Open Access Article
Open Access ArticleInfluence of bicarbonate on the abundance of microbial communities capable of reducing U(VI) in groundwater
Dianxin Li,
Nan Hu,
Yang Sui,
Dexin Ding *,
Ke Li,
Guangyue Li and
Yongdong Wang
*,
Ke Li,
Guangyue Li and
Yongdong Wang
Key Discipline Laboratory for National Defence for Biotechnology in Uranium Mining and Hydrometallurgy, University of South China, 421001 Hengyang, China. E-mail: dingdxzzz@163.com
First published on 25th October 2017
Abstract
In order to investigate the influence of different concentrations of bicarbonate on the abundance of microbial communities capable of reducing U(VI) in groundwater, 7 groups of experiments incubated with lactate and amended with 7 initial concentrations of bicarbonate (0, 5, 10, 15, 20, 25, and 30 mM), respectively, were conducted. It was found that U(VI) concentration failed to decrease below the Chinese uranium wastewater discharge standard (0.05 mg L−1) when the initial bicarbonate concentration was higher than 10 mM. During the nitrate reduction, the abundance of nitrate reduction bacteria (NRB) was 22% in B0, while the abundance of NRB was 3.4% in B10 and B15. The main functional microbial community in B0 was NRB Brevundimonas, whose abundance was over 19%. During the U(VI), sulfate, and Fe(III) reduction, the abundance of sulfate and Fe reduction bacteria (SRB and FRB) was 44% in B0, while the abundance of SRB and FRB were 26% and 27% in B10 and B15, respectively. The main functional microbial communities capable of reducing U(VI) in B0 were Cellulomonas, Desulfosporosinus, and Desulfovibrio, whose abundance was as high as 36%. The main microbial community capable of reducing U(VI) in B10 was Desulfitobacterium, and its abundance was higher than 14%. The main functional microbial communities capable of reducing U(VI) in B15 were Desulfovibrio and Geobacter, and their abundance was over 13%. The overall experimental results indicate that the higher initial bicarbonate concentration leads to the lower abundance of microbial communities capable of reducing U(VI) in groundwater.
Introduction
Uranium mining and processing in the nuclear industry have resulted in contamination of groundwater which was due to the release of uranium into the groundwater.1,2 Uranium commonly appears as soluble U(VI) or sparingly soluble U(IV) phases and exists in the anaerobic conditions of groundwater.3,4 Soluble U(VI) can migrate with the flowing groundwater. Once it accumulates in the human's body, it will be seriously harmful for people's health.5In the alkaline in situ leaching mining operation, bicarbonate is widely used as an agent to extract uranium from the uranium-bearing ores.6 CO32− or HCO3− in groundwater can help to form U–carbonate complexes, such as UO2CO3, UO2(CO3)22−, and UO2(CO3)34−, which resulted in the high mobility of U(VI).7 Van Engelen et al. demonstrated that high concentration of bicarbonate in solution promoted the formation of negatively charged uranyl–carbonate complexes and it inhibited the toxicity of U(VI) to bacteria, but high concentration of bicarbonate reduced the abundance of microbial communities.8 Bicarbonate which presents in the groundwater with neutral pH could decrease the absorption of U(VI) in sediments.9
Previous studies demonstrated that low concentration of bicarbonate was beneficial for U(VI) bioreduction in sediment, while high concentration of bicarbonate and sulfate could inhibit the reduction.10 Philip E. Long et al. found that when low concentration of bicarbonate existed in groundwater, the abundance of Ca–uranyl–carbonate complexes increased, and acetate–bicarbonate treatments could increase the rate of U(VI) reduction in the bicarbonate-impacted sediment.11 Sheng Ling et al. showed that increasing the concentration of sodium bicarbonate in the system would lead to the slower kinetics for U(VI) reduction and it would have profound effects on the speciation of U(VI) in the bacterial cell envelope.12 Obviously, these studies made it clear that high concentration of bicarbonate and sulfate would inhibit the U(VI) bioreduction, acetate–bicarbonate treatments would promote the formation of Ca–uranyl–carbonate complexes. What's more, increasing the concentration of bicarbonate would influence the reduction kinetics and the speciation of U(VI) on the bacterial cell envelope. High concentration of bicarbonate may be able to inhibit U(VI) reduction by influencing the abundance of microbial communities capable of reducing U(VI) in groundwater. However, it is still not clear what concentrations of bicarbonate affect the abundance of microbial communities capable of reducing U(VI) in groundwater. Nitrate can inhibit U(VI) bioreduction.13 Some SRB, including Desulfosporosinus and Desulfovibrio, have the ability of reducing U(VI),14,15 and some FRB, such as Geobacter, can also reduce U(VI).16 U(VI) bioreduction is often after nitrate reduction, and it usually accompanies with sulfate and iron reduction.17 Therefore, in studying what concentrations of bicarbonate affect the abundance of microbial communities capable of reducing U(VI), the impact of NRB, SRB and FRB should also be taken into consideration.
Our previous study demonstrated that high concentration of bicarbonate can inhibit uranium bioreduction, but we did not know what concentrations of bicarbonate in groundwater would affect the bioreduction of uranium and influence the abundance of microbial communities capable of reducing U(VI).18 In this paper, each microcosm was prepared with 50 g sediment and 1000 mL uranium contaminated groundwater. The sediment was taken from a monitoring well near a uranium tailings repository situated in South China, and the groundwater was also taken from the monitoring well. 7 groups of experiments, which were incubated with lactate and amended with 0, 5, 10, 15, 20, 25, and 30 mM initial concentrations of bicarbonate, respectively, were designed. Indigenous microbial communities were stimulated by sodium lactate in the anaerobic microcosms. Functional genes were quantized by qPCR. The abundance of microbial communities was analyzed using Miseq sequencing method.
Materials and methods
Sample site and sample collection
Samples were taken from a monitoring well near a uranium mill tailings repository situated in South China. The repository has a subtropical continental climate with an annual average temperature of 17.9 °C, an annual average rainfall of 1452.0 mm, an annual average evaporation capacity of 1324.5 mm, and the altitude from 210.5 m to 307.0 m above sea level. It covers an area of approximately 1.70 km2, and contains approximately 188 million t of uranium mill tailings produced by a nearby uranium mill where the uranium ore was processed by the acid leaching from 1960s to 1990s.19 Large amounts of waste residue and waste water, which contain nitrate, sulfate, (bi)carbonate, Ca2+, Mg2+, U and other heavy metal ions, and uranium tailings, were discharged into the tailings repository.2 These discharged wastes have heavily polluted the surface environment and underground water.Uranium contaminated groundwater was taken from the monitoring well described above, whose water level was at the depth of 10 m below the surface. The dissolved oxygen (DO) concentration of the contaminated groundwater was below 0.3 mg L−1. The sediment samples were collected from the monitoring well, either. All the samples were immediately taken to the laboratory with ice bags under anaerobic conditions, and they were preserved in an anaerobic glove box at the temperature which was lower than 4 °C until use.20 Geochemical compositions of the sediment and groundwater samples were detected before incubation, and the results were shown in Table 1.
| Physical and chemical characteristics | Sediment (g kg−1) | Groundwater (mg L−1) |
|---|---|---|
| Total Fe | 7.6 | 0.1 |
| Mg | 4.5 | 10.2 |
| Ca | 1.7 | 106.5 |
| Mn | 0.5 | 5.3 |
| K | 2.2 | 26.2 |
| Na | 0.6 | 47.1 |
| Zn | 1.0 | 0.3 |
| Cr | 0.02 | 0.5 |
| U | 0.1 | 6.5 |
| SO42− | — | 2790.5 |
| HCO3− | — | 0.6 |
| DO | — | 0.3 |
| COD | — | 11.0 |
| pH | 6.0 | 6.5 |
Microcosm experiment
Experimental groups were described above. Each group of the experiment was conducted in triplicate.12 For each of 21 sterilized microcosms, 50 g sediment and 1000 mL contaminated groundwater were both added into each of them, respectively,21 and they were divided into 7 groups as previously described, with each group having three microcosms. 0, 0.42, 0.84, 1.26, 1.68, 2.10 and 2.52 g of sodium bicarbonate (AR) were dissolved into each group of microcosms, and each group was named as B0, B10, B15, B20, B25, B30, respectively. To make the microcosms reach balance, they were kept motionless for 24 h. The pH of solutions in all the microcosms was then adjusted to 7.0 with diluted HCl or NaOH.12,22 After that, 2 mL sterile sodium lactate was added into each microcosm, and all microcosms were bubbled with the mixed gas of 95% N2 and 5% H2 for half an hour to remove DO in them. Finally, all microcosms were incubated at 20 °C in dark in the anaerobic glove box.2The interval for taking solution samples was set at 3 days. All the solution samples were collected through the indwelling sampling pump of each microcosm, and they were passed through a 2 μm diameter filter for geochemical analysis. Sediment samples were collected at day 10, day 25, and day 64 after incubation for microbiological analysis.
Analytical techniques
The total U concentration was determined by ICP-MS (Agilent Technologies 7700 Series. USA), and its measurement error was ±2 ng L−1. The total Fe concentration was determined by atomic absorption spectrophotometer (PerkinElmer, PinAAcle900F, USA), and its measurement error was ±0.03 mg L−1. The NO3−, SO42−, and CO32− concentrations were determined by ion chromatography (ICS-900. USA), and their measurement errors were ±0.05 mg L−1, ±0.1 mg L−1 and ±0.1 mg L−1, respectively. The COD concentration was determined by COD rapid determination instrument (AWM. France), and its measurement error was ±0.1 mg L−1. The metal cation concentrations were also determined by atomic absorption spectrophotometer.18 The DO concentration was determined by dissolved oxygen meters (INESA JPSJ-605, China). pH was determined by pH-meter (INESA JPSJ-3F, China).qPCR analyses
The TM1389, dsrB and gltA, qPCR analyses were performed using SYBR green-based detection chemistry, and their primer pairs were BACT1369/PROK1492R, DSRq1F/DSRq1R and CS375F/CS598R, respectively.23,24 25 μL qPCR mixtures contained 12.5 μL of SybrGreen qPCR Master Mix, 0.5 μL of 10 μM concentrations of primer F, 0.5 μL of 10 μM concentrations of primer R, 9.5 μL of ddH2O, and 2 μL cDNA templates. All qPCR experiments were conducted in triplicate. PCR amplification was performed with ABI7500 real-time PCR system.25 The thermal cycling for activation included the initial heating at 95 °C for 10 min, and the 40 cycles with each one including melting at 95 °C for 15 s and annealing or extending at 60 °C for 1 min. Standard curves were drawn using a 10-fold dilution series of quantified plasmid TM1389, dsrB, and gltA gene, respectively.16S rRNA gene amplicon pyrosequencing
The triplicate of 16S rRNA genes was amplified from each DNA extract. Two primers (338F [ACTCCTACGGGAGGCAGCA] and 806R [GGACTACHVGGGTWTCTAAT]) were designed for PCR amplification.26 Cycling was performed using a TransGenAP221-02 (TransStart Fastpfu DNA polymerase, 20 μL). PCRs were performed in a final volume of 20 μL mixture, which was obtained by mixing 4 μL of 5×FastPfu Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of forward Primer (5 μM), 0.8 μL of reverse Primer (5 μM), 0.4 μL of FastPfu Polymerase, and 2 μL of template DNA together, and diluting the mixture with sterile ddH2O to 20 μL. The amplicon process consists of three steps: (a) the initial denaturation at 95 °C for 3 min; (b) 27 cycles with each one including heating at 95 °C for 30 s, at 55 °C for 30 s, and at 72 °C for 45 s; and (c) a final extension at 72 °C for 10 min, and at 10 °C.27 Amplifications were run by PCR instrument (ABI GeneAmp® 9700).Sequences were clustered into OTUs (Operational Taxonomic Units) using Usearch software (version 7.1 http://drive5.com/uparse/). In order to obtain the taxonomic information of each OTU cluster, 97% of similar levels of the OTU were subjected to taxonomic-based analysis using the pipeline initial process of the RDP pyrosequencing pipeline (release 11.3 http://rdp.cme.msu.edu/).28 The community composition of each sample was classified as phylum and genus levels by statistical analysis. Based on the statistical analysis results, diversity and abundance of microbial communities were determined for each sample. Furthermore, the heat-map was drawn to show the consequences.
Results and discussion
Geochemical characteristics of sediment and groundwater samples
To illustrate the geochemical composition of microcosms, the previous sediment and groundwater samples were detected. The measured data were shown in Table 1. Iron was very abundant in sediments, while it was not detected in groundwater. Therefore, microbial communities would reduce ferric ions in sediment to ferrous ones, and ferrous and ferric cations would migrate to groundwater, accompanied by uranium reduction.29 Sediment and groundwater were enriched with calcium, and this resulted in the Ca–uranyl–carbonate complexes with lactate consumption. Thereby, those complexes increased the uranium mobility in groundwater (Table 2).30 As buffer, the low concentration of bicarbonate in groundwater could decrease the toxicity of heavy metals to microbial communities.31 The carbonate of previous groundwater was in very low concentration. However, after 64 days of incubation, the concentration of carbonate was raised to a high level due to the consumption of lactate. The higher concentration of bicarbonate previously added, the higher the carbonate concentration would be generated (Table 2). The variation of nitrate, uranium, sulfate, pH, and COD will be discussed in the text below.| Group | Carbonate concentration (mg L−1) |
|---|---|
| Previous groundwater | 14.7 |
| B0 | 1152.2 |
| B5 | 1564.6 |
| B10 | 1865.4 |
| B15 | 2131.7 |
| B20 | 2334.2 |
| B25 | 2443.4 |
| B30 | 2532.5 |
Variation of nitrate, uranium, sulfate, iron, pH and COD in solution
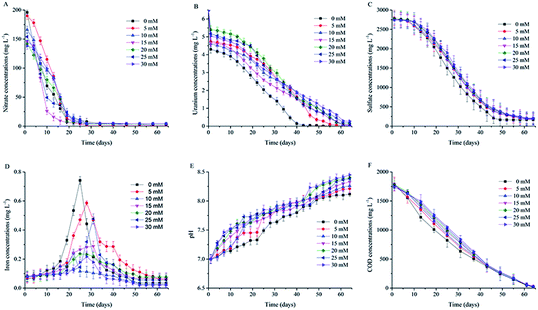
Fig. 1 shows that nitrate concentrations began to decrease after the electron donor was added. In the first 10 days, nitrate concentrations of all groups decreased fast, and the nitrate reduction speed decreased fastest in the group without bicarbonate (B0). This phenomenon indicates that low concentration of bicarbonate is in favor of nitrate reduction.32 Nitrate concentrations declined to the detection limit at day 22 in all groups, and they kept unchanged after that.Uranium concentrations of all experimental groups declined from 6.5 mg L−1 to 4.2–5.4 mg L−1 after all the microcosms were kept motionless for 24 h. The uranium concentration increased with the increase of the initial bicarbonate concentration, but it does not increase proportionally with the bicarbonate concentration. The variation of uranium concentrations in the microcosms is shown in Fig. 1B. It can be observed that U(VI) was reduced slowly in the first 12 days due to nitrate inhibition,33 and since then the concentration of U(VI) decreased fast. For these groups, the uranium concentration decreased faster in the control group (B0) than in the other groups. The final concentrations of U(VI) decreased bellow the maximum contaminant limit for uranium wastewater discharge (0.05 mg L−1) by Chinese Environmental Protection Ministry (GB 23727-2009) in B0, B5, B10, and they remained stable. However, uranium concentrations remained at 0.1 to 0.28 mg L−1 in other groups depending on the bicarbonate concentration. This phenomenon indicates that U(VI) concentrations decreased more quickly in the microcosms amended with low concentrations of bicarbonate than in those amended with high concentrations of bicarbonate; what's more, the U(IV) was more stable in the microcosms amended with low concentrations of bicarbonate. These results obviously illustrated that bicarbonate had the inhibitory effect on U(VI) bioreduction.12,18 The reason why the final concentration of U(VI) was lower in B0 than in other groups was that microbial communities consumed the electron donors and the metabolic CO32− was produced by them. The metabolic CO32− desorbed the U(VI) that was previously adsorbed by sediments, which was also reduced by microbial communities.11 However, bicarbonate concentrations were too high for microbial communities capable of reducing U(VI) in other groups. Their reduction ability was inhibited by high bicarbonate concentrations, and they could not decrease the desorbed U(VI) concentrations to the lower level.
Fig. 1C shows that sulfate concentrations in the microcosms decreased slowly in the first 12 days, and then, they decreased faster. The concentration of sulfate in B0 stopped declining and kept stable at day 49. However, the sulfate concentrations in other groups continued declining until day 58. Fig. 1B and C show that U(VI) reduction accompanied with sulfate reduction. The reason for this was that some kinds of SRB could reduce U(VI) while they were reducing sulfate.34 At the same time, it is clear that the activities of the indigenous microbial communities were inhibited significantly by the high concentration of bicarbonate.35
Total iron concentrations were very low before incubation in all groups. After incubation, they increased to peak values ranging from 0.21 to 0.73 mg L−1 between day 22 and day 31. Then, they began to decline (Fig. 1D). The reason for the increase was that iron was reduced by FRB in sediment; and the reduction resulted in the increase of total iron concentration.36 The subsequent decrease was due to the precipitation of biogenic Fe(II).37 Total iron concentrations increased faster in the low bicarbonate concentration groups (0 mM and 5 mM) than in other groups, and their peak values were higher. This can be attributed to the growth promotion of the low bicarbonate concentration on the indigenous microbial communities.12
It can be seen that the pH of all the microcosms increased to 8.12–8.45 after 64 days of incubation in Fig. 1E. The reason for this was that the microbial communities continuously consumed electron donors and produced carbonate in the solutions (Table 2).11 Significantly, the higher the previous bicarbonate concentrations, the higher the pH would be.
Fig. 1F shows the variation of COD concentrations in the microcosms. COD concentrations decreased quickly as soon as incubation began. The COD was consumed faster in B0 than in other groups since the microbial communities were more active in B0 before the sulfate reduction. After the sulfate was completely reduced, COD was only used to maintain the activities of the microbial communities.38
Based on the above results, we chose the control group (B0), the group whose U(VI) concentration could decrease to 0.05 mg L−1 (B10), and the group whose U(VI) concentration could not decrease to 0.05 mg L−1 (B15) for, qPCR, and PCR amplification analyses.
qPCR results
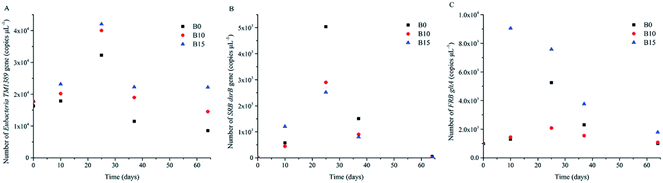
TM1389, dsrB, and gltA are the gene of Eubacteria, SRB, and FRB, respectively. The variations of the numbers of TM1389, dsrB, and gltA can reflect their growth.25,39 In order to quantify TM1389, dsrB and gltA, qPCR was used to analyze sediment samples at day 0, 10, 25, 37, and 64, respectively. Fig. 2 shows that the growth of Eubacteria, SRB and gltA was detected at the five key time points. During uranium, iron and sulfate reducing process, the number of TM1389 increased over time, which indicated that microbial communities were growing during this process.40 Afterwards, it began to decrease (Fig. 2A), and the activities of the microbial communities also decreased. The number of dsrB was detected to be the highest at day 25 in B0, and it reached 5 × 103 copies per μL. However, the numbers of dsrB in B10 and B15 were just half of that in B0, which indicated that the lactate stimulated SRB were more active in B0 than in B10 and B15. The transcriptional peak of dsrB was agreeable with the result of 16S rRNA gene pyrosequencing, and they showed that the dominant genus of SRB, including Desulfitobacterium, Desulfobulbus, Desulfosporosinus, Desulfovibrio, and Desulfurispora, were detected.26,41,42 The number of gltA was detected to be very high at day 10 in B15, while it was tested to be very low in B0 and B10. The transcriptional peak of gltA was in agreement with the result of the 16S rRNA gene pyrosequencing, and they showed that Geobacter and Geothrix were detected at day 10 in B15.25,43 At day 25, the number of gltA increased to 5.2 × 103 copies per μL in B0, while the number of gltA decreased a little in B15. The main function microbial community was Geobacter in B0 and B15. This phenomenon indicated that the lactate stimulated FRB were more active in B15 than in B0 and B10.Abundance of the functionalized indigenous microbial communities during bioreduction
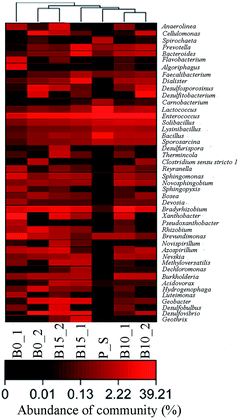
In order to study the abundance of the functionalized indigenous microbial communities in the microcosms amended with lactate, the amplification microarray was used to analyze the preliminary sediment and the sediments of B0, B10 and B15 which were sampled at day 10 and 25, respectively. Fig. 3 illustrates that the compositions of microbial communities whose abundance is more than 1%.44 Microbial communities, including Bacillus, Brevundimonas, Carnobacterium, Enterococcus, Lactococcus, Lysinibacillus, Solibacillus, Spirochaeta, and Sporosarcina, were detected in the preliminary sediment. Among these microbial communities, only Brevundimonas was reported to have the ability of reducing nitrate,45 which indicated that functionalized indigenous microbial communities cannot be stimulated without adding electron donor.At day 10, microbial communities, including Bradyrhizobium, Enterococcus, Sphingomonas, and Xanthobacter, were simultaneously found in the three groups. Among these microbial communities, Sphingomonas is able to bioprecipitate uranium from alkaline solutions,46 and Bradyrhizobium can precipitate metallic cations.47 Microbial communities, including Algoriphagus, Anaerolinea, Azospirillum, Brevundimonas, Devosia, Flavobacterium, Luteimonas, Pseudoxanthobacter, Reyranella, Sphingopyxis, and Rhizobium, were also detected in B0. Of which, Azospirillum, Brevundimonas, and Rhizobium are nitrate reducing bacteria.45,48,49 Some microbial communities were detected in B10, including Bacillus, Bacteroides, Brevundimonas, Burkholderia, Dialister, Lactococcus, Lysinibacillus, Nevskia, Novosphingobium, Prevotella, Solibacillus, and Sporosarcina. Among aforementioned microbial communities, Brevundimonas is a kind of nitrate reducing bacteria which previously mentioned, so is Novosphingobium.50 Bacteroides is a kind of enzyme capable of reducing sulfate,51 and Burkholderia can reduce the toxicity of nickel and uranium.52 And the microbial communities were detected in B15, including Acidovorax, Bacteroides, Burkholderia, Dechloromonas, Desulfovibrio, Dialister, Faecalibacterium, Geobacter, Geothrix, Lactococcus, Methyloversatilis, Nevskia, Prevotella, and Solibacillus. Thereinto, Acidovorax has the ability to remove uranium from solution,53 Bacteroides is a kind of enzyme capable of reducing sulfate which previously mentioned, Burkholderia can reduce nickel and uranium toxicity which is previously stated, Dechloromonas can reduce nitrate and oxidize Fe(II),54 Desulfovibrio is a sulfate reducing bacteria which can reduce sulfate and U(VI),15 Geobacter is an Fe(III)- and U(VI)-reducing bacterium,16 Geothrix is a kind of iron reducing bacteria,55 and Methyloversatilis can reduce nitrate to nitrite.56 During this period, nitrate reduction proceeded faster in B0 than in the other two groups (Fig. 1A). The reason for this was that the abundance of the nitrate reducing bacteria was over 20% in B0, while the abundance of the nitrate reducing bacteria was only 3.4% in B10 and B15. The main functional microbial community was the NRB Brevundimonas in B0, and its abundance was up to 19%. Although SRB and FRB were detected in B15, the nitrate inhibited the reduction of U(VI), sulfate, and Fe(III) (Fig. 1B–D).17
At day 25, many microbial communities, including Azospirillum, Bradyrhizobium, Burkholderia, Cellulomonas, Desulfosporosinus, Desulfovibrio, Enterococcus, and Solibacillus, were simultaneously detected in the three groups. The functional microbial communities including Azospirillum, Bradyrhizobium, and Desulfovibrio have been described above. Cellulomonas is a kind of dissimilatory reduction bacteria capable of reducing Cr(VI), Fe(III), and U(VI),57 and Desulfosporosinus is a kind of sulfate reduction bacteria capable of reducing sulfate and U(VI).14 Some other microbial communities were detected in B0, including Bosea, Brevundimonas, Clostridium_sensu_stricto_1, Desulfitobacterium, Geobacter, Hydrogenophaga, and Rhizobium. The functions of Brevundimonas, Geobacter, and Rhizobium, have been described above. Clostridium_sensu_stricto_1 can reduce nitrate and nitrite,58 and Desulfitobacterium can reduce sulfate and U(VI).59 The abundance of SRB and FRB was 44% in B0. The main functional microbial communities capable of reducing U(VI) were Cellulomonas, Desulfosporosinus, and Desulfovibrio, whose abundance was up to 36%. Many other microbial communities, including Bacillus, Bacteroides, Desulfitobacterium, Dialister, Lactococcus, Nevskia, Novosphingobium, Prevotella, and Sporosarcina, were found in B10. All the functional microbial communities including Bacteroides, Novosphingobium, and Desulfitobacterium, have been described before. The abundance of SRB and FRB was 27% in B10. The main functional microbial community capable of reducing U(VI) was Desulfitobacterium, and its abundance was up to 14%. The microbial communities including Acidovorax, Anaerolinea, Bacillus, Brevundimonas, Desulfobulbus, Desulfurispora, Geobacter, Methyloversatilis, Novispirillum, Rhizobium, Sphingomonas, Thermincola, and Xanthobacter were detected in B15. Among these microbial communities, the functional microbial communities, including Acidovorax, Brevundimonas, Geobacter, Methyloversatilis, Sphingomonas, and Rhizobium, have been described previously. Desulfobulbus and Desulfurispora are sulfate reduction bacteria.42,60 The abundance of SRB and FRB was 44% in B15. The main functional microbial communities capable of reducing U(VI) were Desulfovibrio and Geobacter, and their abundance was up to 13%. During this period, U(VI) reduction proceeded faster in B0 than in B10 and B15 (Fig. 1B). The reason for this was that the abundance of SRB and FRB capable of reducing U(VI) was higher in B0 than in B10 and B15. Sulfate reduction speed was also higher in B0 than in B10 and B15 (Fig. 1C), which was due to the fact that the abundance of SRB was higher in B0 than in the other two groups. Since the activity of FRB was the highest in B0, the concentration of the total iron in the solution was also the highest in it. These results were agreeable with the whole reduction process (Fig. 1).
Conclusions
7 groups of experiments with 7 initial concentrations of bicarbonate (0, 5, 10, 15, 20, 25, and 30 mM) were conducted, and the results showed that the U(VI) concentration could not be decreased below the Chinese uranium wastewater discharge standard (0.05 mg L−1) when the initial bicarbonate concentration was higher than 10 mM. Miseq sequencing results showed that while the nitrate was being reduced, the main functional microbial community was a kind of NRB Brevundimonas in B0, and its abundance was 19%. Moreover, while U(VI), sulfate and Fe(III) were being reduced, it was found that the main functional microbial communities capable of reducing U(VI) were Cellulomonas, Desulfosporosinus, and Desulfovibrio in B0 and their abundance was higher than 36%, that the main functional microbial communities capable of reducing U(VI) was Desulfitobacterium in B10 and its abundance was 14%, and that the main function microbial communities capable of reducing U(VI) were Desulfovibrio and Geobacter in B15 and their abundance was over 13%. The experimental results indicate that the higher initial bicarbonate concentration leads to the lower abundance of microbial communities capable of reducing U(VI) in groundwater.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. U1401231 and 51274124) and the Defense Industrial Technology Development Program (No. B3720132001).References
- C. Hwang, W. Wu, T. J. Gentry, J. Carley, G. A. Corbin, S. L. Carroll, D. B. Watson, P. M. Jardine, J. Zhou, C. S. Criddle and M. W. Fields, ISME J., 2009, 3, 47–64 CrossRef CAS PubMed.
- D. Ding, F. Xu, N. Hu, S. Li, X. Tan and G. Li, J. Radioanal. Nucl. Chem., 2015, 303, 925–933 CrossRef CAS.
- D. M. Singer, J. M. Zachara and G. E. Brown Jr, Environ. Sci. Technol., 2009, 43, 630–636 CrossRef CAS PubMed.
- M. J. Marshall, A. S. Beliaev, A. C. Dohnalkova, D. W. Kennedy, L. Shi, Z. Wang, M. I. Boyanov, B. Lai, K. M. Kemner, J. S. McLean, S. B. Reed, D. E. Culley, V. L. Bailey, C. J. Simonson, D. A. Saffarini, M. F. Romine, J. M. Zachara and J. K. Fredrickson, PLoS Biol., 2006, 4, e268 Search PubMed.
- L. M. Camacho, S. Deng and R. R. Parra, J. Hazard. Mater., 2010, 175, 393–398 CrossRef CAS PubMed.
- E. A. Santos and A. C. Q. Ladeira, Environ. Sci. Technol., 2011, 45, 3591–3597 CrossRef CAS PubMed.
- S. Regenspurg, D. Schild, T. Schäfer, F. Huber and M. E. Malmström, Appl. Geochem., 2009, 24, 1617–1625 CrossRef CAS.
- M. R. VanEngelen, E. K. Field, R. Gerlach, B. D. Lee, W. A. Apel and B. M. Peyton, Environ. Toxicol. Chem., 2010, 29, 763–769 CrossRef CAS PubMed.
- L. Sheng and J. B. Fein, Chem. Geol., 2013, 358, 15–22 CrossRef CAS.
- W. Luo, W. Wu, T. Yan, C. S. Criddle, P. M. Jardine, J. Zhou and B. Gu, Appl. Microbiol. Biotechnol., 2007, 77, 713–721 CrossRef CAS PubMed.
- P. E. Long, K. H. Williams, J. A. Davis, P. M. Fox, M. J. Wilkins, S. B. Yabusaki, Y. Fang, S. R. Waichler, E. S. F. Berman, M. Gupta, D. P. Chandler, C. Murray, A. D. Peacock, L. Giloteaux, K. M. Handley, D. R. Lovley and J. F. Banfield, Geochim. Cosmochim. Acta, 2015, 150, 106–124 CrossRef CAS.
- L. Sheng and J. B. Fein, Environ. Sci. Technol., 2014, 48, 3768–3775 CrossRef CAS PubMed.
- W. Wu, J. Carley, S. J. Green, J. Luo, S. D. Kelly, J. V. Nostrand, K. Lowe, T. Mehlhorn, S. Carroll, B. Boonchayanant, F. E. Löfller, D. Watson, K. M. Kemner, J. Zhou, P. K. Kitanidis, J. E. Kostka, P. M. Jardine and C. S. Criddle, Environ. Sci. Technol., 2010, 44, 5104–5111 CrossRef CAS PubMed.
- E. J. O'Loughlin, S. D. Kelly, R. E. Cook, R. Csencsits and K. M. Kemner, Environ. Sci. Technol., 2003, 37, 721–727 CrossRef.
- R. K. Sani, B. M. Peyton and A. Dohnalkova, Environ. Toxicol. Chem., 2006, 25, 1231–1238 CrossRef CAS PubMed.
- O. Prakash, T. M. Gihring, D. D. Dalton, K. Chin, S. J. Green, D. M. Akob, G. Wanger and J. E. Kostka, Int. J. Syst. Evol. Microbiol., 2010, 60, 546–553 CrossRef CAS PubMed.
- D. M. Akob, L. Kerkhof, K. Küsel, D. B. Watson, A. V. Palumbo and J. E. Kostka, Appl. Environ. Microbiol., 2011, 77, 8197–8200 CrossRef CAS PubMed.
- D. Li, N. Hu, D. Ding, S. Li, G. Li and Y. Wang, J. Radioanal. Nucl. Chem., 2016, 307, 1011–1019 CrossRef CAS.
- G. Li, N. Hu, D. Ding, J. Zheng, Y. Liu, Y. Wang and X. Nie, Bull. Environ. Contam. Toxicol., 2011, 86, 646–652 CrossRef CAS PubMed.
- D. S. Alessi, J. S. Lezama-Pacheco, N. Janot, E. I. Suvorova, J. M. Cerrato, D. E. Giammar, J. A. Davis, P. M. Fox, K. H. Williams, P. E. Long, K. M. Handley, R. Bernier-Latmani and J. R. Bargar, Environ. Sci. Technol., 2014, 48, 12842–12850 CrossRef CAS PubMed.
- Y. Suzuki, S. D. Kelly, K. M. Kemner and J. F. Banfield, Appl. Environ. Microbiol., 2003, 69, 1337–1346 CrossRef CAS PubMed.
- D. S. Alessi, J. S. Lezama-Pacheco, J. E. Stubbs, M. Janousch, J. R. Bargar, P. Persson and R. Bernier-Latmani, Geochim. Cosmochim. Acta, 2014, 131, 115–127 CrossRef CAS.
- B. K. Amos, Y. Sung, K. E. Fletcher, T. J. Gentry, W. Wu, C. S. Criddle, J. Zhou and F. E. Löffler, Appl. Environ. Microbiol., 2007, 73, 6898–6904 CrossRef CAS PubMed.
- D. E. Holmes, L. Giloteaux, A. K. Chaurasia, K. H. Williams, B. Luef, M. J. Wilkins, K. C. Wrighton, C. A. Thompson, L. R. Comolli and D. R. Lovley, ISME J., 2015, 9, 333–346 CrossRef CAS PubMed.
- M. Miletto, K. H. Williams, A. L. N'Guessan and D. R. Lovley, Appl. Environ. Microbiol., 2011, 77, 6502–6509 CrossRef CAS PubMed.
- M. B. Smith, A. M. Rocha, C. S. Smillie, S. W. Olesen, C. Paradis, L. Wu, J. H. Campbell, J. L. Fortney, T. L. Mehlhorn, K. A. Lowe, J. E. Earles, J. Phillips, S. M. Techtmann, D. C. Joyner, D. A. Elias, K. L. Bailey, R. A. Hurt, S. P. Preheim, M. C. Sanders, J. Yang, M. A. Mueller, S. Brooks, D. B. Watson, P. Zhang, Z. He, E. A. Dubinsky, P. D. Adams, A. P. Arkin, M. W. Fields, J. Zhou, E. J. Alm and T. C. Hazen, mBio, 2015, 6, e00326 CrossRef CAS PubMed.
- E. L. Brodie, T. Z. DeSantis, D. C. Joyner, S. M. Baek, J. T. Larsen, G. L. Andersen, T. C. Hazen, P. M. Richardson, D. J. Herman, T. K. Tokunaga, J. M. Wan and M. K. Firestone, Appl. Environ. Microbiol., 2006, 72, 6288–6298 CrossRef CAS PubMed.
- L. Oberauner, C. Zachow, S. Lackner, C. Högenauer, K. Smolle and G. Berg, Sci. Rep., 2013, 3, 1413 CrossRef PubMed.
- E. R. Gilson, S. Huang and P. R. Jaffé, Biodegradation, 2015, 26, 475–482 CrossRef CAS PubMed.
- L. Sheng, J. Szymanowski and J. B. Fein, Geochim. Cosmochim. Acta, 2011, 75, 3558–3567 CrossRef CAS.
- R. K. Sani, B. M. Peyton and A. Dohnalkova, Water Res., 2008, 42, 2993–3002 CrossRef CAS PubMed.
- P. Larese-Casanova, S. B. Haderlein and A. Kappler, Geochim. Cosmochim. Acta, 2010, 74, 3721–3734 CrossRef CAS.
- S. K. Chatterjee, I. Bhattacharjee and G. Chandra, J. Hazard. Mater., 2010, 175, 117–125 CrossRef CAS PubMed.
- E. Cardenas, W. Wu, M. B. Leigh, J. Carley, S. Carroll, T. Gentry, J. Luo, D. Watson, B. Gu, M. Ginder-Vogel, P. K. Kitanidis, P. M. Jardine, J. Zhou, C. S. Criddle, T. L. Marsh and J. M. Tiedje, Appl. Environ. Microbiol., 2010, 76, 6778–6786 CrossRef CAS PubMed.
- J. Luo, F. Weber, O. A. Cirpka, W. Wu, J. L. Nyman, J. Carley, P. M. Jardine, C. S. Criddle and P. K. Kitanidis, J. Contam. Hydrol., 2007, 92, 129–148 CrossRef CAS PubMed.
- M. B. Leigh, W. Wu, E. Cardenas, O. Uhlik, S. Carroll, T. Gentry, T. L. Marsh, J. Zhou, P. Jardine, C. S. Criddle and J. M. Tiedje, Front. Environ. Sci. Eng., 2015, 9, 453–464 CrossRef CAS.
- H. S. Moon, L. McGuinness, R. K. Kukkadapu, A. D. Peacock, J. Komlos, L. J. Kerkhof, P. E. Long and P. R. Jaffé, Water Res., 2010, 44, 4015–4028 CrossRef CAS PubMed.
- M. D. Tucker, L. L. Barton and B. M. Thomson, J. Ind. Microbiol. Biotechnol., 1998, 20, 13–19 CrossRef CAS PubMed.
- M. Barlett, H. S. Moon, A. A. Peacock, D. B. Hedrick, K. H. Williams, P. E. Long, D. Lovley and P. R. Jaffe, Biodegradation, 2012, 23, 535–546 CrossRef CAS PubMed.
- H. S. Moon, L. McGuinness, R. K. Kukkadapu, A. D. Peacock, J. Komlos, L. J. Kerkhof, P. E. Long and P. R. Jaffé, Water Res., 2010, 44, 4015–4028 CrossRef CAS PubMed.
- T. M. Gihring, G. Zhang, C. C. Brandt, S. C. Brooks, J. H. Campbell, S. Carroll, C. S. Criddle, S. J. Green, P. Jardine, J. E. Kostka, K. Lowe, T. L. Mehlhorn, W. Overholt, D. B. Watson, Z. Yang, W. Wu and C. W. Schadt, Appl. Environ. Microbiol., 2011, 77, 5955–5965 CrossRef CAS PubMed.
- Z. Yi, K. Tan, A. Tan, Z. Yu and S. Wang, Int. Biodeterior. Biodegrad., 2007, 60, 258–266 CrossRef CAS.
- D. L. Cologgi, A. M. Speers, B. A. Bullard, S. D. Kelly and G. Reguera, Appl. Environ. Microbiol., 2014, 80, 6638–6646 CrossRef PubMed.
- D. P. Chandler, C. Knickerbocker, L. Bryant, J. Golova, C. Wiles, K. H. Williams, A. D. Peacock and P. E. Long, Appl. Environ. Microbiol., 2013, 79, 799–807 CrossRef CAS PubMed.
- S. Kavitha, R. Selvakumar, M. Sathishkumar, K. Swaminathan, P. Lakshmanaperumalsamy, A. Singh and S. K. Jain, Water Sci. Technol., 2009, 60, 517 CrossRef CAS PubMed.
- K. S. Nilgiriwala, A. Alahari, A. S. Rao and S. K. Apte, Appl. Environ. Microbiol., 2008, 74, 5516–5523 CrossRef CAS PubMed.
- J. Corzo, M. León-Barrios, V. Hernando-Rico and A. M. Gutierrez-Navarro, Appl. Environ. Microbiol., 1994, 60, 4531–4536 CAS.
- H. M. El-Komy, M. A. Hamdia and G. K. Abd El-Baki, Biol. Plant., 2003, 46, 281–287 CrossRef CAS.
- J. A. G. Silveira, J. C. S. Matos, V. M. Cecatto, R. A. Viegas and J. T. A. Oliveira, Environ. Exp. Bot., 2001, 46, 37–46 CrossRef CAS PubMed.
- D. Zhang, Y. Liu and H. Huang, Antonie van Leeuwenhoek, 2017, 110, 19–25 CrossRef CAS PubMed.
- H. Setoyama, A. Imaoka, H. Ishikawa and Y. Umesaki, Microbes Infect., 2003, 5, 115–122 CrossRef PubMed.
- J. D. Van Nostrand, T. J. Khijniak, B. Neely, M. A. Sattar, A. G. Sowder, G. Mills, P. M. Bertsch and P. J. Morris, Environ. Sci. Technol., 2007, 41, 1877–1882 CrossRef CAS PubMed.
- U. Gerber, I. Zirnstein, E. Krawczyk-Bärsch, H. Lünsdorf, T. Arnold and M. L. Merroun, J. Hazard. Mater., 2016, 317, 127–134 CrossRef CAS PubMed.
- A. Chakraborty and F. Picardal, World J. Microbiol. Biotechnol., 2013, 29, 617–623 CrossRef CAS PubMed.
- K. P. Nevin and D. R. Lovley, Appl. Environ. Microbiol., 2002, 68, 2294–2299 CrossRef CAS PubMed.
- I. Mustakhimov, M. G. Kalyuzhnaya, M. E. Lidstrom and L. Chistoserdova, J. Bacteriol., 2013, 195, 2207–2211 CrossRef CAS PubMed.
- R. Sani, B. Peyton, W. Smith, W. Apel and J. Petersen, Appl. Microbiol. Biotechnol., 2002, 60, 192–199 CrossRef CAS PubMed.
- X. F. Hospital, E. Hierro, S. Stringer and M. Fernández, Int. J. Food Microbiol., 2016, 218, 66–70 CrossRef CAS PubMed.
- K. E. Fletcher, M. I. Boyanov, S. H. Thomas, Q. Wu, K. M. Kemner and F. E. Löffler, Environ. Sci. Technol., 2010, 44, 4705–4709 CrossRef CAS PubMed.
- A. H. Kaksonen, S. Spring, P. Schumann, R. M. Kroppenstedt and J. A. Puhakka, Int. J. Syst. Evol. Microbiol., 2007, 57, 1089–1094 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |