2-yl)-8a-(4-methylpyridin-2-yl)-indolizinone (indolizinone) here could selectively recognize DNA MycG4, leading to its greatly enhanced fluorescence intensity. We firstly compared the fluorescence intensity of indolizinone with five oligonucleotides of known G4 structures (Table S1†): intermolecular parallel configuration H7, intramolecular hybrid-type configuration M24, intramolecular anti-parallel configuration A24, intramolecular parallel configuration Kit and Myc, relative to the signal of indolizinone alone. As shown in Fig. 1, indolizinone itself at 4 μM gives a very weak fluorescence signal. While addition of 8 μM DNA MycG4, and then standing for 24 hours, the fluorescence signal of indolizinone lead to an obvious enhancement by 11-fold. Addition of other types of G-quadruplexes, such as intermolecular parallel type H7 led to 0.5-fold enhancement, and for intramolecular types, parallel DNA c-Kit 4.5-fold, hybrid-type M24 1.6-fold and anti-parallel type A24 2.5-fold, respectively. These results indicated that indolizinone preferred to recognize DNA MycG4.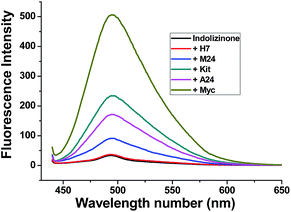 | ||
| Fig. 1 Fluorescence intensity of indolizinone with five varied DNA G-quadruplexes in a 20 mM Tris HCl (40 mM K+, pH 7.0) solution at room temperature, Ex = 423 nm. | ||
Furthermore, fluorescence titrations were carried out to evaluate the binding constants of indolizinone with these five nucleic acid sequences (Fig. S1–S3†). A 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding model for indolizinone with DNA G-quadruplex was confirmed by the job's plot analysis. The typical titration curves and fitting results according to the 1
1 binding model for indolizinone with DNA G-quadruplex was confirmed by the job's plot analysis. The typical titration curves and fitting results according to the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding model are presented. The binding constants for indolizinone with Myc and Kit are (9.91 ± 0.65) × 105 and (5.77 ± 1.60) × 105, respectively. The binding strength to Myc was obviously more strong than to Kit. In comparison with other intramolecular DNAG4s, the binding strength to A24 or M24 was much weaker, with binding constants (2.43 ± 0.37) × 104 and (1.88 ± 0.65) × 104, respectively. Of note, the binding strength to intermolecular H7 was very weak, and the binding constant is less than 10. As shown in Table 1, the trend of the binding constants is consistent with the F/F0 values.
1 binding model are presented. The binding constants for indolizinone with Myc and Kit are (9.91 ± 0.65) × 105 and (5.77 ± 1.60) × 105, respectively. The binding strength to Myc was obviously more strong than to Kit. In comparison with other intramolecular DNAG4s, the binding strength to A24 or M24 was much weaker, with binding constants (2.43 ± 0.37) × 104 and (1.88 ± 0.65) × 104, respectively. Of note, the binding strength to intermolecular H7 was very weak, and the binding constant is less than 10. As shown in Table 1, the trend of the binding constants is consistent with the F/F0 values.
Then we subsequently performed this assay on a larger set of 25 nucleic acid sequences. Structures of these oligonucleotides have been characterized, and these oligonucleotides have been used in the G4-Fluorescence Intercalator Displacement (FID) assay.9 All sequences are listed in Table 2, and results are presented in Fig. 2. Fifteen of the 25 sequences are likely to form G4s. The fluorescence enhancements of indolizinone interacted with the parallel configuration G4 in the promoter region (c-myc, c-kit1, c-kit2, bcl-2, AKT1, VAV1, 35B1, 27Kras) were 11-fold, 4.0-fold, 3.7-fold, 2.5-fold, 4.0-fold, 5.2-fold, 4.3-fold, and 3.4-fold, respectively. And the human telomeric DNA G-quadruplex 22Ag and 45Ag, which adopt anti-parallel configuration in the presence of K+, led to the fluorescence enhancements of indolizinone with the value F/F0 1.5-fold and 2.8-fold, respectively. And meanwhile, in the presence of another five G-quadruplex forming sequences (21CTA, TBA, Ceb25, CT4, CGG12), and other non-G-quadruplex DNA templates including single-stranded (G-tripl, dT30), double-stranded (ds26, 19AT, dx12, PS1c), stem-loop (GCdx), triplex, and trinucleotide (CTG12, CAG12), all these oligonucleotide sequences led to a F/F0 < 2. Indolizinone showed higher response towards intramolecular DNA G-quadruplex with parallel configuration, especially most to DNA MycG4, comparing to other types of G-quadruplex and non-G-quadruplex templates. While it presented very weak response to intermolecular parallel G-quadruplex H7, just leading to 0.5-fold enhancement, probably attributing to its lack of loop-binding sites.
| Name | Type/origin | Sequence (from 5′ to 3′) |
|---|---|---|
| c-myc (Myc) | G4-promoter | TGAG3TG3GAG3TG3GAA |
| c-kit1 (Kit) | G4-promoter | G3AG3CGCTG3AGGAG3 |
| c-kit2 | G4-promoter | G3CG3CGCGAG3AGG3 |
| BCL-2 | G4-promoter | G3CGCG3AG2AATTG3CG3 |
| AKT1 | G4-promoter | G3CG3CGGCTCCG3CGCG3 |
| VAV1 | G4-promoter | G3CAG3AG3AACTG3 |
| 35B1 (Kras) | G4-promoter | AG3CGGTGTG3AAGAG3AAGAG3GGAGGCAG |
| 27 Kras | G4-promoter | G3CG2TGTG3AAGAG3AAGAG3G |
| 7Ag (H7) | G4-humanDNA telomere | TTAG3T |
| 22Ag | G4-humanDNA telomere | AG3TTAG3TTAG3TTAG3 |
| 24Ag (A24) | G4-humanDNA telomere | TTAG3TTAG3TTAG3TTAG3 |
| 24AgA (M24) | G4-humanDNA telomere | TTAG3TTAG3TTAG3TTAG3A |
| 45Ag | G4-human DNA telomere | G3TTAG3TTAG3TTAG3TTAG3TTAG3TTAG3TTAG3 |
| 21CTA | G4-human telomere variant | G3CTAG3CTAG3CTAG3 |
| TBA | G4-aptamer | GGTTGGTGTGGTTGG |
| Ceb25 | G4-minisatellites | AG3TG3TGTAAGTGTG3TG3T |
| CT4 | G4/mixed quartets | G3CTTTTG3C |
| CGG12 | Trinucleotide | (CGG)12 |
| CTG12 | Trinucleotide | (CTG)12 |
| CAG12 | Trinucleotide | (CAG)12 |
| G-tripl | Single strand | GGTTGGTGTGG |
| dT30 | Single strand | T30 |
| Triplex | Triplex | (a) TTTTTTTTTTTTTTTTTTTT |
| (b) AAAAAAAAAAAAAAAAAAAA | ||
| (c) TTTTTTTTTTTTTTTTTTTT | ||
| GCdx | Stem-loop | GCGCGCGCTTTTGCGCGCGC |
| PS1c | Parallel-duplex | (a) TTTTTTTTTTATTAAAATTTATAA |
| (b) AAAAAAAAAATAATTTTAAATATT | ||
| ds26 | Duplex | CAATCGGATCGAATTCGATCCGATTG |
| 19AT | Duplex | (a) ACGTCGATTATAGACGAGC |
| (b) GCTCGTCTATAATCGACGT | ||
| dx12 | Duplex | (a) GCGTGAGTTCGG |
| (b) CCGAACTCACGC |
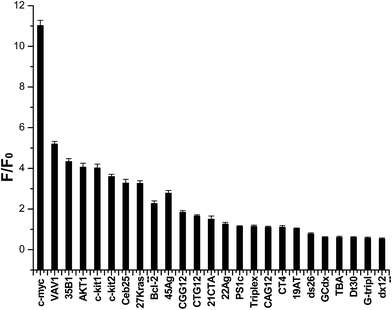 | ||
| Fig. 2 Dependence of indolizinone (4 μM) fluorescence intensity at 494 nm for a variety of DNA sequences (8 μM) in a 20 mM Tris HCl (40 mM K+, pH 7.0) solution. | ||
To evaluate the effect of indolizinone on the specific recognition of DNA MycG4, we applied circular dichroism to monitor the melting temperature (Tm) of DNA MycG4 in the presence of 1 equiv. molar ratio of indolizinone. The change of Tm in the folded and unfolded quadruplex structure upon interacting with ligand provides evidence of thermal stabilization of DNA structure.10 DNA MycG4 was incubated with indolizinone for 24 h, the Tm values for DNA MycG4 were calculated as shown in Fig. 3. Indolizinone shows the preference to stabilize DNA MycG4 by raising the temperature up to 90 °C monitored at 265 nm. In the contrast, indolizinone shows a weak ability obviously to stabilize other types of G-quadruplexes (see Fig. S4†).
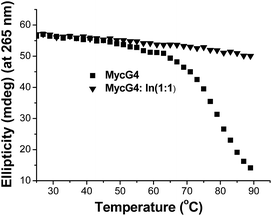 | ||
| Fig. 3 CD melting profile of 4 μM DNA MycG4 alone or in the presence of 1 equiv. molar ratio of indolizinone in a 20 mM Tris HCl (40 mM K+, pH 7.0) solution. | ||
Based on the emission enhancement and the absorption spectral results, fluorescence intensity decay measurements were conducted to demonstrate the strong binding of indolizinone with DNA MycG4. The fluorescence decay of indolizinone was fast (<1 ns) in solution due to the highly feasible torsional relation channel in the excited state. However, the lifetimes increased to the nanosecond level in the presence of DNA MycG4. As shown in Fig. 4, the fluorescence decay was much lower for indolizinone in the presence of 1 equiv. or 2 equiv. molar ratio of DNA MycG4, with the τ value of 3.54 ns and 3.71 ns, respectively. This result indicated that the interaction between indolizinone and DNA MycG4 hinders the rotation of indolizinone and increases its lifetime.
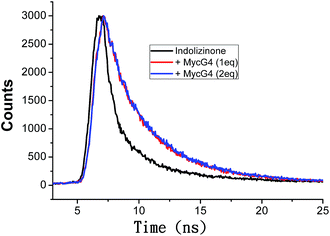 | ||
| Fig. 4 Fluorescence decay traces of indolizinone (4 μM) in the absence and presence of DNA MycG4 in a 20 mM Tris HCl (40 mM K+, pH 7.0) solution. | ||
Oncogene c-myc encodes an important transcriptional regulator c-myc protein, which is involved in cell proliferation, senescence, and apoptosis.11 Because c-myc promoter region contains G-quadruplex forming G-rich sequence, its binding with indolizinone could increase its stability resulting in down-regulation of oncogene c-myc transcription, which would be identified using real-time RT-PCR.12 To test the above hypothesis and understand the effect of indolizinone on oncogene c-myc, two cancer cell lines with different translocation break points within the oncogene c-myc were tested. Human non-small cell line (A549) and human laryngeal epithelial cell line (Hep-2) were obtained from the National Infrastructure of Cell Line Resource (Beijing, China). As shown in Fig. 5, indolizinone could down-regulate the transcription of oncogene c-myc in both A549 and Hep-2 cells, after treated with indolizinone at 0.5, 1.0, 2.0, 4.0, and 8.0 μM for 48 h. The total RNA was extracted and reverse transcripted to cDNA. This cDNA was then used as a template for specific RCR amplification of the c-myc sequence and controlled by β-actin.
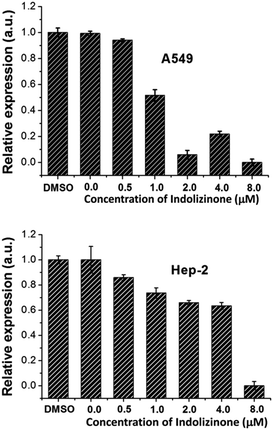 | ||
| Fig. 5 qRT-PCR to determine the transcription of oncogene c-myc in the A549 and Hep-2 cells treated with indolizinone. | ||
Conclusions
Indolizinones are N-heterocycles that have only recently appeared in the chemical literature, which can be prepared from the simple and achievable starting materials efficiently. Here, we firstly reported the indolizinone has the potential ability to recognize DNA MycG4 by fluorescence spectra and varied temperature CD spectra. When indolizinone complexed with DNA MycG4 in the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, and thus it can give an obvious fluorescence enhancement by 11-fold, and meanwhile the DNA MycG4 can be stabilized up to 90 °C. Further, we found indolizinone could down-regulate the transcription of oncogene c-myc in both of human non-small cell line (A549) and human laryngeal epithelial cell line (Hep-2) by real-time RT-PCR experiment. We believed that indolizinone will have a potential application in the exploration of anticancer drugs.
2, and thus it can give an obvious fluorescence enhancement by 11-fold, and meanwhile the DNA MycG4 can be stabilized up to 90 °C. Further, we found indolizinone could down-regulate the transcription of oncogene c-myc in both of human non-small cell line (A549) and human laryngeal epithelial cell line (Hep-2) by real-time RT-PCR experiment. We believed that indolizinone will have a potential application in the exploration of anticancer drugs.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 21472197, 21675162, 21778058, 21302188), the Beijing Natural Science Foundation (Grant No. 7172248), Funding for open access charge: The National Natural Science Foundation of China (21603071).Notes and references
- Selected recent reviews, see: (a) Q. Cao, Y. Li, E. Freisinger, P. Z. Qin, R. K. O. Sigel and Z. W. Mao, Inorg. Chem. Front., 2017, 4, 10–32 RSC; (b) Y. W. Chen and D. Z. Yang, Curr. Protoc. Nucleic Acid Chem., 2012, 17.5.1–17.5.17 CrossRef; (c) K. M. Miller and R. Rodriguez, Expert Rev. Clin. Pharmacol., 2011, 4, 139–142 CrossRef CAS; (d) J. T. Davis, Angew. Chem., Int. Ed., 2004, 43, 668–698 CrossRef CAS; (e) L. H. Hurley, R. T. Wheelhouse, D. Sun, S. M. Kerwin, M. Salazar, O. Y. Fedoroff, F. X. Han, H. Y. Han, E. Izbicka and D. Von Hoff, Pharmacol. Ther., 2000, 85, 141–158 CrossRef CAS; (f) T. M. Ou, Y. J. Lu, J. H. Tan, Z. S. Huang, K. Y. Wong and L. Q. Gu, ChemMedChem, 2008, 3, 690–713 CrossRef CAS.
- Q. Li, J. F. Xiang, Q. F. Yang, H. X. Sun, A. J. Guan and Y. L. Tang, Nucleic Acids Res., 2013, 41, D1115–D1123 CrossRef CAS.
- Selected organic probes: (a) J. Mohanty, N. Barooah, V. Dhamodharan, S. Harikrishna, P. I. Pradeepkumar and A. C. Bhasikuttan, J. Am. Chem. Soc., 2013, 135, 367–376 CrossRef CAS; (b) J. Dai, M. Carver, L. H. Hurley and D. Z. Yang, J. Am. Chem. Soc., 2011, 133, 17673–17680 CrossRef CAS; (c) D. Koirala, S. Dhakal, B. Ashbridge, Y. Sannohe, R. Rodriguez, H. Sugiyama, S. Balasubramanian and H. Mao, Nat. Chem., 2011, 3, 782–787 CrossRef CAS; (d) S. J. Xu, Q. Li, J. F. Xiang, Q. F. Yang, H. X. Sun, A. J. Guan, L. X. Wang, Y. Liu, L. J. Yu, Y. H. Shi, H. B. Chen and Y. L. Tang, Nucleic Acids Res., 2015, 43, 9575–9586 CAS; (e) R. Haudecoeur, L. Stefan, F. Denat and D. Monchaud, J. Am. Chem. Soc., 2013, 135, 550–553 CrossRef CAS; (f) M. Wang, Z. Mao, T. S. Kang, C. Y. Wong, J. L. Mergny, C. H. Leung and D. L. Ma, Chem. Sci., 2016, 7, 2516 RSC.
- (a) R. V. Brown, T. Wang, V. R. Chappeta, G. H. Wu, B. Onel, R. Chawla, H. Quijada, S. M. Camp, E. T. Chiang, Q. R. Lassiter, C. Lee, S. Phanse, M. A. Turnidge, P. Zhao, J. G. Garcia, V. Gokhale, D. Z. Yang and L. H. Hurley, J. Am. Chem. Soc., 2017, 139, 7456–7475 CrossRef CAS; (b) T. M. Ou, Y. J. Lu, C. Zhang, Z. S. Huang, X. D. Wang, J. H. Tan, Y. Chen, D. L. Ma, K. Y. Wong and J. C. Tang, J. Med. Chem., 2007, 50, 1465–1474 CrossRef CAS; (c) K. H. Leung, H. Z. He, B. He, H. J. Zhong, S. Lin, Y. T. Wang, D. L. Ma and C. H. Leung, Chem. Sci., 2015, 6, 2166 RSC.
- L. M. Resar, E. Emison, S. Kim, Q. Li, J. E. Prescott, D. Wonsey and K. Zeller, Exp. Cell Res., 1999, 253, 63–77 CrossRef.
- D. Drygin, A. Siddiqui-Jain, S. O'Brien, M. Schwaebe, A. Lin, J. Bliesath, C. B. Ho, C. Proffitt, K. Trent, J. P. Whitten, J. K. C. Lim, D. Von Hoff, K. Anderes and W. G. Rice, Cancer Res., 2009, 69, 7653–7661 CrossRef CAS.
- A. R. H. Narayan and R. Sarpong, Org. Biomol. Chem., 2012, 10, 70 RSC , and references cited therein.
- B. Q. Hu, L. X. Wang, G. Shen, Y. L. Tang and L. Yang, RSC Adv., 2015, 5, 100097 RSC.
- (a) V. Gabelica, R. Maeda, T. Fujimoto, H. Yaku, T. Murashima, N. Sugimoto and D. Miyoshi, Biochemistry, 2013, 52, 5620–5628 CrossRef CAS PubMed; (b) A. R. Faverie, A. Guédin, A. Bedrat, L. A. Yatsunyk and J. L. Mergny, Nucleic Acids Res., 2014, 1–8 Search PubMed; (c) S. J. Xu, Q. Li, J. F. Xiang, Q. F. Yang, H. X. Sun, A. J. Guan, L. X. Wang, Y. Liu, L. J. Yu, Y. H. Shi, H. B. Chen and Y. L. Tang, Sci. Rep., 2016, 6, 24793–24801 CrossRef CAS.
- L. X. Wang, Q. Shang, Q. Li, J. F. Xiang, Y. Liu, A. J. Guan, H. X. Sun, L. J. Yu and Y. L. Tang, Tetrahedron, 2015, 71, 4982 CrossRef CAS.
- T. Simonsson and M. Henriksson, Biochem. Biophys. Res. Commun., 2002, 290, 11–15 CrossRef CAS.
- T. M. Ou, J. Lin, Y. J. Lu, J. Q. Hou, J. H. Tan, S. H. Chen, Z. Li, Y. P. Li, D. Li, L. Q. Gu and Z. S. Huang, J. Med. Chem., 2011, 54, 5671–5679 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra09870g |
| This journal is © The Royal Society of Chemistry 2017 |
