Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
An in vitro test system for evaluation of SCAP–SREBP pathway inhibitory activities of Traditional Chinese Medicines†
Jianzong Li‡
 a,
Xin Wang‡a,
Yuan Yuan‡a,
Xin Longa,
Jinku Bao*abc and
Xin Li*b
a,
Xin Wang‡a,
Yuan Yuan‡a,
Xin Longa,
Jinku Bao*abc and
Xin Li*b
aKey Laboratory of Bio-Resource and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu 610065, Sichuan, P. R. China. E-mail: baojinku@scu.edu.cn
bState Key Laboratory of Oral Diseases, National Clinical Research Center for Oral Diseases, West China Hospital of Stomatology, Sichuan University, Chengdu, China. E-mail: lixin0914071@126.com
cState Key Laboratory of Biotherapy/Collaborative Innovation Center for Biotherapy, West China Hospital, Sichuan University, Chengdu 610041, China
First published on 23rd October 2017
Abstract
Cellular cholesterol levels are controlled by several transcription factors, including sterol-regulatory element-binding proteins (SREBPs). SREBP cleavage-activating protein (SCAP) acts as a cholesterol sensor responsible for regulating cellular cholesterol balance. We built an in vitro detection system to explore the SCAP–SREBP pathway inhibitory activities of Traditional Chinese Medicine (TCMs). Firstly, a bipartite graph composed of human disease and TCMs linked by medicine-disease binary associations was built. A subnetwork that contained about 100 kinds of medicines concerned with lipid metabolism was identified. Then a stable HepG2 cell line coupled to the firefly luciferase reporter gene was established to evaluate the SREBP pathway inhibitory activities of these medicines. Furthermore, the effects of synthesizing of endogenous cholesterol were investigated using reverse transcription polymerase chain reaction (RT-PCR) assay and amphotericin B cell model. Results revealed that crude extracts of Rhizoma Alismatis and Semen Cassiae showed favorable efforts on inhibiting the activities of SREBP pathway and the biosynthesis of cholesterol. Taken together, Rhizoma Alismatis and Semen Cassiae may be potential source of lipid-lowing drugs in the further drug discovery.
1 Introduction
Cholesterol is an essential cell membrane component and a precursor of steroid hormones and bile acids.1 Maintenance of cholesterol homeostasis is essential to human health. Excess cholesterol is lethal because it deposits in arteries, initiating hyperlipemia, atherosclerosis, or other diseases.2 The sterol-response element-binding proteins (SREBPs) regulate the expression of genes involved in cholesterol biosynthesis.3 Genes associated with cholesterol biosynthetic pathway include HMG-CoA synthase, HMG-CoA reductase, farnesyl diphosphate synthase and squalene synthase.4 When the cholesterol levels are low (<5% of the endoplasmic reticulum lipids), the cholesterol-free form of SREBP cleavage-activating protein (SCAP) is released from the insulin-inducible gene product (Insig) and the SCAP–SREBP complex is transported from the endoplasmic reticulum (ER) to the Golgi in coatomer protein II vesicles.5–7 Once the cholesterol levels are high (>5%), SCAP senses the excess cholesterol through its membranous sterol-sensing domain, changing its conformation in such a way that the SCAP/SREBP complex is no longer incorporated into ER transport vesicles.8–10The biosynthesis of cholesterol is tightly regulated by SREBPs.9,10 Accordingly, many researches have focused on development of new cholesterol-lowing medicines based upon the characteristics of signal transduction mechanism in which SREBPs regulating cholesterol level intracellular.11–13 Masami Shimizu-Albergine et al. demonstrated that SCAP/SREBP pathway was required for maximal steroid hormone biosynthesis.14 Tang et al. suggested that botulin could be a promising lead compound for treatment of hyperlipidemia by inhibition SREBP pathway.15 Inhibition of SREBP pathway could be employed as an alternative strategy to treat metabolic diseases.
Statins are a class of lipid-lowering medications that blocks cholesterol biosynthesis by inhibiting HMG-CoA reductase.16 However, statins have adverse effects including muscle complications and an increased risk of diabetes. To date, numerous HMG-CoA reductase inhibitors have been approved to treat hypercholesterolemia, such as ezetimibe, bile acid sequestrants, omitapide, and mipomersen. Although these drugs confer significant protection against lipid metabolic disorders, a considerable residual risk remains after intensive therapy.
Natural products are potential resources for discovering novel lead compounds.17,18 As a major source of natural products, Traditional Chinese Medicines (TCMs) have attracted much attention to treat hyperlipidemia for many years. TCMs provide multiple sources to identify new candidates for cholesterol-lowering agents. For example, red yeast rice (Monascus purpureus) can depress cholesterol levels by inhibiting HMG-CoA reductase activity.19,20 In addition, studies found that ethanolic extracts of Cassia tora L. seeds and Hibiscus sabdariffa L. leaves exhibited remarkable hypolipidemic activity.21–23 However, mechanisms responsible for this function has not been identified adequately.
The present study aimed to evaluate the SCAP–SREBP pathway inhibitory activities of TCMs. Firstly, a TCM-human disorders interaction network was built to understand the interactions between traditional medicines and human disorders as an integrated system. A sub-network involved in lipid metabolism was built based on the naïve Bayes model. We then established a stable HepG2 cell line transfected with a vector containing the human pGL3-promoter coupled to the firefly luciferase reporter gene to detect the SREBP–SCABP pathway inhibitory activities of TCMs extracts. In addition, the effects of the biosynthesis of endogenous cholesterol were investigated using reverse transcription polymerase chain reaction (RT-PCR) assay and amphotericin B cell model. This work serves as a preliminary study providing a set of combined and detailed screening protocols for screening medicines on lowering lipid in the near future.
2 Materials and methods
2.1 Construct TCM-human disease connections network
The Traditional Chinese Medicines and human disorders connection data manually collected from literatures. We used the search items “Traditional Chinese Medicine”, “TCM”, “Traditional Asian Medicine”, and “Traditional Oriental Medicine” alone and combined with the terms human diseases or human disorders. The TCMs were mapped to Latin names totally, and the lists of human disorders were obtained from the Online Mendelian Inheritance in Man (OMIM) database.24 We classified each disorders to 22 classes based on the physiological system affected (see ESI Table 1 for details†). And the TCMs were manually classified into 64 classes according to Traditional Chinese Medicine theory. We generated a biological relevant network, in which nodes represent human disorders and Latin name of TCMs. Next, naïve Bayes model was used to construct a lowing-lipid related sub-network, a set of potential lipid metabolic related medicine was used to help constructing the screen database.2.2 Promoter-reporter construction, cell culture and transfection
Complementary oligonucleotides containing five copies of classic SREs coupled to a cofactor binding sites of Sp1 were synthesized (5′-GCTAGCAAAATCACCCCACTGCAAACTCCTCCCCCTGCGCTAGC-3′). The double stranded oligonucleotides were inserted into pGL3 based vector (Promega, USA) at KpnI and XhoI sites. The human hepatocellular carcinoma cell line (HepG2) was obtained from ATCC (Manassas, VA, MA). HepG2 cells were cultured in DMEM (Dulbecco's Modified Eagle Medium, Biowhittaker, USA) containing 10% FBS at 37 °C. Translation was carried out in serum free medium in 96 well plates using TransFectin lipid reagent (Bio-Rad, USA) and 100 μg of pGL3–SRE–Sp1 plasmid. For stable transfection, DMEM containing 10% FBS was placed on the wells after transfection, and the cells were cultured at 37 °C in humidified 10% CO2 atmosphere for 24 h before detected. Stably transfected HepG2 cells possessing the cholesterol synthesis responsive elements reporter plasmid in their chromosomal DNA were termed HepG2/pGL3–SRE–Sp1 cells.2.3 Preparation of crude extracts from TCMs
Ninety-three kinds of TCMs ingredients were weighed and crushed into coarse powder (ESI Table 2†). The powder was soaked in water for 30 min, then boiled for 1 h. Combined water extracts, atmospheric evaporation, purified by 75% ethanol, drought the supernatant and concentrated to be the water extracts. The aqueous residues were first soaked with 95% ethanol for 30 min, and then reflux extracted three times, each time for 1 h. Leaching solutions were combined and dried at 50 °C to obtain the ethanol extracts.2.4 Assay SCAP–SREBP pathway inhibitory activity
Cells were grown in low carbohydrates of DMEM with 10% FBS, 2.0 × 104 cells per wells were incubated at 37 °C with 5% CO2 for 12 h, and then transfected the pGL3–pSRE plasmid for 100 μg per well. The negative control and the non-treatment control was also constructed. After cultured for 6 h, extracts of the natural products (100 μg ml−1), vehicle (ethanol, 5 μl) were added to the corresponding wells. The cells were harvested 24 h after sample treatment, lysed and then sent to detect the luciferase activity. The luciferase activities were measured with Wallace Victor II plate reader (Perkin-Elmer, Inc., Wellesley, MA) and normalized to the relative luminescence unit (RLU).2.5 Assay the effects of TCM on the expression of genes involving the biosynthesis of cholesterol
| The primer | Forward primer | Reverse primer |
|---|---|---|
| FAS | 5′-GCTTCTTCGTGCAGCAGTTC-3′ | 5′-TTTCCCGTCGCATACCTG-3′ |
| HMG-CoA reductase | 5′-AGATAGGAACGGTGGGTG-3′ | 5′-TGTCTTCTTGGTGCAAGC-3′ |
| HMG-CoA synthetase | 5′-GACGGTATGCCCTGGTAG-3′ | 5′-CATTTCCCTCTTTCTGCC-3′ |
2.6 Assay the effects of TCMs extracts on the synthesizing of endogenous cholesterol
Briefly, CHO (Chinese hamster ovary) cells were seeded at density 5000 cells per well in 96-well plates in RPMI (Roswell Park Memorial Institute medium) 1640 medium and supplemented with 10% FBS for 1 day. Cells were incubated in medium for 1 day, in which 10% FBS was replaced with 10% lipoprotein-deficient calf serum. After that, cells were treated with 50 μg ml−1 pravastatin, 50 μg ml−1 Rhizoma Alismatis and 50 μg ml−1 Semen Cassiae, respectively. Cells were then treated with different concentration of amphotericin B. Cell viability was assessed using a colorimetric 3-(4,5-dimethyl thiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Cells were incubated with 50 mg of MTT for 6 h, the medium then was removed and the reaction product was dissolved in Me2SO. Plates were read using a molecular dynamics enzyme-linked immunosorbent assay reader at 570/630 nm.3 Results
3.1 Construction of lipid metabolic related network
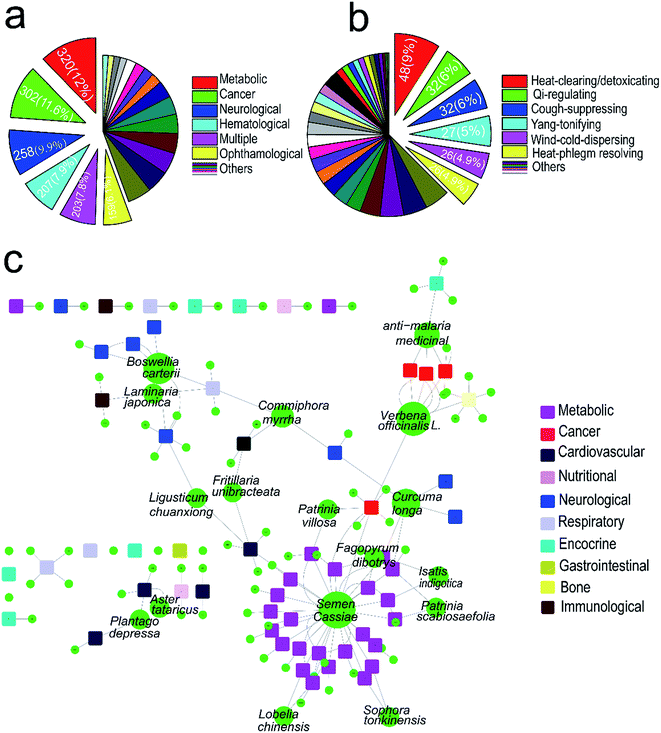
Lists of TCMs and corresponding diseases were obtained from the OMIM database and literatures, covering 529 kinds of TCMs and 2605 kinds of corresponding human diseases (ESI Table 1†). In which, 320 out of 2605 human genetic disorders are metabolic disorders accounted for 12%, followed by cancer (11.6%), neurological (9.9%), hematological (7.9%) and ophthalmological (6.1%), see Fig. 1a. Among 529 kinds of TCMs, 48 TCMs are heat-clearing and detoxicating medicine accounted for 9%, followed by qi-regulating medicine (6%), cough-suppressing (6%) and wind-could-dispersing medicine (4.9%) (Fig. 1b). First we used collected TCMs and their targeting diseases to generate a bipartite graph of TCM–disease interactions in which a TCM and a disease are connected to each other, giving rise to a TCM–disease network. Then a subnetwork contained about 93 kinds of medicines which concern of lipid metabolic was identified based on naïve Bayes model (Fig. 1c). These TCMs were selected for further study.3.2 The SREBP–SCAP pathway inhibitory activities of TCMs extracts assay
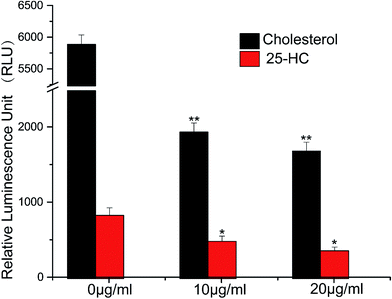
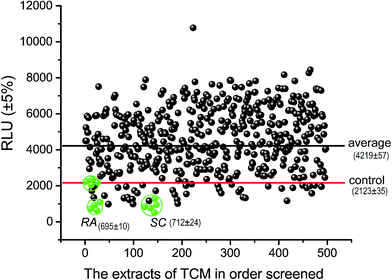
We constructed a luciferase reporter driven by a SRE-containing promoter and established a stable cell line stably expressing this reporter from HepG2 cell line. To test the performance of this in vitro assay system, HepG2/pGL3–SRE–Sp1 cells were treated with various concentrations of cholesterol and 25-Hydroxycholesterol (25-HC). 25-HC is a highly potent compound that inhibit SREBP cleavage, its potency is >100 fold higher than that of cholesterol. Our results showed that RUL values of transfected cells can be brought down through increasing the concentration of cholesterol and 25-HC. As can be seen in Fig. 2, the RLU value of the samples decreased remarkably, indicating that the in vitro system was reliable to detect inhibitory activity of SCAP–SREBP pathway. The HepG2/pGL3–SRE–Sp1 cells based in vitro assay system was then used to detect the SCAP–SREBP pathway inhibitory activities of TCMs extracts. After treating HepG2/pGL3–SRE–Sp1 cells with each TCMs extracts (100 μg ml−1), the luciferase activities were detected, and all the assay results were normalized to the RLU and the untreated HepG2/pGL3–SRE–Sp1 cells were taken as control. Extracts of 93 kinds of TCMs were screened, about 10% of the samples had the RLU value lower than untreated cells. The average RLU values of crude extracts of Rhizoma Alismatis and Semen Cassiae were 695 and 712, respectively, and were about three times lower than that of control (2123), indicating that they might possess favorable SCAP–SREBP pathway inhibitory activities (Fig. 3). Subsequently, a dose depending experiment was performed to determine dose–effect relation of Rhizoma Alismatis and Semen Cassiae extracts. HepG2/pGL3–SRE–Sp1 cells were treated with these samples with a range of concentrations from 200 μg ml−1 to 1 μg ml−1. The results indicated that the RLU values of these samples decreased as the concentration increased. The calculated EC50 of Rhizoma Alismatis extracts was 2.6 μg ml−1, and the EC50 of Semen Cassiae extracts was 4.7 μg ml−1.3.3 Effects of Rhizoma Alismatis and Semen Cassiae extracts on the expression of major target genes of SREBPs
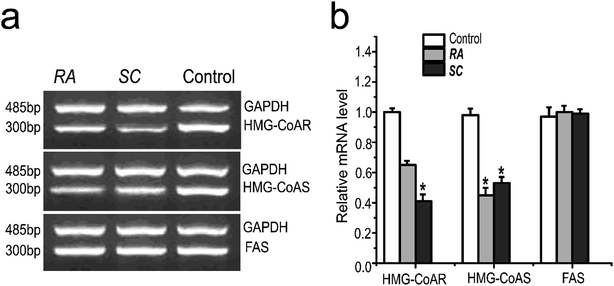
SREBPs pathway plays an important role in cholesterol synthesis, and also regulates the expression of cholesterol synthesis-related genes. The experimental results showed that Rhizoma Alismatis and Semen Cassiae extracts inhibited the gene expression of HMG-CoA reductase and HMG-CoA synthetase remarkably, however there was no significant inhibition effects on fatty acid synthetase (Fig. 4).3.4 Effects of Rhizoma Alismatis and Semen Cassiae extracts on synthesizing of endogenous cholesterol
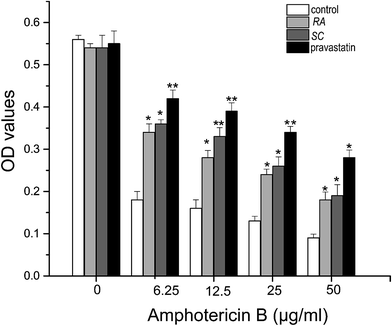
The amphotericin B cell model was applied to detect the affection of positive samples on synthesizing of endogenous cholesterol, with a positive agent pravastatin as positive control. Amphotericin B is a widely used polyene antibiotic that has side effect toxicity on mammalian cells. This drug has known to form pores in cholesterol-rich membranes.25–27 Cells capable of synthesizing of endogenous cholesterol are lysed and killed by amphotericin B treatment, while cells with impaired synthesizing of cholesterol survive.28 CHO cells are incubated in various media and treated with different concentration of amphotericin B, and then cell variability is evaluated using MTT assay.29In Fig. 5, when pravastatin was added along with Rhizoma Alismatis or Semen Cassiae extracts, cell killing was prevented. As can be seen, when the cells were incubated in low concentration of amphotericin B (6.25 μg ml−1), the OD value of control was 0.18, it was obviously lower than that of pravastatin (0.42), Rhizoma Alismatis (0.34) or Semen Cassiae (0.36). And when the concentration of amphotericin B was set to 50 μg ml−1, the cell variability of control group was still lower than that of other experimental groups. The OD values of control, pravastatin, Rhizoma Alismatis and Semen Cassiae treated groups were 0.09, 0.18, 0.19 and 0.28, respectively. Our results indicated that Rhizoma Alismatis and Semen Cassiae extracts prevented endogenous cholesterol from causing amphotericin B-mediated cell killing.
4 Discussion
Hypercholesterolemia is well-known as trigger of cardiovascular diseases. Statins are the mainly treatment drugs of hyperlipemia and hypercholesterolemia, as an inhibitors of HMG-CoA reductase, stain blocks cholesterol biosynthesis.30 However, statins have adverse effects including muscle complications and an increased risk of diabetes.31 SREBPs is a key transcription factor that controls the biosynthesis of cholesterol, triglyceride and fatty acid.10,16,32 Targeting SREBPs pathway critical for cholesterol biosynthesis provides an promising strategy to improve therapeutic option of lipid metabolic diseases.1,14,15In this study, we established a stable HepG2 cell line transfected with a vector containing the human pGL3-promoter coupled to firefly luciferase reporter gene. The activity of luciferase varies with the expression levels of SREBPs in the transfected cells. When cellular sterol levels are high, cholesterol or 25-HC (also known as oxysterols) stimulate the association between SCAP and Insig, SREBP precursors thus are tethered and retained in the ER by the sterol-sensing SCAP–Insig complex. Here, transfected HepG2 cells were treated with various concentrations of cholesterol and 25-HC, respectively. As expected, HepG2 cells treated with cholesterol or 25-HC showed a good differential in RLU compared to control. The RLU value of the treated groups decreased remarkably, indicating that high levels of cholesterol and 25-HC could lower the luciferase activity. This finding demonstrated that our cell line could be used as a reliable screening protocol to detect SREBPs pathway inhibitory activities of TCMs extracts.
The popularity of using TCMs to treat hyperlipidemia is increasing and its role in health care has been recognized by the public at large.33,34 Based on network analysis, 93 kinds of TCMs involved in lipid metabolic diseases were selected for high throughput screening. Crude extracts from Rhizoma Alismatis and Semen Cassiae showed beneficial effects on inhibition of SREBPs pathway activities. Rhizoma Alismatis is a classical TCM originating from the rhizomes of Alisma orientale Juzepczuk (Alismataceae)35 and has been found to possess anti-hyperlipidemic activity. Semen cassiae has long been used to effectively reduce lipid levels, and increase immune function in humans.36
Once SREBPs pathway was inhibited by TCMs, the expression of genes involved in lipid metabolism would be decreased. We then examined the effects of Rhizoma Alismatis and Semen Cassiae extracts on the expression of HMG-CoA reductase, HMG-CoA synthetase and fatty acid synthetase. Interestingly, we found that Rhizoma Alismatis and Semen Cassiae can inhibit the expression of HMG-CoA reductase and HMG-CoA synthetase, however, no experimental evidence involved in inhibition of the expression of fatty acid synthetase was found. We further examined the effects of these TCMs on synthesizing of endogenous cholesterol based on amphotericin B cell model. In which the cell viability was determined by the endogenous cholesterol levels due to the lytic effects of amphotericin B. Encouraged by clear and convincing evidence that Rhizoma Alismatis and Semen Cassiae preserve cell viability, even though the effects are not comparable with pravastatin, indicating that these TCMs can suppress the biosynthesis of endogenous cholesterol. Horton et al. proposed that SREBP possessed three forms in mammals: SREBP-1a, SREBP-1ac and SREBP-2, among which SREBP-2 was responsible for the biosynthesis of cholesterol and low-density lipoprotein receptor (LDLR), the others were responsible for the biosynthesis of fatty acid.37 Therefore, Rhizoma Alismatis and Semen Cassiae may suppress the biosynthesis of endogenous cholesterol through blocking maturation of SREBP-2. However, the molecular basis underlying the activation of the SCAP/SREBP pathway still need to be elucidated by further study.
It is noteworthy that there are several limitations in present study. Although this work showed that crude extracts of Rhizoma Alismatis and Semen Cassiae can inhibit the SREBP pathway activity, in turn suppress the biosynthesis of cholesterol, the effective molecular components of these TCMs were not determined. Therefore, we could not completely exclude the molecular mechanism of inhibition of SREBP pathway. Our studies are based on the cellular experiments in vitro, experimental assays can potentially be subject to interference by a number of naturally occurring non-specific substances in the herbal extracts, thereby providing falsely high results. In order to eliminate the interference results, further studies may be needed, including detecting, removing and accounting for interference compounds. In addition, another limitation is that this work has considered only three genes involved in cholesterol synthetic pathway, that is, HMG-CoA reductase, HMG-CoA synthetase and fatty acid synthetase. There are about ten genes involved in cholesterol synthetic pathway, such as squalene epoxidase, Acetyl-CoA carboxylase alpha. Investigating the effects of TCMs on these genes may provide crucial information for exclude molecular basis of inhibition of SREBP pathway.
5 Conclusions
In conclusion, through an in vitro assay system, we found that the crude extracts of Rhizoma Alismatis and Semen Cassiae showed beneficial effects on SCAP–SREBP pathway inhibitory activities. Results suggested that our assay system was competent to detect the SCAP–SREBP pathway inhibitory activities of Chinese traditional medicines. Rather, the effective components in the positive TCMs extracts still needed to be comprehensively investigated in future studies. Our work serves as a preliminary study providing a set of combined and detailed screening protocols for screening medicines involving lipid metabolism from TCMs. We hope this work could guide people to improve hyperlipidemia or other lipid metabolic disorders.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are grateful to all members of the laboratory for helpful discussions during the course of these studies. This work was supported by grants from the National Natural Science Foundation of China (No. 31300674, 81173093, 30970643, 81373311 and J1103518), the Youth Science Foundation of West China Hospital of Stomatology (No. 2016-3) and the Special Program for Youth Science and the Technology Innovative Research Group of Sichuan Province, China (No. 2011JTD0026).References
- A. Morgan, K. M. Mooney, S. J. Wilkinson, N. Pickles and M. T. Mc Auley, Ageing Res. Rev., 2016, 27, 108–124 CrossRef CAS PubMed.
- M. S. Brown and J. L. Goldstein, Cell, 1997, 89, 331–340 CrossRef CAS PubMed.
- K. F. Tacer, T. Haugen, M. Baltsen, N. Debeljak and D. Rozman, J. Lipid Res., 2002, 43, 82–89 CAS.
- J. D. Horton, J. L. Goldstein and M. S. Brown, J. Clin. Invest., 2002, 109, 1125 CrossRef CAS PubMed.
- A. J. Brown, L. Sun, J. D. Feramisco, M. S. Brown and J. L. Goldstein, Molecular cell, 2002, 10, 237–245 CrossRef CAS PubMed.
- R. A. DeBose-Boyd, M. S. Brown, W.-P. Li, A. Nohturfft, J. L. Goldstein and P. J. Espenshade, Cell, 1999, 99, 703–712 CrossRef CAS PubMed.
- A. Radhakrishnan, J. L. Goldstein, J. G. McDonald and M. S. Brown, Cell Metab., 2008, 8, 512–521 CrossRef CAS PubMed.
- P. J. Espenshade, D. Cheng, J. L. Goldstein and M. S. Brown, J. Biol. Chem., 1999, 274, 22795–22804 CrossRef CAS PubMed.
- P. J. Espenshade, W.-P. Li and D. Yabe, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 11694–11699 CrossRef CAS PubMed.
- J. L. Goldstein, R. A. DeBose-Boyd and M. S. Brown, Cell, 2006, 124, 35–46 CrossRef CAS PubMed.
- T. Grand-Perret, A. Bouillot, A. Perrot, S. Commans, M. Walker and M. Issandou, Nat. Med., 2001, 7, 1332–1338 CrossRef CAS PubMed.
- K. Pahan, Cell. Mol. Life Sci., 2006, 63, 1165–1178 CrossRef CAS PubMed.
- D. Rozman and K. Monostory, Pharmacol. Ther., 2010, 127, 19–40 CrossRef CAS PubMed.
- M. Shimizu-Albergine, B. Van Yserloo, M. G. Golkowski, S.-E. Ong, J. A. Beavo and K. E. Bornfeldt, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E5685–E5693 CrossRef CAS PubMed.
- J.-J. Tang, J.-G. Li, W. Qi, W.-W. Qiu, P.-S. Li, B.-L. Li and B.-L. Song, Cell Metab., 2011, 13, 44–56 CrossRef CAS PubMed.
- M. S. Brown and J. L. Goldstein, J. Lipid Res., 2009, 50, S15–S27 CrossRef PubMed.
- C. R. Pye, M. J. Bertin, R. S. Lokey, W. H. Gerwick and R. G. Linington, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 5601–5606 CrossRef CAS PubMed.
- T. Rodrigues, D. Reker, P. Schneider and G. Schneider, Nat. Chem., 2016, 8, 531–541 CrossRef CAS PubMed.
- R. Y. Man, E. G. Lynn, F. Cheung, P. S. Tsang and O. Karmin, Mol. Cell. Biochem., 2002, 233, 153–158 CrossRef CAS PubMed.
- M. Journoud and P. J. Jones, Life Sci., 2004, 74, 2675–2683 CrossRef CAS PubMed.
- W. Kong, J. Wei, P. Abidi, M. Lin, S. Inaba, C. Li, Y. Wang, Z. Wang, S. Si and H. Pan, Nat. Med., 2004, 10, 1344 CrossRef CAS PubMed.
- U. K. Patil, S. Saraf and V. Dixit, J. Ethnopharmacol., 2004, 90, 249–252 CrossRef PubMed.
- S. Gosain, R. Ircchiaya, P. C. Sharma, S. Thareja, A. Kalra, A. Deep and T. R. Bhardwaj, Acta Pol. Pharm., 2010, 67, 179–184 Search PubMed.
- A. Hamosh, A. F. Scott, J. S. Amberger, C. A. Bocchini and V. A. McKusick, Nucleic Acids Res., 2005, 33, D514–D517 CrossRef CAS PubMed.
- S. Kinsky, Annu. Rev. Pharmacol., 1970, 10, 119–142 CrossRef CAS PubMed.
- A. Norman, R. Demel, B. De Kruyff and L. Van Deenen, J. Biol. Chem., 1972, 247, 1918–1929 CAS.
- B. De Kruijff, Biosci. Rep., 1990, 10, 127–130 CrossRef CAS PubMed.
- N. Dahl, K. Reed, M. A. Daunais, J. Faust and L. Liscum, J. Biol. Chem., 1992, 267, 4889–4896 CAS.
- K. W. Underwood, N. L. Jacobs, A. Howley and L. Liscum, J. Biol. Chem., 1998, 273, 4266–4274 CrossRef CAS PubMed.
- P. Durrington, H. Soran, E. Lammert and M. Zeeb, Metabolism of Human Diseases: Organ Physiology and Pathophysiology, 2014, p. 295 Search PubMed.
- G. Langslet, Eur. Heart J., 2016, 37, 1357–1359 CrossRef PubMed.
- K. Morimoto, H. Itoh and M. Watanabe, Expert Rev. Endocrinol. Metab., 2013, 8, 59–69 CrossRef CAS.
- T.-T. Sham, C.-O. Chan, Y.-H. Wang, J.-M. Yang, D. K.-W. Mok and S.-W. Chan, BioMed Res. Int., 2014, 10, 925302 Search PubMed.
- S. Gu, N. Yin, J. Pei and L. Lai, Mol. BioSyst., 2013, 9, 2696–2700 RSC.
- J.-C. Lee, E. Lee and Y.-C. Lee, Korea J. Herbol., 2008, 23, 113–117 Search PubMed.
- Z. Y. Deng, J. W. Zhang, G. Y. Wu, Y. Yin, Z. Ruan, T. J. Li, W. Y. Chu, X. F. Kong, Y. M. Zhang and Y. W. Fan, J. Sci. Food Agric., 2007, 87, 1868–1873 CrossRef CAS.
- J. D. Horton, N. A. Shah, J. A. Warrington, N. N. Anderson, S. W. Park, M. S. Brown and J. L. Goldstein, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 12027 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra09521j |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |